Abstract
We have used a gene expression array–based strategy to identify the methylation of tissue factor pathway inhibitor 2 (TFPI2), a potential tumor suppressor gene, as a frequent event in human colorectal cancers (CRC). TFPI2 belongs to the recently described group of embryonic cell Polycomb group (PcG)–marked genes that may be predisposed to aberrant DNA methylation in early stages of colorectal carcinogenesis. Aberrant methylation of TFPI2 was detected in almost all CRC adenomas (97%, n = 56) and stages I to IV CRCs (99%, n = 115). We further explored the potential of TFPI2 as a biomarker for the early detection of CRC using stool DNA–based assays in patients with nonmetastatic CRC and average-risk noncancer controls who were candidates for screening. TFPI2 methylation was detected in stool DNA from stage I to III CRC patients with a sensitivity of 76% to 89% and a specificity of 79% to 93%. Detection of TFPI2 methylation in stool DNA may act as a useful adjunct to the noninvasive strategies for screening of CRCs in the future.
Introduction
Epigenetic changes resulting in transcriptional silencing of cancer genes are among the most frequently discovered events in human tumor samples and are marked by aberrant accumulation of 5-methylcytosine and altered chromatin structure in the 5′-regulatory regions (1-3). Hypermethylation of gene promoter–associated CpG islands occurs early during colorectal cancer (CRC) development (4), suggesting its utility as a molecular marker for both early detection and monitoring progression of the disease.
CRCs, although amenable to screening and early detection, still remains the second leading cause of cancer-related mortality in the United States (5). Current screening modalities have resulted in only a modest decrease in mortality and failed to achieve high public participation (<50%; refs. 6, 7). Strategies such as colonoscopy are invasive whereas stool occult blood test has limitations of repeat measurements and interference by dietary components. Stool DNA tests are noninvasive, may have high specificity with single measurements, and may achieve more extensive sampling of colon and has recently been recommended for CRC screening (8, 9). The stool DNA tests tested thus far are frequently based on multiple genetic markers including KRAS, APC, TP53, and BAT26, which may be more costly and difficult to implement (10). Epigenetic events which are seen more frequently than genetic events (11) may have a potential to achieve high sensitivity and specificity using a single gene (9).
We have recently described a novel expression array-based approach to identify the CRC “DNA hypermethylome” estimated to contain ~400 to 600 hypermethylated genes per individual tumor (11). Many of these genes may already be marked for transcriptional repression in embryonic precursor cells by the PcG proteins (11-13), which participate in the three-dimensional structure of nuclear DNA (14) and may target genes with a characteristic “bivalent” promoter chromatin structure containing both active and repressive histone modifications. Such PcG-marked genes are poised to integrate transcriptional signaling during cellular proliferation and differentiation (15). Moreover, such PcG-marked genes may be predisposed to methylation early on in carcinogenesis (12, 13), and such genes may be good targets for investigation as biomarkers for early detection.
One gene that we have identified within the DNA hypermethylome, residing at the intersection of both the hypermethylome and PcG-marked genes, is tissue factor pathway inhibitor (TFPI2), a Kunitz-type serine proteinase inhibitor that protects the extracellular matrix of cancer cells from degradation and inhibits in vitro colony formation and proliferation (16, 17). It is thought that loss of TFPI2 function could predispose cells toward a proinvasive program, consistent with an important role for this protein in later stages of carcinogenesis.
In this study, we investigate the role of TFPI2 in CRC cell lines and tumors. We present data indicating that TFPI2 methylation is a frequent and early event in CRCs. Furthermore, we show that TFPI2 methylation in stool DNA is a potential biomarker for noninvasive detection of colorectal neoplasms.
Materials and Methods
Gene Expression, Methylation Analysis, and Chromatin Immunoprecipitation
Cell culture growth, maintenance, pharmacologic treatment and expression array analysis were performed as previously described (11). Total RNA was isolated as previously described (11) or purchased from Stratagene (normal adult colon). cDNA synthesis and reverse transcriptase PCR (RT-PCR) were performed as described (11). Quantitative RT-PCR was performed with Quantitect SybrGreen PCR Mix (Qiagen) using an iCycler (Bio-Rad). DNA isolation, bisulfite modification, primer design, and methylation analysis were performed as previously described (11). Complete primer sequences and thermocycler variables are shown in Supplementary Table S1. ChIP on Chip experiments and analyses were performed as previously described (18).
Colony Formation Assay and In vitro Growth Rate
We amplified the complete 820 nucleotide TFPI2 coding region with primers 5-ACGA-GCTAGC-GCACCATGGACCCCGCTCG-3 (Nhe I in bold-face) and 5-AGCT-GAATTC-TGTTTAAAATTGCTTCTTCCG-3 (Eco RI in boldface) from normal human colon cDNA (Stratagene), purified, restriction enzyme digested, and cloned into the Nhe I and Eco RI site of pIRES-Neo3 (Invitrogen). Transfection, colony formation, and scoring were as previously described (11). In vitro growth rate was performed using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. MTT assay was performed in HCT116 cell lines transfected with plasmid carrying the TFPI-2 gene (1 μg/μL), or empty vector (1 μg/μL) alone as a control. The ATCC-MTT cell proliferation assay kit was used and all experiments were performed as previously described (19).
Primary Human Tissue Samples
Primary tissue samples were obtained from the archives of the Department of Pathology, Johns Hopkins University with Institutional Review Board approval and Health Insurance Portability and Accountability Act compliance. This study compiles data from formalin-fixed and paraffin-embedded primary CRCs of tumor stages I to IV (N = 115; n = 35 stage I, n = 36 stage II, n = 28 stage III, n = 16 stage IV; mean age, 65.2 years) and corresponding adjacent nonneoplastic colon tissue in a subset of 22 of these CRC patients (n = 22), colorectal adenomas from patients without an associated invasive CRC (N = 56; n = 17 serrated, n = 17 tubular, n = 22 villous) and normal colon controls of patients without any colorectal neoplasms (n = 48). Within this subset of normal colon controls, 16 samples were age-matched for CRCs (mean age, 63.4 years; P = not significant). For expression analysis, fresh-frozen CRCs (n = 12) and corresponding adjacent normal colon tissue (n = 3) were used.
Stool Studies
Study population and sample size
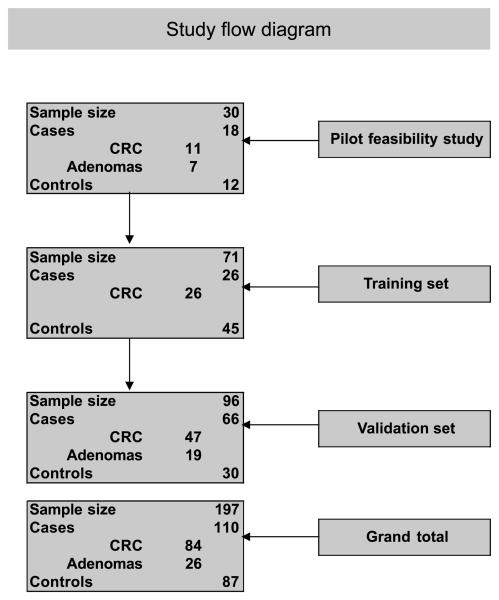
Colonoscopy-negative control stool samples were obtained from healthy, average-risk subjects >50 years of age who were screened for CRC within the framework of a workplace-based community CRC study at the Maastricht University Medical Center. Stool samples from colonoscopy-confirmed CRC/adenoma patients covering all CRC stages and matched CRC tissue were collected at the VU University Medical Center in Amsterdam. The Medical Ethical Committee of the Maastricht University Medical Center and the Dutch Health Council approved this study. Written informed consent was obtained for all stool samples. All control stool samples and a subset of the CRC stool samples were collected within 2 weeks prior to colon purgation and colonoscopy. Some CRC stool samples were collected 5 to 7 days following colonoscopy. All stool samples were homogenized in stool stabilization buffer (Exact Sciences), aliquoted in portions containing the equivalent of 4 g and processed within 48 h after defecation. Stool samples were handled and analyzed in a blinded fashion during storage, DNA isolation, and PCR analysis. A pilot feasibility study with a sample size of 30, including 18 cases (11 CRCs and 7 adenomas) and 12 controls, was performed to investigate the potential utility of TFPI2 methylation in stool DNA for the detection of CRC. Following the pilot feasibility study, a double blind study with a training set and validation set was designed. The training set consisted of 71 samples whereas the validation set consisted of 96 samples. The training set included 26 stage I to III CRCs and 45 controls. The validation set, which had a sample size of 96, included 66 cases and 30 controls. Cases in the validation set targeted the screen-relevant neoplasms including advanced adenomas (i.e., >1 cm size, high-grade dysplasia, or villous histology; n = 19) and stage I to III CRCs (n = 47). Figure 1 depicts the schematic representation of the study design.
Figure 1.
Schematic representation of the stool DNA study design: 30 samples including 11 CRCs, 7 adenomas, and 12 controls were analyzed in the initial pilot study. Independent training and validation set with sample sizes of 71 and 96, respectively, were then further tested.
Stool Sample Collection and Processing
Stool DNA extraction
Samples were homogenized in excess volume (1:7) of stool homogenization buffer (Amresco) and aliquoted in portions of 32 mL (containing ~4 g of stool). Single aliquots were centrifuged and the supernatants were incubated with RNase A for 60 min at 37°C. For the pilot feasibility stool DNA studies, total DNA was precipitated with sodium acetate (pH 5.2)-isopropanol, washed with 70% ethanol and resuspended in 4 mL of 1× TE (pH 7.4). Following the addition of 400 μL of 10× buffer [240 mmol/L EDTA (pH 8.0), 750 mmol/L NaCl], 400 μL of 10% SDS, and 20 μL of proteinase K (20 mg/mL), samples were incubated for 16 h at 48°C with shaking. After the addition of 5 mL of phenol/chloroform/isoamyl alcohol, the samples were incubated for 10 min at room temperature before centrifugation. The aqueous layer was transferred to a new tube, DNA was precipitated, washed, and pellets were resuspended in 2 mL of LoTE (pH 8.0).
The stool DNA extractions for the training and validation studies were performed using the QIAamp DNA stool mini kit protocol for ease of DNA extraction. The initial homogenization steps are similar. After RNase A treatment, total DNA was precipitated and resuspended in 1× TE. Half of this resuspended DNA was then used as input for the QIAamp DNA stool midi test kit (Qiagen, user-developed protocol). ASL buffer (1.5 mL) and an InhibitEX tablet were added and the mixture was centrifuged. Next, 150 μL of proteinase K was added to 2 mL of supernatant, which was mixed with 2.4 mL of buffer AL and incubated for 10 min at 70°C. Ethanol (2 mL) was added and loaded onto the column in portions of 3.3 mL maximum. After a double washing step, the column was dried and DNA was eluted using 200 μL of buffer AE.
Stool DNA Methylation Studies
Pilot feasibility study
Genomic DNA (2 μg) was modified using Zymo Kit according to the instructions of the manufacturer. A nested strategy, as previously described, was used as DNA obtained from stool is limited in amount (20). An initial unbiased amplification of bisulfite-modified DNA was done using external primers located in the flanking region for 35 cycles. Each PCR was carried out in a volume of 25 μL with 1 μL of JumpStart Red Taq DNA polymerase (Sigma), 10 pmol of each external primer, and 4 μL of bisulfite-modified DNA. PCR conditions included an initial denaturation at 95°C for 5 min followed by 35 cycles of (95°C × 30 s, annealing temp × 30 s, 72°C × 30 s) and final elongation at 72°C for 5 min. In the second stage, 1:1,000 dilutions of first-stage products were used and reamplified for 30 cycles using an internal primer set. Except for the annealing temperatures and cycle number, PCR cycling conditions were the same for the first and second stages. Primer sequences are provided in Supplementary Table S1.
For the training and validation sets, a quantitative approach was pursued. Quantitative methylation-specific PCR was performed using a 7900HT real-time PCR system (Applied Biosystems). DNA (2.4 μL) was added to a PCR mix containing buffer [16.6 mmol/L (NH4)2SO4, 67 mmol/L Tris, 6.7 mmol/L MgCl2, 10 mmol/L β-mercaptoethanol], deoxynucleotide triphosphates (5 mmol/L), forward primer (6 ng/μL), reverse primer (18 ng/μL), molecular beacon (0.16 μmol/L), BSA (0.1 μg), and Jumpstart Taq polymerase (0.4 units; Sigma Aldrich). The PCR program was as follows: 5 min at 95°C, followed by 45 cycles of 30 s at 95°C, 30 s at 57°C, and 30 s at 72°C, followed by 5 min at 72°C. A standard curve (2 × 106–20 copies) was included to determine copy numbers of unknown samples by interpolation of their Ct values to the standard curve. Primer and beacon sequences are provided in Supplementary Table S1.
Statistical Analysis
Statistical analysis was performed using the STATA 9.2 software package. Receiver operator characteristic (ROC) curve analysis, using the area under the curve, was used to determine the best cutoff value for the highest sensitivity and specificity in the training set. Wherever mean ages of the cases and controls differed significantly, a ROC-GLM regression model was used to assess the accuracy of TFPI2 promoter methylation after adjustment for age (21). TFPI2 methylation was considered positive if a methylation value was higher than the cutoff (10 copies).
Results
The TFPI2 gene showed a ~50-fold increase in gene expression in HCT116 (Fig. 2A) after 5-aza-2′-deoxycytidine (DAC) treatment in our array-based hypermethylome analyses (11), generating further interest in the potential role of this gene in CRCs. We first tested TFPI2 expression by RT-PCR (Fig. 2B, top) and DNA methylation by MSP (Fig. 2B, bottom) before and after DAC treatment in HCT116 colon cancer cells. Consistent with our microarray results, TFPI2 was silenced and methylated in untreated HCT116 cells and exhibited re-expression with concurrent demethylation of the TFPI2 promoter (Fig. 2B and C) after drug treatment with DAC. Bisulfite sequencing of a region spanning the promoter from −286 bp to +76 bp confirmed dense methylation of TFPI2 in HCT116, whereas DKO a control cell line, harboring almost no cytosine methylation, showed a complete absence of methylation (Fig. 2C, bottom), again consistent with the microarray data (Supplementary Fig. S1). These results suggest an association between TFPI2 promoter methylation and expression in HCT116 cells, as predicted by our hypermethylome array analyses. Additionally, TFPI2 promoter showed an enrichment peak for the repressive histone mark, H3K27me3, along with an enrichment peak for the active mark, H3K4me2, over the transcription start site in HCT116 cells, where it is methylated and silenced. Although in DKO cells, where TFPI2 is unmethylated and expressed, there is a further increase in enrichment of H3K4me2, accompanied by rearrangement of this mark to now exhibit a dip at the transcription start site, a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells (Fig. 2D; ref. 18). These data show the bivalent chromatin structure marked by the presence of both active and repressive histone marks near the TFPI2 promoter in cancer cells consistent with it's identification as a PcG target gene (22-24).
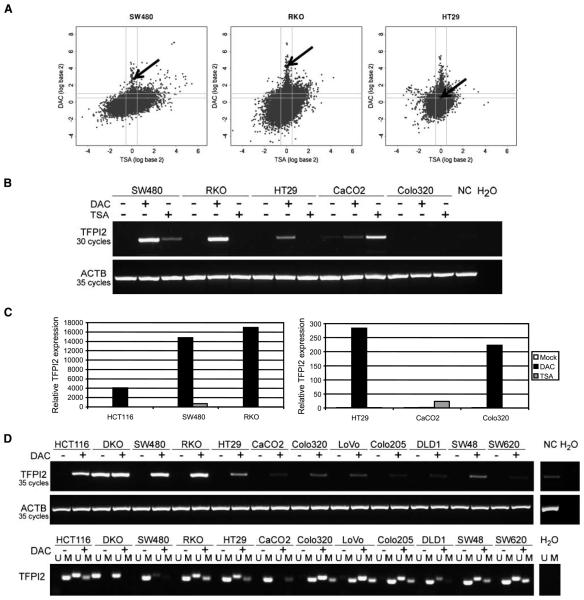
Figure 2.
Epigenetic inactivation of TFPI2 in CRC cell line HCT116. A, gene expression changes are plotted by fold change (log scale) in HCT116 after DAC (Y-axis) or trichostatin A (TSA; X-axis) treatment. TFPI2 expression increases 5.24 in log 2 ratio (37.7-fold) in HCT116 after DAC treatment compared with HCT116 Mock and shows no response to TSA. ◆ and ↑, location of TFPI2 in the scatter plot. B, expression status (top) of TFPI2 in HCT116 without any treatment (Mock) and after DAC or TSA treatment, respectively, versus DKO. MSP analyses (bottom) in cell lines without (Mock) and after DAC treatment for unmethylated (U) and methylated (M) DNA. C, MSP tiling of the TFPI2 promoter and exon 1 along −118 bp to +284 bp from TSS using four different MSP primer sets (MSP1–4) shows unequivocal methylation status (top). IVD, in vitro methylated DNA, positive control; NL, normal lymphocytes from healthy individuals; TFPI2 gene map with the promoter region (gray box), and subsequent genomic sequence including exons 1 to 5 (black boxes). The primer binding sites for MSP and BSS are indicated along the CpG island (middle). The MSP2 primer set was used for methylation studies in primary formalin-fixed and paraffin-embedded and fresh-frozen tissues. MSP5 primer set was used for nested MSP analysis for fecal DNA methylation studies. Bisulfite sequencing of the TFPI2 promoter in HCT116 and DKO cells (bottom). ○, unmethylated CpG residues; ●, methylated CpG residues; arrow, transcription start site. D, histone marks for H3K4me2 and H3K27me3 at the TFPI2 promoter of HCT116 (left) and DKO (right) cells as determined by ChIP on ChIP assay.
We further tested the expression of TFPI2 in a panel of CRC cell lines. Scatter plot analyses indicated significant re-expression of TFPI2 after DAC treatment in SW480 (6.9-fold) and RKO (19.3-fold) cells, consistent with data obtained in HCT116 cells. However, only a modest re-expression (1.5-fold) was seen in HT29 cells (Fig. 3A). RT-PCR (Fig. 3B and C) and MSP (Fig. 3D) confirmed re-expression of TFPI2 and concomitant demethylation, following DAC treatment, in nearly all CRC cell lines (Fig. 3D). Methylation of TFPI2 was also detected in other tumor cell lines including gastric, esophageal, pancreatic, and breast cancer cell lines (Supplementary Fig. S2).
Figure 3.
Epigenetic inactivation of TFPI2 in CRC cell lines. A, gene expression changes for the indicated cell lines are plotted by fold change (log scale) after DAC (Y-axis) or TSA (X-axis) treatment. ◆ and ↑, the location of TFPI2. B, RT-PCR results for TFPI2 in CRC cell lines without any (−) and after DAC or TSA (+) treatment, respectively, as indicated. Normal colon (NC) was used as positive control for TFPI2 expression. β-Actin (ACTB) was used as control for equal amplification. C, real-time RT-PCR results for HCT116, SW480, and RKO (left) and HT29, CaCO2, and Colo320 (right) without any (Mock) and after DAC or TSA treatment, respectively. Please note the differing scale used for the two graphs. White columns, mock; black columns, DAC; gray columns, TSA. D, expression (top) and MSP status (bottom) for various CRC cell lines before (−) and after (+) DAC treatment (1 μmol/L, 72 h).
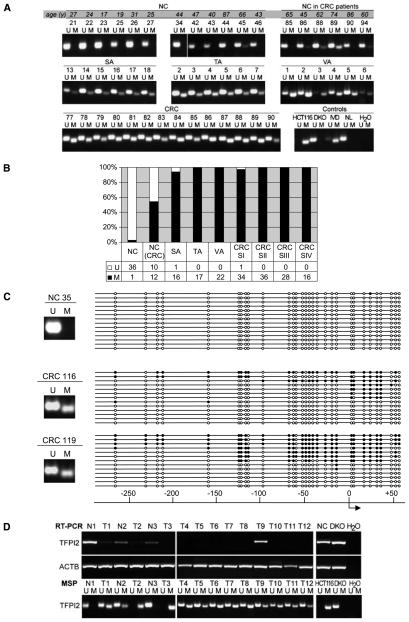
We next tested for the methylation of TFPI2 in normal colonic tissue from cancer-free patients sorted by age (Fig. 4A, top; and Fig. 4B). TFPI2 methylation was seen infrequently in normal colon from noncancerous patients (3 of 48; 6.2%; Fig. 4). Within this subset of normal colon controls, 16 samples were age-matched for the CRCs (mean age, 63.4 years; P = not significant), and methylation was observed only in this subset of patients. However, surrounding nonneoplastic colonic tissue of patients with CRC showed much more frequent TFPI2 methylation (12 of 22; 54%; Fig. 4A, top; Fig. 4B). Interestingly, TFPI2 methylation was observed in 94% of serrated adenomas (16 of 17), 100% of tubular adenomas (17 of 17), and 100% of villous adenomas (22 of 22), all of which represent preinvasive stages of colorectal carcinogenesis and in 99% of invasive CRC stages I to IV (114 out of 115; Fig. 4A, middle and bottom; Fig. 4B). Bisulfite sequencing of TFPI2 showed dense DNA methylation in primary tumor samples (Fig. 4C). Interestingly, the expression of TFPI2 in fresh-frozen primary samples also correlates with promoter DNA methylation status. We found a lack of TFPI2 expression in virtually all CRC samples (11 out of 12 CRC samples), and RT-PCR analysis of three paired normal cancer samples show that the gene is still expressed in surrounding normal colon tissue (Fig. 4D, top; the comparative DNA methylation status of these samples is shown in Fig. 4D, bottom).
Figure 4.
Epigenetic inactivation of TFPI2 in primary human tissues. A, methylation status by MSP in representative samples of normal colon (NC), adjacent normal colon from CRC patients, adenomas (serrated, SA; tubular, TA; villous, VA), and CRCs. For normal colons, note the age in years (y) as indicated in shaded box. U, unmethylated; M, methylated; NL, normal lymphocytes. B, bar graph showing MSP results of all primary samples (n = 230). C, bisulfite sequencing results for TFPI2 of one normal colon (NC 35) and two representative CRC samples (CRC 116 and CRC 119), with the location relative to the TSS as indicated. ● methylated CpG, ○, unmethylated CpG. Insets, methylation status by MSP. D, TFPI2 expression and methylation status in fresh-frozen primary human CRCs (T1–12) and corresponding normal colon (N1–3).
We also performed studies to detect the putative tumor suppressor role of TFPI2 in CRC tumorigenesis. We used in vitro colony formation assays to determine the effects of full-length TFPI2 transfected into HCT116 cells lacking TFPI2 expression. Overexpression of full-length TFPI2 induces a nearly 10-fold reduction in colony number of HCT116 cells, with surviving clones showing a severely depleted size, comparable to results obtained with the bona fide tumor suppressor p53 (Fig. 5A and B). These studies suggest a putative tumor suppressor role for TFPI2 in CRC. MTT assay showed a modest decline in cell growth in TFPI2-transfected cells as compared with empty vector (P = not significant). There was no significant difference in cell growth in the mock-treated and empty vector–treated cells. Supplementary Fig. S3 shows the results for MTT assay in detail.
Figure 5.
A, colony formation assay in HCT116 cells transfected with either full-length TFPI2 (left) or p53 (right) versus corresponding empty vector (middle) or no vector control (bottom) after hygromycin selection (top). Resulting colonies show a significant decrease in colony size in HCT116 with TFPI2 (left) as well as p53 (right) containing vector versus corresponding empty vector below (bottom). B, bar graph representing quantification of colony number after transfection of the indicated plasmids.
All of the above data indicate that TFPI2 is a potential tumor suppressor gene which is expressed and unmethylated in colonic epithelium from cancer-free individuals, and early methylation may occur during carcinogenesis. With this in mind, we tested the potential viability of using TFPI2 DNA methylation to detect CRC in stool DNA. We conducted a pilot feasibility study with a blinded case-control design to analyze stool DNA extracted from 30 patient samples: 18 cases → mean age = 58.5 years, including 11 patients with CRC and 7 patients with adenomas with dysplasia; and 12 controls → mean age = 54.2 years (Table 1A). 60% of the subjects were males whereas 40% were females. In 8 of 11 stool samples from patients with CRC and 3 of 7 samples from patients with preinvasive neoplastic lesions we detected methylated TFPI2 alleles whereas none of 12 (0%) samples of control patients were positive for methylated TFPI2 (sensitivity, 73%; specificity, 100% for cancers; sensitivity, 43%; specificity, 100%, for adenomas; Table 1B).
Table 1.
Patient characteristics
| (A) Study population characteristics | |||
|---|---|---|---|
| Pilot study | Training set | Validation set | |
| Number of study subjects (N) | 30 | 71 | 96 |
| Age (y) | Mean (y) | ||
| CRC | 60.7 | 69.3 | 71.1 |
| Adenoma | 55.1 | 61.4 | |
| Controls | 54.2 | 55 | 52.3 |
| Sex | Percentage | ||
| Male | 60 | 52 | 45 |
| Female | 40 | 46 | 54 |
| Unknown | 2 | 1 | |
| (B) Summary of test performance | |||
| Pilot study (n = 30) | Training set (n = 71) | Validation set (n = 96) | |
|
| |||
| CRC | n = 11 | n = 26 | n = 47 |
| Sensitivity (95% CI) | 73 (39–94) | 89 (70–98) | 76 (60–88) |
| Specificity (95% CI) | 100 (74–100) | 79 (64–90) | 93 (77–99) |
| PPV (95% CI) | 100 (63–100) | 72 (53–86) | 94 (80–99) |
| NPV (95% CI) | 80 (52–96) | 92 (78–98) | 73 (56–86) |
| Adenoma | n = 7 | n = 19 | |
| Sensitivity (95% CI) | 43 (10–82) | 21 (6–46) | |
| Specificity (95% CI) | 100 (74–100) | 93 (78–99) | |
| PPV (95% CI) | 100 (29–100) | 67 (22–96) | |
| NPV (95% CI) | 75 (48–93) | 65 (49–79) | |
Abbreviations: PPV, positive predictive value; NPV, negative predictive value; 95% CI, 95% confidence interval.
After the promising initial pilot feasibility study, we further verified the utility of TFPI2 stool DNA methylation for detection of CRC using a training and validation set. Initial pilot feasibility study had a limitation of using phenol chloroform extraction protocol for DNA extraction from stool which is a time-consuming and labor-intensive process. Envisioning the potential utility of TFPI2 to design stool assays for screening purposes in the future, we used QIAamp DNA extraction stool mini kit protocol which is a faster and more efficient platform for DNA extraction and similarly adapted to a real-time platform. Methodologies and analyses were consistent for the training set and validation set. In the training set, overall mean age for all subjects including cases and controls was 60.5 years. Mean ages for stage I to III CRCs and controls were 69.3 and 55 years, respectively (Table 1A); 52% of the subjects were males whereas 46% were females (Table 1A). In the training set, TFPI2 stool DNA methylation had a sensitivity of 89% for detection of stage I to III CRCs with a specificity of 79% (Table 1B). Because the mean age of the cases and controls differed, a ROC-GLM regression model was used to assess the accuracy of TFPI2 promoter methylation after adjustment for age. This analysis indicated that age did not significantly influence the accuracy (P = 0.58, ROC-GLM regression model). In the validation set, mean age for all subjects including cases and controls was 63.3 years. Mean ages for cases and controls were 68.3 years (71.1 years for CRC and 61.4 years for adenomas) and 52.3 years, respectively. 45% of the study subjects were males whereas 54% were females (Table 1A). In the validation set, TFPI2 stool DNA methylation had a sensitivity of 76% for detection of stage I to III CRCs with a specificity of 93%. However, only 4 out of 19 advanced adenomas (sensitivity 21%) could be detected with a specificity of 93%. Table 1B summarizes the results in detail.
Discussion
Previous studies have shown TFPI2 hypermethylation in other cancer types (16, 17, 25-28), suggesting an important role for this gene in the etiology of human malignancies. Based on the high level of gene re-expression measured in our array analysis, frequent methylation and silencing of TFPI2, which responded to DAC treatment in CRC cell lines, and the presence of the PcG mark, which predisposes genes to methylation early on during carcino-genesis, we speculated that TFPI2 might represent a candidate for the detection of CRC. We detected the methylation of TFPI2 in nearly all primary adenomas and CRC samples. For stool DNA tests, the sensitivity was found to be 76% to 89% for stage I to III CRCs, with a specificity of 79% to 93%. Evidence from our study and others indicate that TFPI2 may be an important tumor suppressor gene in CRC and other malignancies (16, 17, 25-28).
Our present findings have potential clinical implications. To date, it seems that the most reliable early detection of cancer is provided by a signature combining multiple gene mutations, each of which individually has a low sensitivity (10, 29, 30). This is further supported by the current genome-wide sequencing efforts which have shown that each of these mutational event is seen in a small number of tumors (<15-50%; ref. 31). Interestingly, some of these studies have reported high sensitivity for stool DNA–based mutational assays but did not comment on specificity (32). Nonetheless, these studies have played an important role in spreading the idea of stool DNA–based tests and provided insightful data on mutations present in the stool DNA of CRC patients. Detection of CRC-specific methylation in stool samples using a single epigenetic marker has also been explored recently (33-35). Ultimately, the goal is to design an ideal screening tool which is noninvasive, sensitive, specific, cost-effective, and easy to implement across large populations. This is particularly of importance, given the recent guidelines by the American Cancer Society, recommending stool DNA testing for CRC detection (8).
Epigenetic studies which focus on analyzing single genes may be more cost-effective for screening. In general, the available results of single marker epigenetic studies are difficult to compare because study designs and patient populations differ. However, these studies have provided impressive preliminary data about these single epigenetic markers and their potential utility as stool DNA–based assays. Zhang and colleagues reported a sensitivity of 86% and specificity of 89% for SFRP1 using a cohort of 36 patients, of which 9 had undergone neoadjuvant therapy and most cases (31 of 36, 88%) were predominantly rectal or left-sided (35). Our results are not comparable with theirs as they had a smaller sample size, no validation cohort, lacked a blinded study design, and included stage IV patients and patients who had undergone neoadjuvant chemotherapy. Moreover, inclusion of rectal adenomas and predominantly rectal and left-sided carcinomas may result in higher sensitivity. Other single marker methylation-based stool studies using Vimentin (33) and SFRP2 (34, 36, 37) have reported a sensitivity of 52% and 87% to 94%, respectively, and a specificity of 90% and 85% to 90%, respectively. Muller and colleagues reported a sensitivity of 77% to 90% and a specificity of 77% for SFRP2 in a set of 49 patients (38). Some of these studies included a substantial number of stage IV patients or did not mention the stage of tumors (34, 36). Conversely, the strengths of our study include its double-blinded design, exclusion of stage IV patients, and independent training and validation sets which were lacking in previous studies. Another interesting finding is the discovery of frequent methylation of TFPI2 in >95% of colorectal adenomas and cancers which has not been investigated earlier. Our data indicate that TFPI2 stool DNA methylation has a sensitivity of 76% to 89% for the detection of stage I to III CRCs and specificity of 79% to 93%. However, for adenomas, although >95% methylation was seen in primary tissues, only 21% to 43% sensitivity was achieved for stool samples. This could potentially be explained by fewer cells shed by adenomas and the lack of high potential for anchorage-independent growth in the shed adenoma cells, which could have resulted in their degradation (9). Other stool DNA methylation–based studies have reported a sensitivity of 42% to 77% for the detection of CRCs and a relatively low sensitivity of 31% to 48% for the detection of adenomas with a specificity of 73% to 100% for both CRCs and adenomas (33-35, 39).
Although a total of 197 stool DNA samples were analyzed in the current study, the sample size is still smaller as compared with multicenter trials and there is a need for studies with larger sample size for cost-effective analyses and better measurement of sensitivity and specificity. Another limitation is the lack of methylation data on tumors of patients whose stool was analyzed in the current study. This may have provided better correlation and understanding of factors which may have resulted in reduced sensitivity for adenomas.
In conclusion, TFPI2 methylation is an early and frequent event in colorectal adenomas and carcinomas which is seen infrequently in the mucosa of normal noncancer patients. We have reported, for the first time, the bivalency of the TFPI2 promoter and its potential role as a tumor suppressor in CRC. Our data on stool DNA of CRC and adenoma patients suggests that TFPI2 promoter methylation is a feasible epigenetic marker for the detection of CRC and may be useful for CRC screening in future.
Supplementary Material
Acknowledgments
Grant support: The study was supported by NIHK23CA127141, the Ross Clinician Scientist Award and the Wendy Will Foundation.
We would like to thank Sharon Metzger-Gaud and the Johns Hopkins Cancer Registry for assistance with the primary cancer databases.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
G.A. Meijer and M. Van Engeland: commercial research grant and ownership interest, Oncomethylome Sciences. The other authors disclosed no potential conflicts of interest.
References
- 1.Ohm JE, Baylin SB. Stem cell chromatin patterns: an instructive mechanism for DNA hypermethylation? Cell Cycle. 2007;6:1040–3. doi: 10.4161/cc.6.9.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 4.Shames DS, Minna JD, Gazdar AF. DNA methylation in health, disease, and cancer. Curr Mol Med. 2007;7:85–102. doi: 10.2174/156652407779940413. [DOI] [PubMed] [Google Scholar]
- 5.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 6.Heresbach D, Manfredi S, D'Halluin PN, Bretagne JF, Branger B. Review in depth and meta-analysis of controlled trials on colorectal cancer screening by faecal occult blood test. Eur J Gastroenterol Hepatol. 2006;18:427–33. doi: 10.1097/00042737-200604000-00018. [DOI] [PubMed] [Google Scholar]
- 7.Meissner HI, Breen N, Klabunde CN, Vernon SW. Patterns of colorectal cancer screening uptake among men and women in the United States. Cancer Epidemiol Biomarkers Prev. 2006;15:389–94. doi: 10.1158/1055-9965.EPI-05-0678. [DOI] [PubMed] [Google Scholar]
- 8.Levin B, Lieberman DA, McFarland B, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. CA Cancer J Clin. 2008;58:130–60. doi: 10.3322/CA.2007.0018. [DOI] [PubMed] [Google Scholar]
- 9.Osborn NK, Ahlquist DA. Stool screening for colorectal cancer: molecular approaches. Gastroenterology. 2005;128:192–206. doi: 10.1053/j.gastro.2004.10.041. [DOI] [PubMed] [Google Scholar]
- 10.Ahlquist DA, Sargent DJ, Loprinzi CL, et al. Stool DNA and occult blood testing for screen detection of colorectal neoplasia. Ann Intern Med. 2008;149:441–50, W481. doi: 10.7326/0003-4819-149-7-200810070-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–23. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohm JE, McGarvey KM, Yu X, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–42. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Widschwendter M, Fiegl H, Egle D, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–8. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 14.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–56. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 15.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 16.Wong CM, Ng YL, Lee JM, et al. Tissue factor pathway inhibitor-2 as a frequently silenced tumor suppressor gene in hepatocellular carcinoma. Hepatology. 2007;45:1129–38. doi: 10.1002/hep.21578. [DOI] [PubMed] [Google Scholar]
- 17.Sato N, Parker AR, Fukushima N, et al. Epigenetic inactivation of TFPI-2 as a common mechanism associated with growth and invasion of pancreatic ductal adenocarcinoma. Oncogene. 2005;24:850–8. doi: 10.1038/sj.onc.1208050. [DOI] [PubMed] [Google Scholar]
- 18.McGarvey KM, Van Neste L, Cope L, et al. Defining a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells. Cancer Res. 2008;68:5753–9. doi: 10.1158/0008-5472.CAN-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim MS, Chang X, Nagpal JK, et al. The N-methyl-D-aspartate receptor type 2A is frequently methylated in human colorectal carcinoma and suppresses cell growth. Oncogene. 2008;27:2045–54. doi: 10.1038/sj.onc.1210842. [DOI] [PubMed] [Google Scholar]
- 20.van Engeland M, Weijenberg MP, Roemen GM, et al. Effects of dietary folate and alcohol intake on promoter methylation in sporadic colorectal cancer: the Netherlands Cohort Study on Diet and Cancer. Cancer Res. 2003;63:3133–7. [PubMed] [Google Scholar]
- 21.Janes H, Pepe MS. Adjusting for covariates in studies of diagnostic, screening, or prognostic markers: an old concept in a new setting. Am J Epidemiol. 2008;168:89–97. doi: 10.1093/aje/kwn099. [DOI] [PubMed] [Google Scholar]
- 22.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–36. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nobeyama Y, Okochi-Takada E, Furuta J, et al. Silencing of tissue factor pathway inhibitor-2 gene in malignant melanomas. Int J Cancer. 2007;121:301–7. doi: 10.1002/ijc.22637. [DOI] [PubMed] [Google Scholar]
- 26.Rollin J, Iochmann S, Blechet C, et al. Expression and methylation status of tissue factor pathway inhibitor-2 gene in non-small-cell lung cancer. Br J Cancer. 2005;92:775–83. doi: 10.1038/sj.bjc.6602298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shames DS, Girard L, Gao B, et al. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. PLoS Med. 2006;3:e486. doi: 10.1371/journal.pmed.0030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sova P, Feng Q, Geiss G, et al. Discovery of novel methylation biomarkers in cervical carcinoma by global demethylation and microarray analysis. Cancer Epidemiol Biomarkers Prev. 2006;15:114–23. doi: 10.1158/1055-9965.EPI-05-0323. [DOI] [PubMed] [Google Scholar]
- 29.Mixich F, Ioana M, Voinea F, Saftoiu A, Ciurea T. Noninvasive detection through REMS-PCR technique of K-ras mutations in stool DNA of patients with colorectal cancer. J Gastrointestin Liver Dis. 2007;16:5–10. [PubMed] [Google Scholar]
- 30.Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704–14. doi: 10.1056/NEJMoa033403. [DOI] [PubMed] [Google Scholar]
- 31.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 32.Diehl F, Schmidt K, Durkee KH, et al. Analysis of mutations in DNA isolated from plasma and stool of colorectal cancer patients. Gastroenterology. 2008;135:489–98. doi: 10.1053/j.gastro.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen WD, Han ZJ, Skoletsky J, et al. Detection in fecal DNA of colon cancer-specific methylation of the non-expressed vimentin gene. J Natl Cancer Inst. 2005;97:1124–32. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- 34.Wang DR, Tang D. Hypermethylated SFRP2 gene in fecal DNA is a high potential biomarker for colorectal cancer noninvasive screening. World J Gastroenterol. 2008;14:524–31. doi: 10.3748/wjg.14.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang W, Bauer M, Croner RS, et al. DNA stool test for colorectal cancer: hypermethylation of the secreted frizzled-related protein-1 gene. Dis Colon Rectum. 2007;50:1618–26. doi: 10.1007/s10350-007-0286-6. discussion 1626–1617. [DOI] [PubMed] [Google Scholar]
- 36.Huang Z, Li L, Wang J. Hypermethylation of SFRP2 as a potential marker for stool-based detection of colorectal cancer and precancerous lesions. Dig Dis Sci. 2007;52:2287–91. doi: 10.1007/s10620-007-9755-y. [DOI] [PubMed] [Google Scholar]
- 37.Oberwalder M, Zitt M, Wontner C, et al. SFRP2 methylation in fecal DNA—a marker for colorectal polyps. Int J Colorectal Dis. 2008;23:15–9. doi: 10.1007/s00384-007-0355-2. [DOI] [PubMed] [Google Scholar]
- 38.Muller HM, Oberwalder M, Fiegl H, et al. Methylation changes in faecal DNA: a marker for colorectal cancer screening? Lancet. 2004;363:1283–5. doi: 10.1016/S0140-6736(04)16002-9. [DOI] [PubMed] [Google Scholar]
- 39.Petko Z, Ghiassi M, Shuber A, et al. Aberrantly methylated CDKN2A, MGMT, MLH1 in colon polyps and in fecal DNA from patients with colorectal polyps. Clin Cancer Res. 2005;11:1203–9. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.