Abstract
Conjugated linoleic acids (CLA) are found naturally in dairy products. Two isomers of CLA, that differ only in the location of cis and trans double bonds, are found to have distinct and different biological effects. The cis 9 trans 11 (C9T11) isomer is attributed to have the anti-carcinogenic effects, while the trans 10 cis 12 (T10C12) isomer is believed to be responsible for the anti-obesity effects. Since dietary CLA are incorporated into membrane phospholipids, we have used Molecular Dynamics (MD) simulations to investigate the comparative effects of the two isomers on lipid bilayer structure. Specifically, simulations of phosphatidylcholine lipid bilayers in which the sn-2 chains contained one of the two isomers of CLA were performed. Force field parameters for the torsional potential of double bonds were obtained from ab initio calculations. From the MD trajectories we calculated and compared structural properties of the two lipid bilayers, including areas per molecule, density profiles, thickness of bilayers, tilt angle of tail chains, order parameters profiles, radial distribution function (RDF) and lateral pressure profiles. The main differences found between bilayers of the two CLA isomers, are (1) the order parameter profile for C9T11 has a dip in the middle of sn-2 chain while the profile for T10C12 has a deeper dip close to terminal of sn-2 chain, and (2) the lateral pressure profiles show differences between the two isomers. Our simulation results reveal localized physical structural differences between bilayers of the two CLA isomers that may contribute to different biological effects through differential interactions with membrane proteins or cholesterol.
Keywords: Conjugated linoleic acids, molecular dynamic simulation
1 Introduction
Isomers of linoleic acid that contain conjugated double bonds (conjugated linoleic acids, CLA) occur naturally in dairy products and meats, and are known to exert several biological effects in experimental animals, including anti-carcinogenic effects, reduction in adiposity, and improvement of immune function (1; 2; 3). Although 16 isomers of CLA have been identified in natural products, only two, cis 9 trans 11 (80%) and trans 10 cis 12 isomers (10%) dominate in dairy products, whereas man-made CLA preparations which are used as nutritional supplements generally contain equal amounts of the same two isomers. Even though both these isomers contain one trans double bond and one cis double bond, they differ significantly from each other in their biological effects, suggesting that the location of the cis and trans double bonds in the acyl chain plays a critical role in the biological function. Thus, the cis 9 trans 11 isomer appears to be responsible for most of the anti-carcinogenic effects, while the trans 10 cis 12 isomer appears to be responsible for the anti-obesity and anti-atherogenic effects (4), as well as the insulin-resistance effects in mice (5). The mechanism of action, and the molecular basis for the divergent effects of CLA isomers remain unclear, despite numerous studies in experimental animals and cultured cells. Some of the biological effects of CLA are due to their regulation of transcription factors such as PPARs, LXR, SREBPs, and NF kappa B (2; 6). While the two CLA isomers have also been shown to differentially regulate the transcription of selected genes in cultured cells (7; 8), the pleiotropic effects of CLA cannot be completely explained by these mechanisms. Furthermore CLA, like other dietary fatty acids, do not remain as free fatty acids in the cells, but are incorporated into membrane phospholipids (9; 10; 11; 12; 13). Therefore it is possible that at least some of the effects of CLA are through modification of membrane structure and function, as demonstrated for the omega 3 fatty acids which alter the distribution of lipids and proteins between raft and non-raft fractions of the membranes, and thereby affect cellular functions (14; 15; 16; 17). To test this hypothesis, we conducted a molecular dynamics (MD) simulation study of bilayers of phosphatidylcholine (PC) which contained a palmitic acid at sn-1 position, and cis 9 trans 11 CLA or trans 10 cis 12 CLA at sn-2 position. MD simulations of phospholipid bilayers are widely used to study the physical properties of specific bilayers with different lipid compositions, with cholesterol, or with embedded peptide/proteins. Current MD studies of lipid bilayer structure predict bilayer structural properties that are in excellent agreement with experimental data. Given this agreement, simulations can add atomic level insight into interactions between lipids, cholesterol, and water (18; 19; 20; 21; 22; 23; 24). For example, Pandit et al (25) used atomistic simulations to compare the effect of chain unsaturation on lipid-cholesterol interactions in binary mixtures, and thereby providing insight into differential lipid interactions related to the formation of “raft”–like domains. In this paper, we focus on two linoleic acid isomers, cis 9 trans 11 and trans 10 cis 12, as these two isomers have important physiological functions. Although the effect of isolated double bonds has been studied by MD simulation (26), no studies have been conducted on the effect of conjugated double bonds on phospholipid structure.
2 MD Method
In MD simulations it is necessary to specify the “force field”, a listing of all interaction parameters between all pairs of molecules, both bonded and non-bonded. Force fields are generally calculated by independently fitting MD simulations to well characterized simpler compounds such as liquid alkanes, water, or various “model compounds” that resemble other fragments of lipid molecules. The force-field parameters used for this work were taken from the newly developed GROMOS 43A1-S3 parameter set (27). This set provides significant improvement over earlier GROMOS parameter sets (28; 29) through improved parameters for hydrocarbon chain van der Waals potentials, and for polar group dihedral potentials. Our 43A1-S3 force field was designed with simulations using 16 angstrom cutoffs. The choice of 16 angstroms was based on simulations of hydrocarbons, where we found that the was the necessary cutoff to eliminate a strong cutoff-dependence of computed specific volume. The van der Waals parameters were set by matching specific volume data for hydrocarbons (for the lipid tails) and for small molecule analogues of the head groups. They were then validated by showing that when used in lipid bilayers, they match the diffraction data, which is the raw data from which cross-sectional areas are derived. As Pabst et al(30) point out, recent advances in the computational tools for interpreting diffraction data (31) have resulted in reducing the experimental estimates of lipid cross-sectional area by as much as 10%. Thus the parallel advances in simulation technology and technology for data analysis have resulted in a reduction in our best estimates for lipid cross-sectional area.
For CLA simulations it was necessary to calculate additional dihedral parameters for the conjugated double bonds for this work. The importance of these parameters has been emphasized in a recent study by Martinez-Seara et al (32)This was done following the procedure used for the 43A1-S3 force field (27), using the model compound trans, cis (EZ)-2,4-hexadiene for both isomers (since the cis/trans order in C9T11 and T10C12 is simply reversed). For bonding parameters not part of the -CH=CH-CH=CH- dihedral we used existing parameters in the 43A1-S3 force field. For the conjugated double bond dihedral energy parameterizations, in both cases, non bonding parameters were set to existing parameters in 43A1-S3 and ab initio calculations were done at the MP2=full/6-31G** level using Gaussian. Figure 2 shows that the potential energy profile of E, Z-2,4 hexadiene (CH3 - CH = CH - CH = CH - CH3) as a function of the central dihedral angle ( - CH = CH - CH = CH -) is quantitatively the same for the quantum mechanical calculation and the molecular mechanical calculation: both have primary minima at 180 ° and secondary minima at about 50 °. The parameters were further validated using MD simulations of liquid EZ-2–4-hexadiene with the simulated results: density (298 K) = 0.7074 g/l.ΔHvap = 36 kJ/mol with same protocols used for other parameters (27). Experimental data are: density (298 K) = 0.7185 (Chem & Phys Handbook). ΔHvap = 33.9 kJ/mol (Chemical properties Handbook, Mcgraw-Hill).
Figure 2.
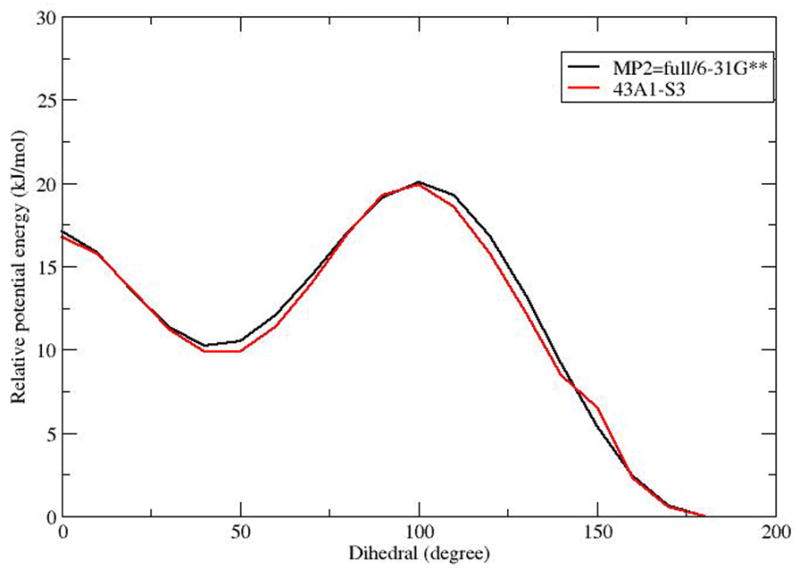
Potential energy profile of E, Z-2,4 hexadiene (CH3 - CH = CH -CH = CH - CH3) as a function of the central dihedral (- CH = CH - CH = CH -).
Figure 1 shows the structures and numbering schemes for each of the CLA-PC isomers. The total number of lipids in each bilayer was 200, or 100 per leaflet. The bilayers were hydrated with 6800 SPC-E water molecules, for a hydration level of 34 water molecules per lipid. All the simulations were performed using the GROMACS 4.05 package (33). The LINCS algorithm was used to constrain all the bonds in the system (34) allowing an integration time step of 2 fs. Periodic boundary conditions were applied in all three dimensions and long range electrostatics were calculated using the PME algorithm (35) with a real space cutoff of 10 Å. As noted above, a cutoff of 16 Å was employed for van der Waals interactions. All simulations were performed at 298 K using the Nose-Hoover temperature coupling scheme. The system was simulated in an NPT ensemble using the Parrinello-Rahman pressure coupling scheme at a constant pressure of 1 bar.
Figure 1.
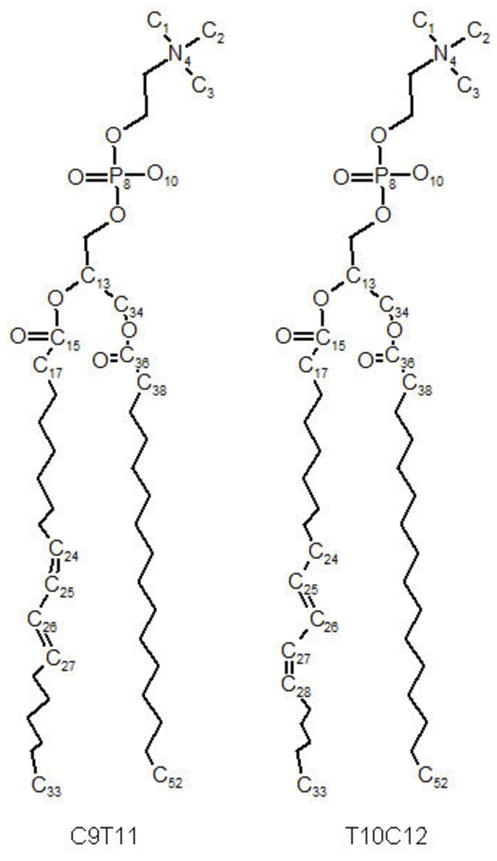
Structures of CLA-PCs molecules, with the numbering schema used in the simulations.
The only difference between CLA-PC and palmitoyl-oleoyl phosphatidylcholine (POPC) molecules is in the conjugated double bonds on the sn-2 chain. Therefore we built CLA-PC molecules for both C9T11 and T10C12 from a POPC molecule, and then did energy minimization on the resulting structure. Initial configurations of bilayers were subsequently generated by random placement of 100 CLA-PC molecules per leaflet using Gromacs utilities. The systems were energy minimized to remove bad contacts. A 200 ps MD simulation was performed on each system at 500 K. This was done to ensure proper disordering of the hydrocarbon chains. Then the temperatures were brought down to the target temperature of 298 K in steps of 50 K. At each temperature step a small 200 ps MD simulation was performed on each system. At this point the systems were simulated for 180 ns of MD with regeneration of velocities from a Maxwellian distribution at 298 K after every 200 ps. Then 100 ns continuous MD trajectories were generated and used for analysis. To verify that simulation results were not dependent on initial conditions, we built two more systems with different initial configuration of bilayer, larger system size and used a longer annealing time. Calculated properties from those runs are quantitatively identical to results presented here.
3 MD Results and discussion
Dihedral distributions of conjugated double bonds
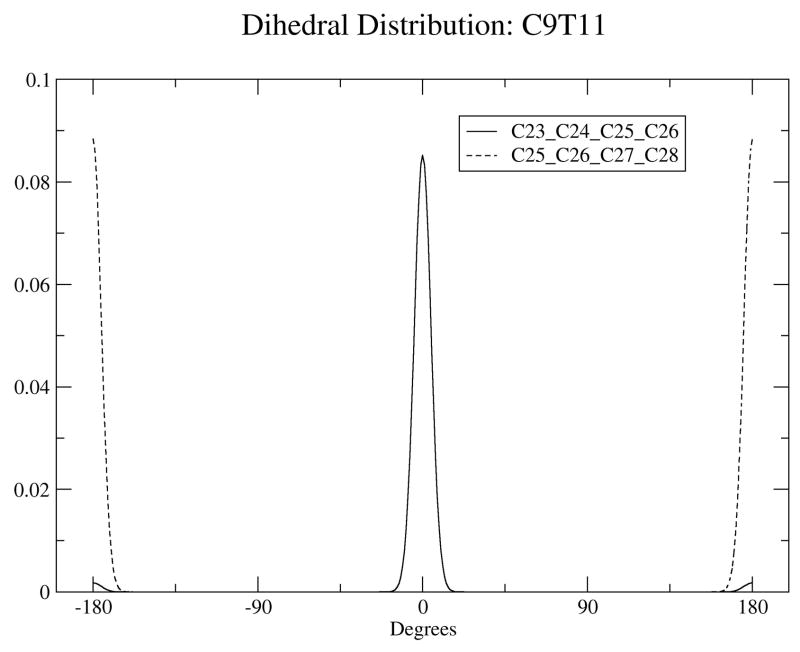
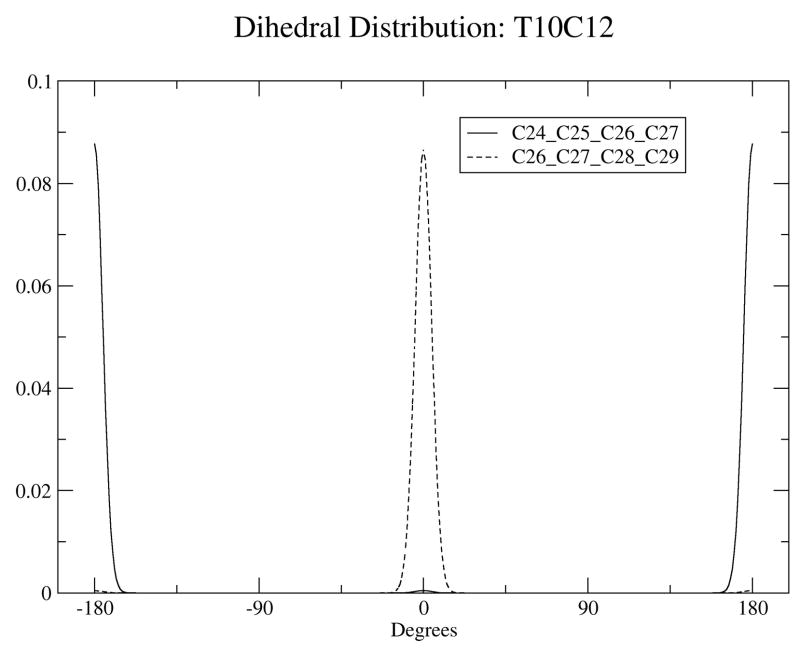
To further validate the calculated conjugated bond dihedral potential functions, dihedral angle distributions were monitored during MD simulations. Trajectory average distributions of conjugated dihedral angles for the two dihedrals involving double bonds in the two bilayer systems are plotted in Figure 3. Angles around 0° indicate the four carbons of the dihedral are in the cis configuration, while angles around 180° indicate the trans configuration. For C9T11, the cis double bond lies between C24 and C25, the solid line in the middle shows the bond remains in the cis conformation, while the dotted line shows the trans double bond, between C26 and C27 remains in that conformation. For T10C12, the cis/trans order is reversed compared to C9T11, with the trans bond between C25 and C26 and the cis bond between C26 and C28 as shown in Figure 3. We calculated reorientational correlation times for the dihedrals for which the middle C-C bond is the single bond (the dihedrals of Figure 3(c)) for both isomers, using GROMACS utilities. For C9T11 we obtained a correlation time of 0.177 ns, and for T10C12 we obtained a correlation time of 0.174 ns, so that the rates are basically the same. For comparison, for the single cis double bond in di-oleyol phosphatidylcholine (DOPC) the orientational relaxation time, from Deuterium NMR experiments, is about 0.11 ns at 40° C. (36)
Figure 3.
The dihedral distribution of double bonds for C9T11 (a) and T10C12 (b) isomers. (c): Dihedral distrubituon for the central carbon atoms for both isomers.
Structural and thermodynamic properties
In MD studies of lipid bilayers, the calculated area per molecule is often used to assess the validity of a simulation. We obtained molecular areas by dividing the xy area of the simulation box by the number of lipids per leaflet, 100. The initial fluctuations in the area per lipid were large, but decreased over time. After 100 ns the areas became stable with small fluctuations, so we began production runs at that point. Figure 4 shows the calculated areas per lipid over the full simulations for C9T11 and T10C12. The values for the mean area per lipid are: AC9T11 = 63.06 ± 0.007Å2, and AT10C12 = 62.43 ± 0.005Å2. The area per molecule for the isomers are nearly identical for the two simulated bilayers at 298 K, in agreement with experimental results for lipid monolayers spread on a purified water substrate at 25° C and a surface pressure of 20 mN/m. (37) More generally, the simulated areas for the two isomers are consistent with experimental isotherm data lipid monolayers (37) for full isotherms of C9T11 and T10C12 at room temperature (24° C). In these data, isotherms of C9T11 and T10C12 monolayers are nearly identical. The small area fluctuations in the simulations indicate good equilibration of the systems after 280 ns simulation time. For comparison to a well studied phospholipid bilayer, the experimental value of the area per lipid of a POPC bilayer, from the X-ray scattering experiments, is 68.3 ± 1.5 (38) at 303K. Bachar et al (26) reported, in simulations run at at 300K under a constant pressure of one atmosphere (NPT conditions) an area per lipid for cis 9 cis 12 (C9C12) of 70.3 ± 0.1. For further comparison, the area for phospholipid containing docosahexaenoic acid (DHA) was also reported to be about 70 Å2. (39). CLAs have two double bonds in their sn-2 chain compared to one in POPC, but with one double bond in a trans configuration. Conformationally, the trans double bond acts like saturated bond which makes the sn-2 chain more trans-like in its conformation, but also somewhat more rigid. The smaller surface area for both CLA simulations indicates that the presence of the conjugated trans double bond decreases the surface area, as observed in monolayer experiments. (37).
Figure 4.
The average areas per lipid versus time for C9T11 and T10C12 isomers.
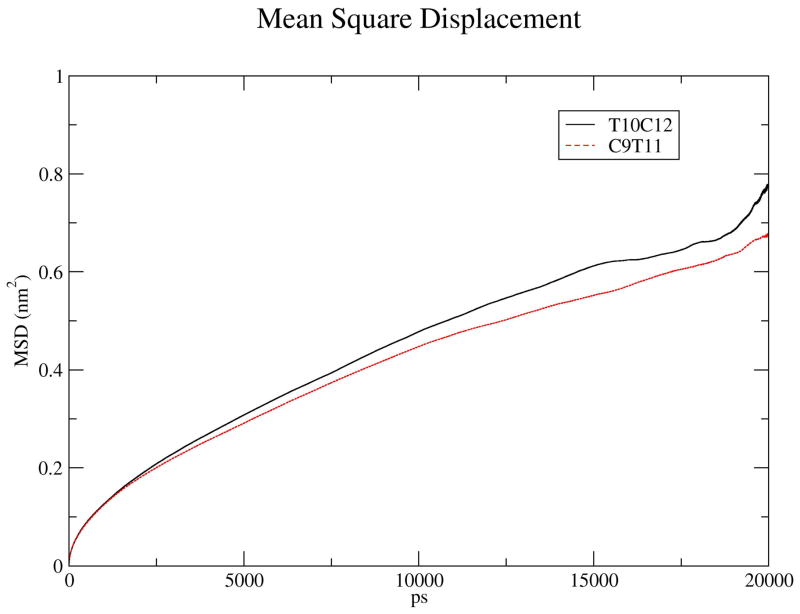
Lipids move diffusively in the membrane plane with a experimentally determined lateral diffusion coeffcients on the order of 8 μm2=s at 298 K for POPC (40), and from simulations for DHA at 310 K (41). For the CLA bilayer simulations, we calculated mean square displacements (MSD) of CLA lipids over simulation production runs. Fig. 5 shows lateral MSD curves on the xy plane of the simulation boxes for C9T11 and T10C12. Both MSD curves are nearly linear, with the curve for C9T11 exhibiting a smaller slope. Using Einstein’s relation
Figure 5.
The lateral mean square displacement (MSD) of the centers of mass of C9T11 and T10C12 isomers, averaged over 3ve trajectories of 20 ns duration each, versus the relative trajectory time. Windows for the five trajectories were 10–30 ns, 30–50 ns, 50–70 ns, 60–80 ns, and 80–100 ns. The centers of mass motion of the full monolayers were removed before analysis.
| (1) |
we calculated the lateral diffusion coeffcients (D||) by fitting a straight line to the MSD traces over windows 20 ns in width in the MD trajectories. Windows were used between 10 ns − 30 ns, 30 ns − 50 ns, 50 ns − 70 ns, 60 – 80 ns, and 80 – 100 ns. Fig. 5 shows the lateral MSD of the centers of mass of C9T11 and T10C12 isomers, averaged over the five 20 ns trajectory windows for each isomer, versus the relative trajectory time. Diffusion constants were calculated from slopes for each window, and averaged. The results are: D|| for C9T11 = 4.12 ± 1.9μm2/s; D|| for T10C12 = 5.76 ± 3:5μm2/s. The diffusion constant for T10C12 is larger than that for C9T11 but the difference is within the error bars.
Lipid chain conformational flexibility is commonly described by profiles of “order parameters”. The definition of the order parameter for the m’th carbon in a chain is
| (2) |
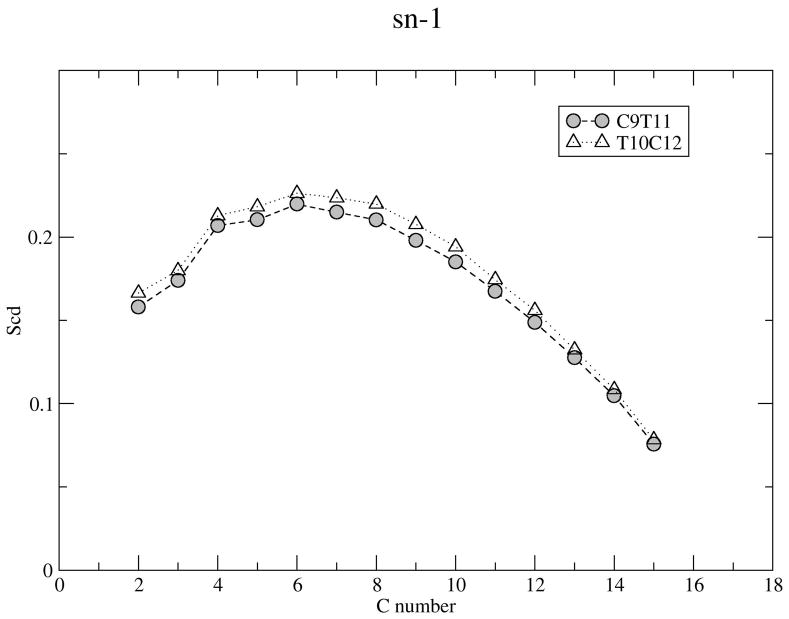
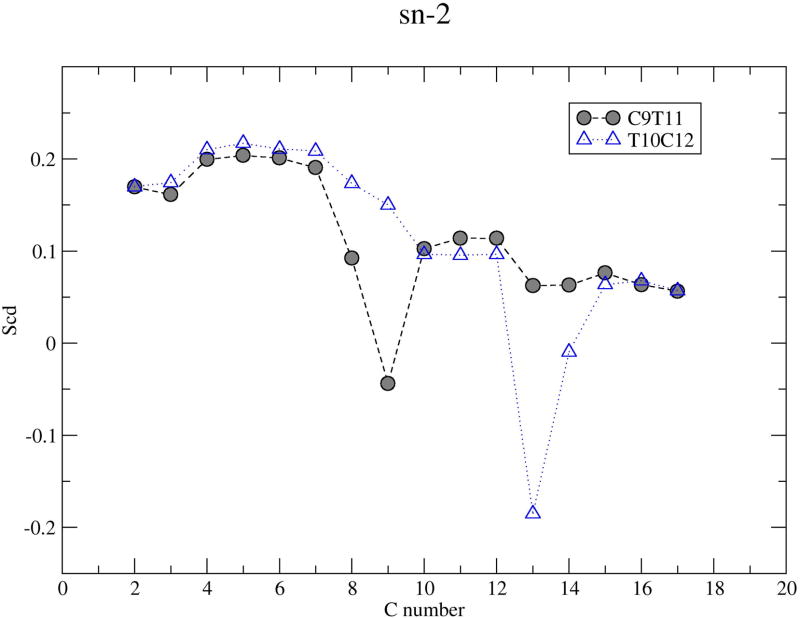
where θm is the angle between the C-H bond vectors and the bilayer normal for carbon m. (42) In general, if a segment can freely rotate, its order parameter will be close to 0 whereas if it is completely rigid and the chain is oriented parallel to the bilayer normal, its order parameter will be close to 0.5 in absolute value. Figure 6 shows the segmental deuterium order parameters for C9T11 and T10C12 saturated and unsaturated chains. The SCD for T10C12 saturated chain are slightly higher than those of C9T11, reaching a maximum of about 0.22. There are currently no experimental measurements of segmental order parameters for CLA but we can compare our results with simulations of polyunsaturated chains in bilayers. Bacher et al. (26) find maximum values for order parameters of about 0.18–0.19 for C9C12. Figure 6 shows that the palmitic acid (sn-1) chain carbons are more ordered than CLA (sn-2) tail carbons and the cis double bond creates a characteristic dip in the profiles for the unsaturated chains. The order parameter profile for C9T11 has a dip at carbon 9, while that for T10C12 has a lower dip at carbon 13 and the middle carbons are less ordered in this chain. By comparison, the simulated order parameter profile for a C9C12 unsaturated chain Bachar et al (26) contains two dips, a narrow dip at carbon 10 and another broad dip at carbon 13. The fact that cis double bonds enhance the disorder to a far greater extent that than do trans double bonds indicates that the ordering of the cis and trans double bonds in CLA isomers contribute to differing levels of disorder within the membrane. Lipid order parameter profiles tend to reflect fluidity of membrane interior. Salem et al (43) studied the effects of the number of double bonds on lipid fluidity by making small unilamellar vesicles (SUV) from PCs containing stearic acid (18:0) in the sn-1 position and long chain fatty acids with 0 to 6 double bonds in the sn-2 position. Their room temperature results showed a large increase in fluidity upon the addition of the first double bond, a smaller further increase with the second and a still smaller increase in fluidity with up to 4 double bonds. Docosahexaenoic acid (DHA) is the longest and most unsaturated fatty acid commonly found in membranes. Some room temperature dietary studies (44; 45) have observed an increase in membrane fluidity in animals/cells upon incorporation of DHA. In the present simulations, both CLA isomers have two double bonds which may also influence membrane fluidity.
Figure 6.
Deuterium order parameters for C9T11 and T10C12 lipid chains at 298K
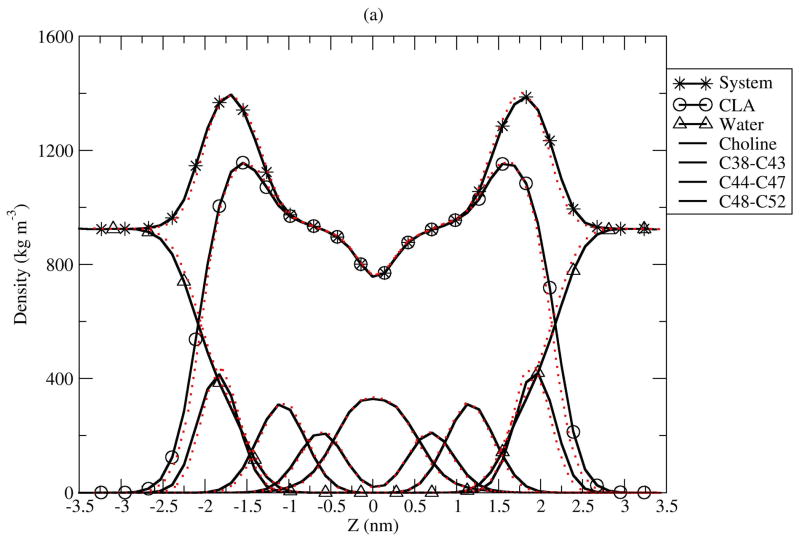
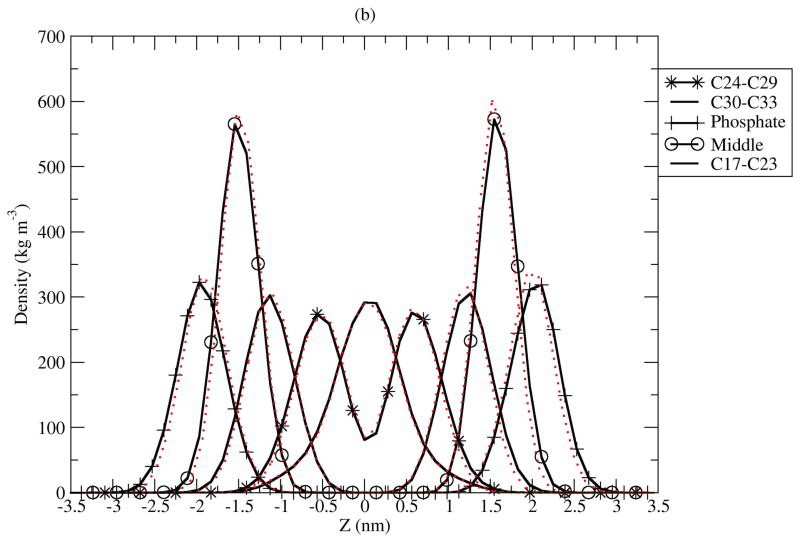
Atomic density profiles across the bilayers present a picture of the distributions of specific molecular subgroups within the membrane. Figure 7 shows calculated mass density profiles from the simulations. The decomposition of the total density into contributions from CLA, water, choline, phosphate and double bond (C24 to C29) groups shows little difference between the two isomers. We measured the thickness of the bilayer as the peak to peak distance between phosphate groups in the density profiles. The bilayer thickness for C9T11 is about 39.4 Å, which is about 0.7 Å thicker than that of T10C12. The thickness difference is within the estimated uncertainties in our simulations, but both bilayers are significantly thicker than that of C9C12 reported by Bachar et al, 35 Å, under simulation conditions described above. (26) Membrane permeability and oxidizablity are related to thickness of bilayer, and therefore these properties should be the same for bilayers composed of either CLA isomer, but would be different from non-conjugated C9C12. Experiments shows acyl chains of phospholipids containing polyunsaturated DHA, with 22 carbons and 6 double bonds on the sn-2 chain, tend to fold back (upturn) to make the bilayer thinner. The mass density profiles in Fig. 7 show no significant “upturn” in these membranes. It is also of interest to examine the average tilt of the sn-1 and sn-2 chains for the C9T11 and T01C12 isomers. Given that there are no upturns in the chains on average, tilt angles were calculated from the angle between the bilayer normal and vector between C33 and C15 carbons for sn-2, and between C52 and C36 for sn-1 respectively, for both CLA isomers. The results (not shown) reveal nearly identical distributions of tilt angle for both CLA isomers. The distributions have peaks around 22 degrees for sn-1 and sn-2 chains fand widths of about 35° at half maximum. For both isomers, the sn-2 chains exhibit a slightly larger tilt angle distribution than the sn-1 chains. The maximum tilt angles for the CLA isomers are are smaller than the tilt distribution calculated for C9C12 (26), 34.8 ± 18.3°.
Figure 7.
Mass density profiles for C9T11 and T10C12 isomers. The solid line is C9T11, the dotted line is T10C12. (a): under 400 kg=m3, the order of curves from left=right to middle (0) are: Choline group, C38 to C43, C44 to C47 and C48 to C52. (b): the order of curves from left/right to middle are: PO4 group, middle atoms (C12, C13, OS14, C15, O16, C34, OS35, C36, O37), C17 to C23, C24 to C29 (sn-2 double bonds), C30 to C33.
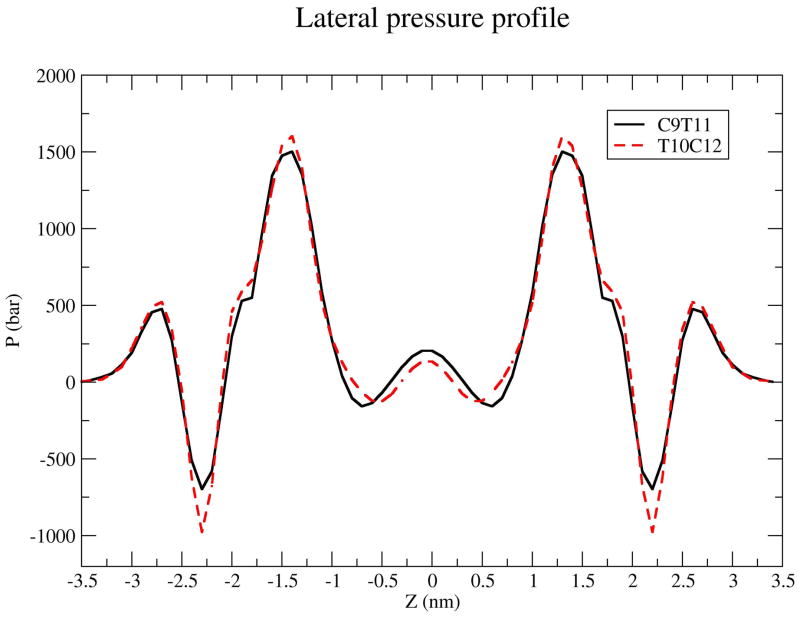
A sensitive indicator of the state of the interior of a bilayer is the lateral pressure profile, P(z). The lateral pressure in a membrane interior can affect interactions between lipids and membrane proteins, and can thereby influence the organization of the proteins, which can in turn affect signalling and other biological functions. (46) In MD simulations lateral pressure profiles are calculated from the diagonal components of the pressure tensor which are in turn calculated from the virial function for the membrane. (47) Figure 8 shows our calculated pressure profiles for the two CLA isomers. The profiles are qualitatively similar but with differences in the membrane near the choline groups, where T10C12 has a lower (more negative) pressure, which is indicative of a relatively larger surface tension in this region. In the bilayer central region, T10C12 has a lower pressure. T10C12 has a slightly higher pressure in the region of the membrane interior between the location of the first double bond and the aqueous interface. While the differences on a relative scale are small, we note that the scale on Figure 8 is hundreds of bars. We also note that the data for Figure 8 have been symmetrized by averaging over the two leaflets for each membrane. We can compare our calculated profiles with profiles calculated by MD simulation for polyunsaturated lipid bilayers by Ollila et al. (48) They also find the expected minimum in the lateral pressure near the membrane choline groups, and they find that the magnitude of the negative pressure in this region increases with the degree of unsaturation of the membrane. They also find that the pressure in the bilayer center decreases with increased unsaturation. In order to identify subtle differences between the pressure profiles for the two isomers, that may contribute to differences in mechanical and thermodynamical properties. We also calculated the first two moments of the pressure profiles:
| (3) |
Figure 8.
Lateral pressure profiles for C9T11 (solid line) and T10C12 (dashed line) isomers. Data have symmetrized by averaging over the two leaflets.
For this calculation we integrate from the bilayer center out to the polar group region. We found that the moments are again nearly identical: For c9t11 M1/kBT = 3.5 Å−1 and M2/kBT = 57.7 while for T10C12 M1/kBT = 3.6Å−1 and M2/kBT = 59.6. By comparison Gullingsrud and Schulten (49) obtain values of M1/kBT = −.59 ± 0.05 Å−1 and M2/kBT = −3.7 ± 0.2 for POPC at the fixed area per molecule of 64.0 Å2. Direct comparison is difficult due to Gullingsrud and Schulten’s (1) use of a constant area constraint, (2) use of a different force field (CHARMM 22), and (3) the large fluctuations in the local pressure tensor, (4) the physical difference between CLA and POPC chains. Thus the primary conclusion we draw is that the pressure profiles and the moments are nearly identical for both CLA isomers in this study.
Discussion
While the biological effects of the C9T11 and T10C12 are markedly different, current experimental data do not reveal large scale structural differences. In addition to the monolayer isotherm data discussed earlier (37), thermal properties of the two isomers are also similar. (50) the melting temperature of C9T11 is 14.9 °C and the enthalpy of fusion is 38.7 kJ/mol. For T10C12, the melting temperature of C9T11 is 19.8 °C and the enthalpy of fusion is 35.6kJ/mol. (50) In this paper we have used MD simulations to examine atomic level differences in structure and interactions in CLA bilayers composed of the two isomers C9T11 and T10C12. In our CLA simulations, we have found that the different degrees of chain segmental ordering in the two isomers does not appear to significantly affect certain large scale bilayer structural properties. Our simulated molecular areas, chain tilt, and mass distributions are very similar for C9T11 and T10C12 CLA. Differences were found in lipid chain order parameter profiles, lipid diffusion constants, and bilayer lateral pressure profiles. We conclude from the MD simulations that the distinctly different biological effects attributed to the two isomers cannot be directly related to large scale structural differences in pure bilayers of the isomers. However, our results do reveal that the two structurally similar bilayers have different distributions of internal chain order (order parameters) induced by the cis double bond. This raises the possibility that the CLA isomers may affect biological properties through differential interactions with membrane proteins or other molecules, such as cholesterol, that depend on the location of the cis double bond-induced disorder. The biological environment is of course highly heterogeneous compared to our simulations of structural and dynamical properties of pure, homogeneous lipid bilayers composed of C9T11 and T10C12 CLA isomers, respectively. In a biological membrane, any incorporated CLA will interact primarily with other membrane lipids, membrane proteins, and cholesterol, and these interactions have the possibility of depending strongly on the location of the cis double bond in CLA. For example, Subbaiah et al (37) have found that, compared to the C9T11 CLA-PC, T10C12 CLA-PC interacts more strongly with cholesterol at low cholesterol concentration, decreases membrane permeability, and is more oxidizable by free radicals. Additionally, there are numerous studies of the effect of membrane fluidity on biological function. For example, the role of membrane fluidity on gene expression has been recently reviewed by Los and Zinchenko. (51) To begin to examine these interactions, we are currently running simulations that consider cholesterol effects in CLA-PC bilayers of C9T11 and T10C12, respectively.
MD simulation results are of course products of the force field parameters that they use, and past experience has shown that small changes in some force field parameters can have large affects on bilayer properties. Thus it is imperative that force fields be as accurate as possible. For this simulation we have, as discussed earlier, made use of the state-of-the-art GROMACS 43a1-S3 force field that has been shown to directly reproduce sensitive properties of lipid bilayers including X-ray form factors. For this reason we believe that the conclusions regarding the similarities between the C9T11 and T10C12 bilayers are not artifacts of the MD force fields.
Acknowledgments
HLS, GJ, SC, and EJ were supported by NIH Grant Number PHS 2 PN2 EY016570B from the National Institutes of Health through the NIH Roadmap for Medical Research. PVS was supported by NIH Grant Numbers DK78165 and HL 68585
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pariza M. Perspective on the safety and effectiveness of conjugated linoleic acid. American Journal of Clinical Nutrition. 2004;79(6):1132S–1136S. doi: 10.1093/ajcn/79.6.1132S. [DOI] [PubMed] [Google Scholar]
- 2.Wahle K, Heys S, Rotondo D. Conjugated linoleic acids: are they beneficial or detrimental to health? Progress in Lipid Research. 2004;43(6):553–587. doi: 10.1016/j.plipres.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Beruly M. Dietary conjugated linoleic acid in health: physiological effects and mechanisms of action. Annu Rev Nutr. 2002;22:505–531. doi: 10.1146/annurev.nutr.22.021302.121842. [DOI] [PubMed] [Google Scholar]
- 4.Churruca I, Fernandez-Quintela A, Portillo M. Conjugated linoleic acid isomers: Differences in metabolism and biological effects. BioFactors. 2009;35:105–111. doi: 10.1002/biof.13. [DOI] [PubMed] [Google Scholar]
- 5.Taylor C, Zahradka P. Dietary conjugated linoleic acid and insulin sensitivity and resistance in rodent models. American Journal of Clinical Nutrition. 2004;79(6):1164S–1168S. doi: 10.1093/ajcn/79.6.1164S. [DOI] [PubMed] [Google Scholar]
- 6.House R, Cassady J, Eisen E, McIntosh M, Odle J. Conjugated linoleic acid evokes de-lipidation through the regulation of genes controlling lipid metabolism in adipose and liver tissue. Obesity reviews. 2005;6(3):247–258. doi: 10.1111/j.1467-789X.2005.00198.x. [DOI] [PubMed] [Google Scholar]
- 7.Brown J, Boysen M, Jensen S, Morrison R, Storkson J, Lea-Currie R, Pariza M, Mandrup S, McIntosh M. Isomer-specific regulation of metabolism and PPARγ signaling by CLA in human preadipocytes. Journal of lipid research. 2003;44(7):1287–1300. doi: 10.1194/jlr.M300001-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown J, Boysen M, Chung S, Fabiyi O, Morrison R, Mandrup S, McIntosh M. Conjugated linoleic acid induces human adipocyte delipidation: autocrine/paracrine regulation of MEK/ERK signaling by adipocytokines. Journal of Biological Chemistry. 2004;279(25):26735–26747. doi: 10.1074/jbc.M401766200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ha Y, Storkson J, Pariza M. Inhibition of benzo (a) pyrene-induced mouse forestomach neoplasia by conjugated dienoic derivatives of linoleic acid. Cancer Research. 1990;50(4):1097–1101. [PubMed] [Google Scholar]
- 10.Al-Madaney M, Kramer J, Deng Z, Vanderhoek J. Effects of lipidesterified conjugated linoleic acid isomers on platelet function: evidence for stimulation of platelet phospholipase activity. BBA-Molecular and Cell Biology of Lipids. 2003;1635(2–3):75–82. doi: 10.1016/j.bbalip.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Amarù D, Field C. Conjugated Linoleic Acid Decreases MCF-7 Human Breast Cancer Cell Growth and Insulin-Like Growth Factor-1 Receptor Levels. Lipids. 2009;44(5):449–458. doi: 10.1007/s11745-009-3288-4. [DOI] [PubMed] [Google Scholar]
- 12.Cao Y, Chen J, Yang L, Chen Z. Differential incorporation of dietary conjugated linolenic and linoleic acids into milk lipids and liver phospholipids in lactating and suckling rats. The Journal of Nutritional Biochemistry. 2009;20:685–693. doi: 10.1016/j.jnutbio.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Sébédio J, Angioni E, Chardigny J, Gregoire S, Juaneda P, Berdeaux O. The effect of conjugated linoleic acid isomers on fatty acid profiles of liver and adipose tissues and their conversion to isomers of 16: 2 and 18: 3 conjugated fatty acids in rats. Lipids. 2001;36(6):575–582. doi: 10.1007/s11745-001-0759-8. [DOI] [PubMed] [Google Scholar]
- 14.Stulnig T, Huber J, Leitinger N, Imre E, Angelisova P, Nowotny P, Waldhausl W. Polyunsaturated eicosapentaenoic acid displaces proteins from membrane rafts by altering raft lipid composition. Journal of Biological Chemistry. 2001;276(40):37335–37340. doi: 10.1074/jbc.M106193200. [DOI] [PubMed] [Google Scholar]
- 15.Liu S, Baracos V, Quinney H, Clandinin M. Dietary omega-3 and polyunsaturated fatty acids modify fatty acyl composition and insulin binding in skeletal-muscle sarcolemma. Biochemical Journal. 1994;299(Pt 3):831–837. doi: 10.1042/bj2990831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q, Wang M, Tan L, Wang C, Ma J, Li N, Li Y, Xu G, Li J. Docosahexaenoic acid changes lipid composition and interleukin-2 receptor signaling in membrane rafts. The Journal of Lipid Research. 2005;46(9):1904–1913. doi: 10.1194/jlr.M500033-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Ma D, Seo J, Davidson L, Callaway E, Fan Y, Lupton J, Chapkin R. n-3 PUFA alter caveolae lipid composition and resident protein localization in mouse colon. The FASEB Journal. 2004;18(9):1040–1042. doi: 10.1096/fj.03-1430fje. [DOI] [PubMed] [Google Scholar]
- 18.Chiu SW, Jakobsson E, Scott HL. Combined monte carlo and molecular dynamics simulation of hydrated lipid-cholesterol lipid bilayers at low cholesterol concentration. Biophys J. 2001;80(3):1104–1114. doi: 10.1016/S0006-3495(01)76088-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tu K, Klein ML, Tobias DJ. Constant–pressure molecular dynamics investigation of cholesterol effects in a dipalmitoylphosphatidylcholine bilayer. Biophys J. 1998;75:2147–2156. doi: 10.1016/S0006-3495(98)77657-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smondyrev AM, Berkowitz ML. Structure of dipalmitoylphos-phatidylcholine/cholesterol bilayer at low and high cholesterol concentrations: Molecular dynamics simulation. Biophys J. 1999;77:2075–2089. doi: 10.1016/S0006-3495(99)77049-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasenkiewicz-Gierula M, Róg T, Kitamura K, Kusumi A. Cholesterol effects on the phosphatidylcholine bilayer polar region: A molecular simulation study. Biophys J. 2000;78(3):1376–1389. doi: 10.1016/S0006-3495(00)76691-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott HL. Modeling the lipid component of membranes. Curr Op Struct Biol. 2002;12(4):495–502. doi: 10.1016/s0959-440x(02)00353-6. [DOI] [PubMed] [Google Scholar]
- 23.Pandit SA, Bostick DL, Berkowitz ML. Complexation of phosphatidylcholine lipids with cholesterol. Biophys J. 2004;86(3):1345– 1356. doi: 10.1016/S0006-3495(04)74206-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandit S, Chiu S-W, Jakobsson E, Grama A, Scott HL. Cholesterol surrogates: a comparison of cholesterol and 16:0 ceramide in popc bilayers. Biophys J. 2007;92:920–927. doi: 10.1529/biophysj.106.095034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandit S, Chiu S-W, Jakobsson E, Grama A, Scott HL. Cholesterol packing around lipids with saturated and unsaturated chains: A simulation study. Langmuir. 2008;24:6858–6865. doi: 10.1021/la8004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bachar M, Brunelle P, Tieleman D, Rauk A. Molecular dynamics simulation of a polyunsaturated lipid bilayer susceptible to lipid peroxidation. J Phys Chem B. 2004;108(22):7170–7179. [Google Scholar]
- 27.Chiu S, Pandit S, Scott H, Jakobsson E. An Improved United Atom Force Field for Simulation of Mixed Lipid Bilayers. J Phys Chem B. 2009;113(9):2748–2763. doi: 10.1021/jp807056c. [DOI] [PubMed] [Google Scholar]
- 28.Chiu S-W, Clark MM, Jakobsson E, Subramaniam S, Scott HL. Optimization of hydrocarbon chain interaction parameters: Application to the simulation of fluid phase lipid bilayers. J Phys Chem B. 1999;103:6323–6327. [Google Scholar]
- 29.Berger O, Edholm O, Jähnig F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pabst G, Kucerka N, Neih N-P, RMC, Katsaras J. Applications of neutron and X-ray scattering to the study of biologically relevant model membranes. Chem Phys Lipids. 2010;163:460–479. doi: 10.1016/j.chemphyslip.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 31.Kučerka N, Tristram-Nagle S, Nagle J. Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. Journal of Membrane Biology. 2006;208(3):193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- 32.Maartinez-Seara H, Rot T, Karttunen M, Reigada R, Vattulainen I. Influence of cis double-bond parametrization on lipid membrane properties: How seemingly insignificant details in force-field change even qualitative trends. J Chem Phys. 2008;129:105103. doi: 10.1063/1.2976443. [DOI] [PubMed] [Google Scholar]
- 33.Hess B, Kutzner C, van der Spoel D, Lindahl E. Gromacs 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4(3):435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 34.Hess B, Bekker H, Berendsen HJC, Fraaige JGEM. LINCS: A linear constraint solver for molecular simulations. J Comp Chem. 1997;18(12):1463–1472. [Google Scholar]
- 35.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103(19):8577–8593. [Google Scholar]
- 36.Tamm L, Seelig J. Lipid Solvation of Cytochrome c Oxidase. deuterium, Nitrogen-14, and Phosphorous-31 Nuclear Magnetic Resonance Studies of the Phosphocholine Head Group and on Cis-Unsaturated Fatty Acyl Chains. Biochemistry. 1983;22:1474–14838. doi: 10.1021/bi00275a023. [DOI] [PubMed] [Google Scholar]
- 37.Subbaiah P, Sircar D, Aizezi B, Mintzer E. Differential effects of conjugated linoleic acid isomers on the biophysical and biochemical properties of model membranes. Biochimica et Biophysica Acta. 2010;1798(12):506–514. doi: 10.1016/j.bbamem.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kučerka N, Liu Y, Chu N, Petrache H, Tristram-Nagle S, Nagle J. Structure of fully hydrated fluid phase DMPC and DLPC lipid bilayers using X-ray scattering from oriented multilamellar arrays and from unilamellar vesicles. Biophysical journal. 2005;88(4):2626–2637. doi: 10.1529/biophysj.104.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wassall S, Stillwell W. Polyunsaturated fatty acid–cholesterol interactions: Domain formation in membranes. BBA-Biomembranes. 2009;1788(1):24–32. doi: 10.1016/j.bbamem.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 40.Filippov A, Oradd G, Lindblom G. The effect of cholesterol on the lateral diffusion of phospholipids in oriented bilayers. Biophysical journal. 2003;84(5):3079–3086. doi: 10.1016/S0006-3495(03)70033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pittman M, Suits F, MacKerrell JA, Feller S. Molecular Level Organization of Saturated and Polyunsaturated Fatty Acids in a Phosphatidylcholine Bilayer Containing Cholesterol. Biochemistry. 2004;43:15318–15329. doi: 10.1021/bi048231w. [DOI] [PubMed] [Google Scholar]
- 42.Douliez J-P, Léonard A, Dufourc EJ. Restatement of order parameters in biomembranes: Calculation of C-C bond order parameters for C-D quadrupolar splittings. Biophys J. 1995;68:1727–1739. doi: 10.1016/S0006-3495(95)80350-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salem N, Niebylski C. The nervous system has an absolute molecular species requirement for proper function. Molecular Membrane Biology. 1995;12(1):131–134. doi: 10.3109/09687689509038508. [DOI] [PubMed] [Google Scholar]
- 44.Ernst E. Effects of n-3 fatty acids on blood rheology. Journal of internal medicine. 1989;(Supplement 731):129–32. doi: 10.1111/j.1365-2796.1989.tb01446.x. [DOI] [PubMed] [Google Scholar]
- 45.Brown ER, Subbaiah PV. Differential effects of eicosapentaenoic acid and docosahexaenoic acid on human skin fibroblasts. Lipids. 1994;29(12):825–829. doi: 10.1007/BF02536249. [DOI] [PubMed] [Google Scholar]
- 46.Cantor R. Lateral pressures in cell membranes: a mechanism for modulation of protein function. J Phys Chem B. 1997;101(10):1723–1725. [Google Scholar]
- 47.Ollila OHS, Risselada HJ, Louhivuori M, Lindahl E, Vattulainen I, Marrink SJ. 3d pressure field in lipid membranes and membrane-protein complexes. Phys Rev Lett. 2009;102(7):078101. doi: 10.1103/PhysRevLett.102.078101. [DOI] [PubMed] [Google Scholar]
- 48.Ollila S, Hyvonen M, Vattulainen I. Polyunsaturation in lipid membranes: dynamic properties and lateral pressure profiles. J Phys Chem B. 2007;111(12):3139–3150. doi: 10.1021/jp065424f. [DOI] [PubMed] [Google Scholar]
- 49.Gullingsrud J, Schulten K. Lipid Bilayer Pressure profiles and Mechanosensitive Channel Gating. Biophys J. 2004;86:3496–3509. doi: 10.1529/biophysj.103.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uehara H, Suganuma T, Negishi S, Uda Y, Furukawa Y, Ueno S, Sato K. Physical Properties of Two Isomers of Conjugated Linoleic Acid. J Am Oil Chem Soc. 2008;85:29–36. [Google Scholar]
- 51.Los D, Zinchenko V. Regulatory Role of Membrane Fluidity in Gene Expression. Lipids in Photosynthesis. :329–348. doi: 10.1007/s11120-013-9823-4. [DOI] [PubMed] [Google Scholar]