Abstract
Two recent papers, one from the Staley laboratory (Koodathingal and colleagues) and the other from the Cheng laboratory (Tseng and colleagues), show that the RNA-dependent ATPase Prp16, which is required for the second step of splicing, acts to reject slowly splicing pre-mRNAs immediately before the first catalytic reaction in pre-mRNA splicing. The results answer long-investigated questions about the actions of Prp16 and provide a wealth of molecular details on the proofreading process in pre-mRNA splicing. The discussion here reviews and integrates the results of the two papers and describes the implications for proofreading in splicing.
Keywords: Prp16, splicing fidelity, RNA helicase
The studies discussed here address two of the central questions in understanding the mechanism of pre-mRNA splicing: What is the role of ATP, and how is splicing fidelity achieved? Different approaches are used by the two groups, one involving time-resolved analysis of the interactions of a set of proteins with the spliceosome and the other following a slowly spliced substrate through its rejection pathway. The conclusions that the two groups reach are similar in form and combining their findings yields a remarkably detailed picture of the events surrounding the first catalytic reaction in splicing (Fig. 1).
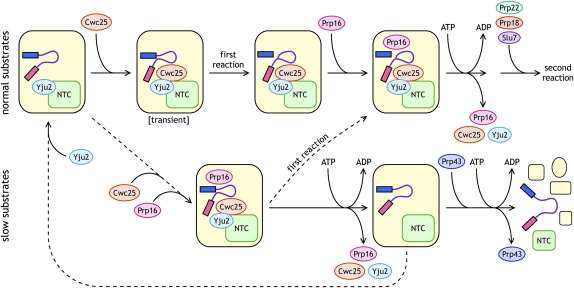
FIGURE 1.
Schematic of proteins joining and leaving the spliceosome surrounding the actions of Prp16. Two pathways, one for normal (wild-type) substrates (upper) and one for slow (mutant) substrates (lower), are shown. Steps that occur relatively quickly are shown with unbroken arrows, whereas those that occur relatively slowly are shown with dashed arrows. The spliceosome is shown in yellow; exon 1 as a blue rectangle; the intron as a purple line; exon 2 as a magenta rectangle; the NTC (Nineteen Complex) in green; Yju2 in light blue; Cwc25 in orange; Prp16 in pink; Prp43 in blue; Prp22 in green; Prp18 in red; and Slu7 in purple. ATP hydrolysis-requiring steps are indicated. Yju2 can, but does not necessarily, join the spliceosome early, but it must join before Cwc25. Cwc25 may join as shown here, but the spliceosomal species labeled “transient” has not been detected; Cwc25 may join after the first step, but it must join before Prp16. For slow substrates, the overall pathway is likely slower than for normal substrates (as indicated by the first dashed arrow), and the “first reaction” for slow substrates must be slower than the Prp16-rejection pathway.
The main character in the two papers, Prp16, was discovered more than two decades ago in a screen of yeast for suppressors of a branchpoint A to C mutant (brC) (Couto et al. 1987). brC mutation appeared to block splicing during the first step. Prp16, however, was found to be an RNA-dependent ATPase that is required only for the second step of splicing and does not detectably interact with the spliceosome before the first reaction (Schwer and Guthrie 1991). ATP hydrolysis by Prp16 fuels a conformational change in the spliceosome that paves the way for the second reaction (Schwer and Guthrie 1992). The finding that Prp16 suppressors of brC have reduced ATPase activity led to the hypothesis that Prp16 preferentially rejects brC splicing intermediates during the second step (Burgess and Guthrie 1993). More recently, genetic studies again suggested a first-step role for Prp16 in the splicing of mutant substrates (Query and Konarska 2004; Villa and Guthrie 2005).
The papers by Koodathingal et al. (2010) (from the Staley laboratory) and Tseng et al. (2011) (from the Cheng laboratory) examine the effect of Prp16 on splicing when splicing is blocked at the time of the first chemical reaction in vitro. Koodathingal et al. block the first step with a mutant U6 snRNA. The Lin laboratory showed that U6 snRNA bearing an Sp phosphorthioate bridging nucleotides 79 and 80 (U6-sU80) specifically blocks first-step catalysis when Mg+2 is the only metal available and that thiophilic metals relieve the inhibition (Yean et al. 2000). Koodathingal et al. affinity-purify U6-sU80 spliceosomes arrested prior to the first reaction. They show that blocking the first step with Mg+2 depends on ATP and the first reaction in U6-sU80 spliceosomes can proceed, albeit slowly, if ATP is absent. Inhibition is an active process because the depletion of ATP from reactions in which the first step was blocked allows the first step to proceed. Further, the first step will occur in the presence of ATP if an ATPase-deficient Prp16 is used, implying that the ATPase activity of Prp16 is required to block the first-step activity of U6-sU80 spliceosomes. Finally, Prp16, which is not needed for the first reaction of normal (fast) splicing (Schwer and Guthrie 1991), does bind to U6-s80 spliceosomes prior to the first step. The results support a model in which Prp16 can bind to spliceosomes before the first reaction and will hydrolyze ATP to block splicing if the first reaction is slow, thereby “proofreading” the rate of the first reaction. The same Prp16-catalyzed ATP hydrolysis will move splicing forward if the first reaction has already occurred.
Tseng et al. (2011) examine the role of Prp16 using an array of tools developed in the Cheng laboratory. Functional studies showed that the proteins Yju2 and Cwc25 are required for the first step of splicing (Liu et al. 2007; Chiu et al. 2009), consistent with proteomic analyses that found that Yju2 and Cwc25 join the spliceosome late in the first step and leave during the second step (Fabrizio et al. 2009; Lardelli et al. 2010). Yju2 joins the spliceosome early with the Nineteen Complex (Fig. 1) and is required for the binding of Cwc25, which joins after ATP hydrolysis by Prp2, just prior to the first reaction (Chiu et al. 2009; Warkocki et al. 2009). With normal substrates, the first reaction is rapid and Cwc25 is not readily detected bound to spliceosomes with pre-mRNA. In the new work, Tseng et al. show first that ATP hydrolysis by Prp16 removes both Yju2 and Cwc25 from normal spliceosomes following the first step (Fig. 1). This conformational change allows binding of Slu7, Prp18, and Prp22, which are the only new proteins needed for the second step (Schwer and Guthrie 1992; James et al. 2002; Warkocki et al. 2009).
Tseng et al. (2011) investigate substrate selectivity using the branchpoint mutations that were originally used to identify Prp16. Branchpoint mutant pre-mRNAs assemble slowly into spliceosomes, suggesting an early fidelity check, and Yju2 associates normally with these spliceosomes. Following assembly, splicing of the brC pre-mRNA is inhibited, primarily during the first step. Depletion of Prp16 reduces the amount of brC splicing from some to none, showing that Prp16 plays a role in the first step of brC splicing. ATP hydrolysis by Prp16 is not required for promoting the first reaction. Under normal splicing conditions (in the presence of ATP and Prp16), neither Cwc25 nor Prp16 is stably associated with brC spliceosomes; however, in the absence of ATP, both Cwc25 and Prp16 bind to pre-mRNA-containing brC spliceosomes. Prp16 and Cwc25 require each other for stable association with brC spliceosomes. The “slow substrates” branch of Figure 1 depicts the binding of Cwc25 and Prp16 to a spliceosome, forming a stable spliceosome. With both proteins bound, the slow splicing reaction can then occur, resulting in rejoining the “normal” pathway, or ATP hydrolysis by Prp16 will release Cwc25, Prp16, and Yju2 from the spliceosome. The results show clearly how the original slow-ATPase Prp16 proteins (Burgess and Guthrie 1993) enhance brC splicing by increasing the lifetime of the Cwc25-Prp16 spliceosome and thereby promoting the first reaction.
The splicing of the brG pre-mRNA is useful for understanding how Cwc25 might work. The first step of splicing with brG is faster than that of brC. Nevertheless, Cwc25 is not stably associated with brG spliceosomes, even in the absence of Prp16 (a condition that stabilizes Cwc25 binding to spliceosomes with wild-type splicing intermediates). This result may mean that Cwc25 binding is sensitive to the identity of the branchpoint base, but other interpretations are also possible.
The fate of spliceosomes rejected by Prp16 is addressed by Koodathingal et al. whose results show that rejected spliceosomes can still carry out splicing if ATP is depleted (a requirement for the first reaction in their system). The results of Tseng et al. imply that Yju2, Cwc25, and Prp16 must be added back to those spliceosomes. Combining the results generates the cycle shown in Figure 1. This cycle is driven forward by ATP hydrolysis by Prp16 and is not thermodynamically reversible. To escape this cycle, the spliceosomes bearing the rejected pre-mRNAs must be disassembled. Koodathingal et al. show that the RNA helicase Prp43 is required for the disassembly following rejection by Prp16. Prp43 disassembles the spliceosome both at the end of splicing and along the splicing pathway to discard slow spliceosomes (Arenas and Abelson 1997; Pandit et al. 2006; Mayas et al. 2010). Rejection by Prp16 is required for Prp43 disassembly, but the mechanistic basis for this dependence is not known.
The two papers use different methods to conclude that Prp16 is involved in the first catalytic reaction of splicing for slowly splicing substrates and that the ATPase of Prp16 is a proofreading activity for poor substrates. Incorrect substrates can be identified for proofreading either because they splice slowly and are rejected by Prp16 based on the rate of the first reaction or because Prp16 recognizes a specific structure in the slow spliceosomes that it disapproves. This distinction and the resulting mechanisms are discussed extensively by Koodathingal et al. (2010). At first glance, the U6-sU80 and brC mutations used in the two studies appear to be distinct, supporting a “timing” model. However, in the model proposed by Yean et al. (2000) U6-U80 and the branchpoint base are spatially close, bridged by a metal ion, and the “structure-recognition” model for proofreading cannot be excluded on this basis. Koodathingal et al. show that speeding up the first reaction of U6-sU80 spliceosomes by using the thiophilic metal Mn+2 instead of Mg+2 makes the first reaction independent of ATP (because the first reaction now occurs faster than rejection by Prp16), supporting the idea that Prp16 is a timer and that fidelity in the first reaction results from differences in rates. Tseng et al. also interpret their results in terms of a timing model. This model is similar to that proposed for Prp22 proofreading during the second step (Mayas et al. 2006).
The results of Tseng et al. also provide support for an alternative model that I describe here in which splicing fidelity during the first reaction is ensured by Cwc25 and Yju2, not by Prp16. In the absence of Prp16, wild-type pre-mRNAs are spliced through the first step normally, as expected from the entry of Prp16 into the pathway after the first reaction (Fig. 1; (Schwer and Guthrie 1991; Tseng et al. 2011). The first step of splicing of brC and brG pre-mRNAs is very slow (undetectable for brC) in the absence of Prp16. Adding Prp16 does not affect wild-type splicing; however, addition of Prp16 increases the amounts of first-step products for both brC and brG (cf. Fig. 5A in Tseng et al. 2011). That is, Prp16 reduces the fidelity of the first reaction of splicing, even in the presence of ATP. The spliceosome can choose the correct splice sites and execute the first step without Prp16, as long as Yju2 and Cwc25 are present. The role of Prp16 is not to enhance fidelity but to move splicing forward by removing Cwc25 and Yju2 (Fig. 1). (The spliceosome's specificity presumably requires the binding energy of Cwc25, which has to be pushed off by Prp16.) Perhaps as a consequence of its required activity, Prp16 stabilizes the interaction of Cwc25 with the spliceosome permitting brC and brG splicing to go forward. The combination of Yju2, Cwc25, and Prp16 enhance fidelity, but the real enforcer of specificity in this view is Cwc25, which does not bind to spliceosomes with mutant pre-mRNAs (described previously); Prp16 is an expediter. This model and those presented in the previous paragraph, which focus on Prp16 as the guarantor of fidelity, are not mutually exclusive, and a combination of them may be used to ensure fidelity during the first reaction.
The results of Tseng et al. show that the DExH proteins Prp2, Prp16, and Prp22 act in analogous ways during splicing. In each case, the action of the DExH protein removes a small set of proteins from the spliceosome and sets the stage for binding of the subsequent set of proteins together with its DExH protein (see Fig. 1 in Tseng et al. 2011). For Prp2, the binding of Spp2 allows Prp2 to join the spliceosome, and ATP hydrolysis by Prp2 releases Spp2, SF3a/b, and Prp2 (Silverman et al. 2004; Warkocki et al. 2009; Lardelli et al. 2010). For Prp22, the binding of Slu7 and Prp18 is required before Prp22 binding, whose action releases them all following the second step (James et al. 2002). Prp16 is now seen to act in a very similar way. The binding of Yju2 and Cwc25 is needed for Prp16 binding, and ATP hydrolysis by Prp16 releases them all from the spliceosome (Fig. 1). For these three DExH proteins, the common action established is the release of proteins from the spliceosome. Whether this results from direct action on the proteins or indirectly from an RNA rearrangement is not known. Only for Prp22 is a duplex RNA substrate—between loop 1 of U5 and the mRNA—known (Schwer 2008). For Prp16, mutations in motif III, which are expected to uncouple RNA unwinding from ATPase activity, do not affect its splicing activity (Schneider et al. 2002), perhaps hinting that the primary result of Prp16 action is the removal of the Cwc25 and Yju2 proteins that Tseng et al. have demonstrated. Continuing along this line, if the removal of proteins from the spliceosome is the primary action of Prp16, then reduced-function alleles of Prp16 should be rescued by mutations that destabilize these proteins from the spliceosome, as proposed originally by Madhani and Guthrie (1994), and mutations that rescue cold-sensitive alleles of PRP16 may not identify sites of direct Prp16 action.
The papers by Koodathingal et al. (2010) and Tseng et al. (2011) elucidate the dynamics of the splicing pathway at the time of the first catalytic reaction. The papers demonstrate that Prp16 acts during the first step for slowly spliced substrates to divert them from the splicing pathway into the disassembly pathway. At the same time, Prp16 does not act until the second step for wild-type substrates, propelling the spliceosome forward to the second step. These divergent actions, carried out with the Yju2 and Cwc25 proteins, ensure the fidelity of splicing during the first reaction.
ACKNOWLEDGMENTS
I thank Soo-Chen Cheng, Jonathan Staley, Prakash Koodathingal, Daniel Semlow, and Luciana Crotti for their comments on the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2577511.
REFERENCES
- Arenas JE, Abelson JN 1997. Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci 94: 11798–11802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Guthrie C 1993. A mechanism to enhance mRNA splicing fidelity: The RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell 73: 1377–1391 [DOI] [PubMed] [Google Scholar]
- Chiu YF, Liu YC, Chiang TW, Yeh TC, Tseng CK, Wu NY, Cheng SC 2009. Cwc25 is a novel splicing factor required after Prp2 and Yju2 to facilitate the first catalytic reaction. Mol Cell Biol 29: 5671–5678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto JR, Tamm J, Parker R, Guthrie C 1987. A trans-acting suppressor restores splicing of a yeast intron with a branch point mutation. Genes Dev 1: 445–455 [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Luhrmann R 2009. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell 36: 593–608 [DOI] [PubMed] [Google Scholar]
- James SA, Turner W, Schwer B 2002. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA 8: 1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koodathingal P, Novak T, Piccirilli JA, Staley JP 2010. The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5′ splice site cleavage during pre-mRNA splicing. Mol Cell 39: 385–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardelli RM, Thompson JX, Yates JR 3rd, Stevens SW 2010. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 16: 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Chen HC, Wu NY, Cheng SC 2007. A novel splicing factor, Yju2, is associated with NTC and acts after Prp2 in promoting the first catalytic reaction of pre-mRNA splicing. Mol Cell Biol 27: 5403–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C 1994. Genetic interactions between the yeast RNA helicase homolog Prp16 and spliceosomal snRNAs identify candidate ligands for the Prp16 RNA-dependent ATPase. Genetics 137: 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayas RM, Maita H, Staley JP 2006. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol 13: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayas RM, Maita H, Semlow DR, Staley JP 2010. Spliceosome discards intermediates via the DEAH box ATPase Prp43p. Proc Natl Acad Sci 107: 10020–10025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S, Lynn B, Rymond BC 2006. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc Natl Acad Sci 103: 13700–13705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query CC, Konarska MM 2004. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell 14: 343–354 [DOI] [PubMed] [Google Scholar]
- Schneider S, Hotz HR, Schwer B 2002. Characterization of dominant-negative mutants of the DEAH-box splicing factors Prp22 and Prp16. J Biol Chem 277: 15452–15458 [DOI] [PubMed] [Google Scholar]
- Schwer B 2008. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell 30: 743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Guthrie C 1991. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature 349: 494–499 [DOI] [PubMed] [Google Scholar]
- Schwer B, Guthrie C 1992. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J 11: 5033–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman EJ, Maeda A, Wei J, Smith P, Beggs JD, Lin RJ 2004. Interaction between a G-patch protein and a spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol Cell Biol 24: 10101–10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C-K, Liu H-L, Cheng S-C 2011. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA 17: 145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa T, Guthrie C 2005. The Isy1p component of the NineTeen complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes Dev 19: 1894–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warkocki Z, Odenwalder P, Schmitzova J, Platzmann F, Stark H, Urlaub H, Ficner R, Fabrizio P, Luhrmann R 2009. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol 16: 1237–1243 [DOI] [PubMed] [Google Scholar]
- Yean SL, Wuenschell G, Termini J, Lin RJ 2000. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature 408: 881–884 [DOI] [PMC free article] [PubMed] [Google Scholar]