Abstract
Delivering small interfering RNA (siRNA) to tumors is the major technical hurdle that prevents the advancement of siRNA-based cancer therapy. One of the difficulties associated with the development of clinically relevant delivery systems is the lack of reliable tools for monitoring siRNA delivery to tumors in vivo. We describe here a novel, positive-readout system where siRNA-mediated target knockdown elicits a rapid and robust increase of reporter activity. Using the positive-readout system, we created (1) β-galactosidase-based tumor models that allow the detection of target knockdown in 1%–2% of tumor cells and can distinguish between tumor areas where effective target knockdown occurs versus tumor areas that are not accessible to delivery, and (2) luciferase-based tumor models that allow the quantitative assessment of a large number of delivery systems. Using these positive-readout models, we screened a number of literature-described siRNA delivery systems and identified lipid nanoparticles as a promising delivery platform for siRNA-based cancer therapy.
Keywords: siRNA, RNAi, therapy, delivery, cancer
INTRODUCTION
RNA interference (RNAi) holds promise for use in many therapeutic applications. Using this technology, almost any proteins involved in mediating disease can be selectively targeted by simply designing a complementary small interfering RNA (siRNA) to the mRNA that encodes the protein of interest. It is widely recognized that the efficient delivery of siRNA to target cells remains a significant challenge and is the primary technical hurdle limiting the advancement of RNAi-based therapies. A number of novel siRNA delivery strategies have been explored, including the use of nucleic-acid-delivery vehicles that involve lipid or polymer-based nanoparticles or the direct chemical modification of siRNA duplexes (Schiffelers et al. 2004; Takei et al. 2004; Hu-Lieskovan et al. 2005; Landen et al. 2005; Song et al. 2005; Takeshita et al. 2005; Santel et al. 2006; Pirollo et al. 2007). Despite some encouraging proof-of-concept successes, efficient delivery of siRNA to tumors via clinically viable vehicles has still not been achieved. In some cases, systemically delivered siRNAs only have access to the tumor vasculature or to a small percentage of tumor cells (Santel et al. 2006). To fully capture the potential of siRNA-based cancer therapy, low-toxicity siRNA formulations that can reach the majority of tumor cells need to be developed.
One of the difficulties associated with the identification of suitable siRNA delivery systems for therapeutic applications is the lack of reliable assays that enable the comparison and optimization of various delivery approaches in vivo. Although it is theoretically feasible to monitor siRNA delivery to tumors using Western, PCR, or Northern analysis, these methods often fail to produce reliable results unless robust target knockdown occurs in the majority of the tumor cells. In this study, we report the development of a novel positive-readout assay that circumvents some of the limitations associated with the conventional knockdown assays. This positive-readout assay is much more robust and sensitive compared to the conventional assays and can be used to monitor functional siRNA delivery both in cultured cells in vitro and in xenograft tumors in vivo. Using these positive-readout models, we identified lipid nanoparticles as a promising delivery platform for siRNA-based cancer therapy.
MATERIALS AND METHODS
Molecular cloning
To create the TetR-ODC fusion protein, the coding sequence of TetR and ODC were amplified by PCR and linked together via restriction enzyme digestion and ligation to create the following TetR-ODC coding sequence:
Atgtctagattagataaaagtaaagtgattaacagcgcattagagctgcttaatgaggtcggaatcgaaggtttaacaacccgtaaactcgcccagaagctaggtgtagagcagcctacattgtattggcatgtaaaaaataagcgggctttgctcgacgccttagccattgagatgttagataggcaccatactcacttttgccctttagaaggggaaagctggcaagattttttacgtaataacgctaaaagttttagatgtgctttactaagtcatcgcgatggagcaaaagtacatttaggtacacggcctacagaaaaacagtatgaaactctcgaaaatcaattagcctttttatgccaacaaggtttttcactagagaatgcattatatgcactcagcgctgtggggcattttactttaggttgcgtattggaagatcaagagcatcaagtcgctaaagaagaaagggaaacacctactactgatagtatgccgccattattacgacaagctatcgaattatttgatcaccaaggtgcagagccagccttcttattcggccttgaattgatcatatgcggattagaaaaacaacttaaatgtgaaagtgggtccgcgtacagcggatcccgggaattcagatcttatagccatggcttcccgccggcggtggcggcgcaggatgatggcacgctgcccatgtcttgtgcccaggagagcgggatggaccgtcaccctgcagcctgtgcttctgctaggatcaatgtgtaa. The TetR-ODC coding sequence was further cloned into pcDNA6 (Invitrogen) and the resulting construct was verified by sequencing. Plasmids that express the TetR shRNA were created using PCR-based cloning as previously described (Scacheri et al. 2004; Li et al. 2005).
Cell culture and reagents
All MDA-MB435-derived cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% tet system approved fetal bovine serum (Clontech). The H1299 cells were maintained in RPMI-1640 media supplemented with 10% human serum (Invitrogen). To generate TetR-ODC-expressing clones, the parental MDA-MB435 cells were transfected with the TetR-ODC expressing plasmid using lipofectamine 2000 (Invitrogen). Twenty-four hours post-transfection, the transfected cells were passaged at dilutions from 1:10 to 1:100 into 10-cm plates and maintained in the selection medium containing 10 μg/mL of blasticidin (Invitrogen). To generate cell lines in which the expression of LacZ or Luciferase reporter is under the control of TetR-ODC, the tet-responsive LacZ or Luciferase reporter was transfected into the MDA-MB435-TetR-ODC c11 cells, and the cells were selected with 350 μg/mL zeocin (Invitrogen). All siRNAs used in this study were purchased from Dharmacon. Lipofectamine2000 (Invitrogen) was used to transfect siRNAs into cells according to the manufacturer's suggested protocol. For all siRNA experiments, the siRNAs were transfected at a final concentration of 20 nM, or with the indicated final concentrations in dose-response experiments. Cyclohexamide (CHX, Sigma) was used to treat cells at a final concentration of 10 μg/mL. Doxycycline (Dox, Sigma) was used at a final concentration of 1 μg/mL in cells or 1 mg/mL in drinking water in the animal studies.
Western blotting
Cells were directly lysed in six-well plates in 1X Laemmli sample buffer (Bio-Rad). Proteins were separated by SDS-PAGE, transferred to a PVDF membrane, and Western blotting was performed according to standard procedures using a monoclonal antibody against TetR (MoBiTec) or a monoclonal β-actin antibody (Sigma).
Determine β-galactosidase expression using the activity assay or staining
The β-galactosidase activity was measured using the β-galactosidase enzyme assay system (Promega). The in situ β-galactosidase staining kit (Stratagene) was used to measure the β-galactosidase expression in cells via staining. The manufacturer-suggested protocols were followed in both the activity and the staining assays.
Tumor models
The animal studies were carried out in accordance with internal Institutional Animal Care and Use Committee guidelines at Abbott Laboratories. All animals were obtained from Charles River Laboratory. Scid female mice at 6–8 wk of age were used for subcutaneous and intraliver tumor models. Female nude rats were used for brain tumors. All cell lines used for creating xenograft tumors were subjected to the IMPACT profile I test (18 agents) at the University of Missouri Research Animal Diagnostic and Investigative Laboratory, and all cell lines were found negative for the 18 infectious agents tested. For flank tumors, cancer cells were suspended in a 1:1 mixture of S-MEM (Invitrogen) and matrigel (BD Bioscience) and inoculated subcutaneously into the hindquarter of mice. For liver tumors, tumor cells were directly injected into the mouse liver. For brain tumors, tumor cells were inoculated intracranially into the right cerebral hemisphere 3 mm in depth, 2.5 mm lateral, and 2 mm anterior to the bregma using the stereotaxic instrument.
Animal dosing and sample harvesting
For in vivo knockdown evaluation in positive-readout models, treatments were started 3–4 wk after tumor inoculation for liver and brain tumors, or when tumors reached 200 mg for subcutaneous tumors. For doxycycline treatment in the TetR-ODC-Luc model, doxycycline was supplied in drinking water at a concentration of 1 mg/mL for 4 d before imaging. For the doxycycline treatment in the TetR-ODC-LacZ model, doxycycline was supplied for 7 d before collecting samples for IHC analysis of β-galactosidase expression. For siRNA treatment in the positive-readout models, formulated or unformulated siRNAs were administered via tail vein (i.v), or intraperitoneal (i.p) injection, or direct injection into tumors at the indicated doses and schedules. For hydrodynamic delivery of siRNA, 1.8 mL of siRNA in saline solution was injected into tail vein within 6 sec.
IHC analysis
IHC was carried out as previously described (Li et al. 2005). Briefly, tumors were excised, cut into pieces of <3 mm in thickness and immediately fixed in buffered formalin solution with neutral pH (Sigma). The formalin-fixed and paraffin-embedded tumor sections were then used for staining. The mouse anti-β-galactosidase mAb (Promega) was used to detect β-galactosidase in tumor sections. IHC analysis of tumor hypoxia was carried out using the Hypoxyprobe-1 kit (Natural Pharmacia International Inc.) according to the manufacturer's suggested protocol. Briefly, 0.2 mL pimonidazole HCl solution (6 mg/mL in saline) was administered via intravenous injection 1 h before tumor harvesting. The hypoxic areas within tumor sections were detected using mouse antibody recognizing the protein adduct formed by hypoxyprobe. For all IHC studies, DAB (3,3′-diaminobenzidine) was used as the chromogen, and IHC images were acquired using the Nikon TE2000 inverted microscope.
Bioluminescence imaging and analysis
In vivo bioluminescence imaging and analysis were conducted on the IVIS 200 system using the Living Image acquisition and analysis software (Caliper Life Science). After an intraperitoneal injection of luciferin (Promege) at 150 mg/kg, mice were anesthetized with isofluorane. Four minutes after the injection of luciferin, a series of time-lapse images were acquired at 2-min intervals in a total of 10 min. Regions of interest (ROI) were drawn around the tumors and signal intensity was quantified as the sum of photon counts per second within the ROI after the subtraction of background luminescence. The peak reading during the 10-min imaging period was used for calculating the signal ratio before and after siRNA delivery.
Liposome formulations for siRNA delivery
1,2-Dioleoyl-3-trimethylammonium-propane (DOTAP), 1,2-dioleoyl-3-(dimethylamino) propane (DODAP), 3β-[N-(N′,N′-dimethylaminoethane)-carbamoyl]cholesterol (DC-Chol) and cholesterol (Chol) were purchased from Avanti Polar Lipids. Dioleoyl phosphatidylethanolamine (DOPE) and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[carboxy(polyethylene glycol)2000] were purchased from Genzyme. The compositions for the DOTAP-, DODAP-, and DC-Chol-based liposomes are DOTAP/DOPE/Chol/PEG-DSPE (40/39/20/1, molar %), DODAP/DSPC/Chol/PEG-DSPE (40/10/48/2, molar %), and DC-Chol/DOPE/DPPC/PEG-DSPE (40/20/39/1, molar %) respectively. To prepare the liposomes, lipids were dissolved in tert-butanol at a final concentration of 10 mg/mL. siRNA was dissolved in dH2O and further mixed in an acidic citrate buffer. After brief heating in a 60°C water bath, the lipid solution was injected into the siRNA solution through a 28-gauge needle, while the siRNA solution was under magnetic stirring to form emulsion. Resulted emulsions were further buffered using a pH 7.4 phosphate buffered saline (PBS) at room temperature, and subjected to diafiltration (Pellion ultrafiltraiton unit, MW 100K, PES membrane, Millipore) against PBS before being concentrated to the desired volume. The final liposome formulations have a siRNA concentration of 250 μg/mL. The resulted liposomes exhibited the follow characteristics: DODAP-based liposome (size = 139 nm, PDI = 0.09, zeta potential = −0.735 mV), DOTAP-based liposome (size = 169 nm, PDI = 0.17, zeta potential = 4.32 mV), and DC-Chol-based liposome (size = 166 nm, PDI = 0.27, zeta potential = 6.01 mV). Samples were sterile filtrated through 0.22 μm syringe filters before further biological evaluations. The SNALP liposomes were prepared according to the literature-described procedure (Zimmermann et al. 2006). D-Lin-DMA and PEG-C-DMA were synthesized in-house and formulated with DSPC, Cholesterol, and siRNA using a 25:1 lipid/siRNA ratio and a 48/40/10/2 molar ratio of Cholesterol/D-Lin-DMA/DSPC/PEG-C-DMA. The resulting SNALP liposomes have similar characteristics to what was described in the literature (80–100 nm in size, PDI < 0.1, and a near neutral zeta potential).
Nonliposome formulations for siRNA delivery
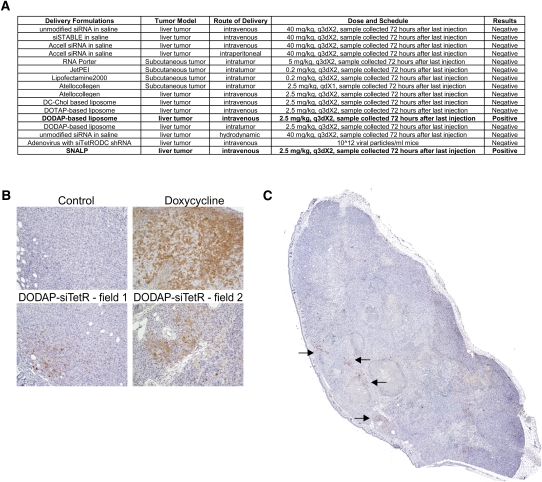
The commercially available siRNA delivery reagents Lipofectamine2000 (Invitrogen), RNA Porter (Bioo Scientific), in vivo Jet-PEI (Polyplus-transfection), and Atellocollegen (Cohesion) were obtained from each vendor. The siRNAs were formulated according to vendors' instructions or the literature-described procedures, diluted in saline and administered at 0.2 mL/mouse according to the dosage and schedule indicated in Fig. 4A, below.
FIGURE 4.
Evaluate siRNA delivery methods using the positive-readout model. (A) Various siRNA delivery methods were used to deliver the TetR siRNA to the positive-readout, TetR-ODC-LacZ-derived subcutaneous or liver tumors in vivo using the indicated dose and administration schedule. Each delivery method was tested in three to five tumor-bearing mice. (B) Mice bearing the TetR-ODC-LacZ-derived liver tumors were fed with water (control), or doxycycline (Doxycycline), or administered with DODAP liposomes containing the control siRNA (Control) or the TetR siRNA (DODAP-siTetR) (i.v., 2.5 mg siRNA/kg on day 1 and day 4), and samples were collected on day 7 for IHC analysis of β-galactosidase expression. n = 5 for each cohort. Pictures represent selected fields from each treatment group. The brown color represents the β-galactosidase staining. (C) The picture of an entire section of a tumor from DODAP-treated mice. Arrows point to areas with positive β-galactosidase staining.
Adenovirus-mediated delivery of shRNAs
Adenovirus particles expressing shRNA were produced by Invitrogen using their BLOCK-iT virus production service. Briefly, the best TetR shRNA sequence and a negative control shRNA (GGACATCACTTACGCTGAGTTCAAGAGACTCAGCGTAAGTGATGTCCTT) were cloned into pENTR/U6 vector and transferred to pAd/BLOCK-iT-DEST to create the pAd-GW/U6-shRNAs vectors. The pAd-GW/U6-shRNA vectors were then used to transfect 293A cells to produce adenovirus. The resulted viral stock was then amplified and concentrated to produce the working viral stock.
RESULTS
Establishing a TetR-based positive-readout system for monitoring siRNA delivery
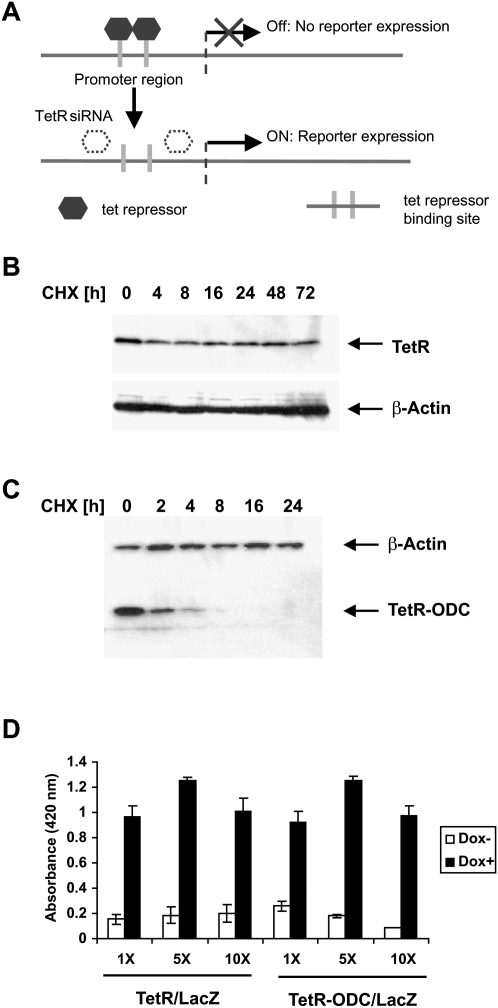
Our strategy for developing a positive-readout of functional siRNA delivery involves the use of the tet repressor-based inducible expression systems, in which binding of the TetR homodimers to the tet operators impedes the binding of TATA binding protein and other accessory proteins to the promoter, resulting in an inhibition of reporter gene transcription. A potent siRNA against TetR, when introduced into cells, serves as an inducing agent by knocking down the TetR protein, thus allowing up-regulation of the reporter (Fig. 1A). However, unlike the small molecule inducers, tetracycline and doxycycline, which trigger the immediate onset of gene expression; the onset of TetR siRNA-induced reporter transcription depends on the half-life of the TetR protein because degradation of pre-existing TetR is required to induce reporter transcription. Our initial studies of the wild-type TetR protein suggested that it is extremely stable, with a half-life that far exceeds 72 h (Fig. 1B). Indeed, a reporter system that is regulated by the wild-type TetR protein does not exhibit an adequate response to siRNA delivery (results not shown). To shorten the half-life of TetR, we engineered a fusion protein with the PEST domain from ornithine decarboxylase (ODC) attached to the C-terminus of TetR (TetR-ODC). The PEST domain of ODC has been shown previously to promote protein degradation (Zhang et al. 2008). In contrast to the wild-type TetR, the TetR-ODC protein is quickly degraded after protein synthesis is inhibited using cyclohexamide, indicating that the ODC PEST domain is able to mediate rapid protein degradation in the context of the TetR-ODC protein (Fig. 1C). Importantly, when cotransfected into cells with a tet-responsive LacZ reporter, TetR-ODC retained the full activity of the wild-type TetR in regulating a tet-responsive promoter (Fig. 1D).
FIGURE 1.
Establish a positive-readout system for monitoring siRNA delivery. (A) Schematics of the positive-readout system. (B) Cells that express the wild-type TetR protein were treated with cycloheximide (CHX). Cells were collected at indicated time points and the intracellular levels of TetR protein were determined by Western analysis. (C) Cells that express the TetR-ODC expression construct were treated with cycloheximide, collected at the indicated time points, and the protein level of tet-RODC was determined by Western analysis. (D) H1299 cells were transfected with a tetracycline responsive LacZ reporter (LacZ) and a plasmid expressing the wild-type TetR or the TetR-ODC protein at a 1:1, 1:5, or 1:10 molar ratio (LacZ reporter: TetR or TetR-ODC expressing plasmid). Twenty-four hours after transfection, the cells were switched to doxycycline containing medium (Dox +) or kept in regular medium without doxycyline (Dox −) for an additional 48 h, and the lacZ reporter activity was determined using a β-galactosidase assay kit.
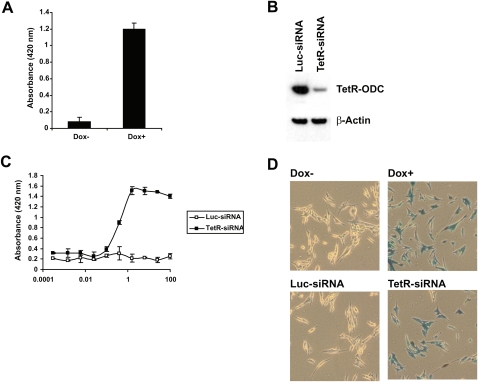
To create cancer cell lines that will respond to the delivery of a TetR siRNA with an increase of lacZ reporter activity, we first created a MDA-MB435SLM-derived cell line that carries a stably integrated TetR-ODC expression cassette and then introduced a tet-responsive LacZ reporter. Upon isolating and testing a large number of stable clones for their responses to doxycycline, one of the clones (TetR-ODC-LacZ) was found to have minimal β-galactosidase activity in the absence of doxycycline and exhibited a significant increase of β-galactosidase activity in the presence of doxycycline (Fig. 2A).
FIGURE 2.
Create positive-readout cell lines. (A) An MDA-MB-435-derived cell line with stably integrated TetR-ODC expression cassette and the tet-responsive lacZ reporter (TetR-ODC-LacZ) was cultured in the presence (Dox +) or absence (Dox −) of doxycycline for 72 h, and the lacZ reporter activity was determined. (B) The TetR-ODC-LacZ cells were transfected with a control siRNA (Luc-siRNA) or a TetR siRNA (TetR-siRNA). The TetR-ODC protein levels were determined 48 h after siRNA transfection by Western analysis using an antibody against TetR. (C) The TetR-ODC-LacZ cells were transfected with different amounts of siTetRODC (TetR-siRNA) or a luciferase siRNA (Luc-siRNA), and the β-galactosidase activity was determined using a β-gal assay kit. (D) The TetR-ODC-LacZ cells were cultured in the presence (Dox +) or absence (Dox −) of doxycycline for 72 h (top panel) or transfected with a control siRNA (Luc-siRNA) or a TetR siRNA (TetR-siRNA) and cultured in the absence of doxycycline for 72 h (bottom panel). The β-galactosidase activity in these cells was then determined by x-gal staining.
In addition to creating cell lines that exhibit a tightly regulated reporter activity, a potent siRNA against TetR-ODC is also required in order to establish a sensitive assay for monitoring siRNA delivery. We have shown previously that functional shRNAs and siRNAs share extensive similarities in nucleotide preference along the duplex (Li et al. 2007). Therefore, siRNA duplexes derived from functional shRNAs are likely to be functional siRNAs. After screening more than 20 shRNAs against TetR-ODC, we selected the most potent shRNA and used this sequence to design a conventional TetR siRNA (Sense stand: 5′GUUGCGUAUUGGAAGAUCA 3′). When transfected into the TetR-ODC-LacZ cells using Lipofectamine2000, this TetR siRNA triggered an efficient knockdown of the TetR-ODC protein and a robust induction of β-galactosidase activity in a dose-dependent manner (Fig. 2B,C, TetR-siRNA). The estimated EC50 of the TetR siRNA in the reporter assay is ∼1 nM. β-gal staining of the TetR-ODC-LacZ cells further demonstrated that the TetR siRNA induced a strong β-galactosidase expression in the majority of the cells (TetR-siRNA) (Fig. 2D). As controls, cells transfected with the luciferase siRNA exhibited very low levels of β-galactosidase expression, as judged by both the activity assay and β-gal staining (Luc-siRNA) (Fig. 2C,D). These results collectively demonstrate that a robust and tightly regulated reporter expression can be achieved in the TetR-ODC-LacZ cells upon the delivery of a siRNA targeting the TetR-ODC protein.
Creating positive-readout tumor models
The tumorogenic potential of the parental cell line was retained in the TetR-ODC-LacZ cells as judged by their ability to form subcutaneous, liver, or brain tumors when inoculated at these sites, respectively (data not shown). Upon feeding the tumor-bearing mice with doxycycline for 7 d, a robust increase of β-galactosidase activity was observed in sections of subcutaneous, liver, and brain tumors. In contrast, sections of the control tumors only had very weak staining (Fig. 3A). These results suggest that a robust induction of β-galactosidase expression can be achieved in tumors regardless of the inoculation sites. It is noteworthy that, except for a few areas where no β-galactosidase staining was observed, the majority of the tumor cells in the doxycycline-treated group exhibited strong β-galactosidase staining, suggesting that the positive-readout system, when induced by doxycycline, is functional in cancer cells in vivo. We suspect that the few negative-staining areas likely represent host-derived matrix/stroma components.
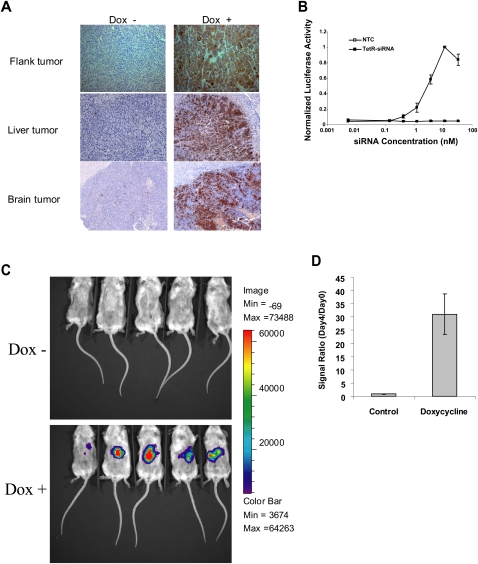
FIGURE 3.
Establish positive-readout tumor models. (A) The TetR-ODC-LacZ cells were used to form flank or liver tumors in scid females via subcutaneous inoculation or intraheptic injection, respectively (top, middle panels). In addition, the TetR-ODC-LacZ cells were also directly injected into the rat brain to form brain tumors (bottom panel). Tumors from animals fed with water (Dox −) or doxycycline (Dox +) for 7 d were collected and the β-galactosidase expression in tumors were determined by IHC analysis using a β-galactosidase specific antibody. (B) An MDA-MB-435-derived cell line with stably integrated TetR-ODC expression cassette and the tet-responsive luciferase reporter (TetR-ODC-Luc) was transfected with different amounts of a control siRNA (NTC) or a potent siRNA targeting TetR (TetR-siRNA) using Lipofectamine2000. Luciferase activity in cells was determined 72 h after siRNA transfection. (C) The TetR-ODC-Luc cells were used to form liver tumors in scid females via intraheptic inoculation. Mice were imaged before the start of doxycycline treatment (day 0) and 4 d after the start of doxycycline treatment (day 4). (D) The signal ratio of day 4/day 0 from doxycycline-treated and control mice. Each bar in the graph represents the average signal ratio ± SEM for each treatment group.
Because a positive-staining area can be easily distinguished from the surrounding negative-staining cells, IHC analysis of β-galactosidase expression in tumor sections is ideal for the detection of localized target knockdown in tumors. However, IHC analysis has a relatively low throughput, and it is often difficult to obtain a quantitative comparison of the target knockdown produced by different delivery systems. To overcome this limitation, we extended the positive-readout model to include luciferase as a reporter, thereby enabling bioluminescence imaging analysis. Bioluminescent signals are easy to quantify and can be acquired in live animals. Therefore, the imaging-based method is expected to have a better quantification and higher assay throughput compared with the IHC method. We introduced a tet-responsive luciferase reporter into a TetR-ODC-expressing cell line and isolated a stable clone (TetR-ODC-Luc) that exhibited more than a 20-fold increase of luciferase signal upon doxycycline treatment. Lipofectamine2000-mediated transfection of the TetR siRNA triggered a dose-dependent increase of the luciferase activity in this cell line (Fig. 3B), and the estimated EC50 of the TetR siRNA in the luciferase reporter assay is ∼5 nM. Similar to the TetR-ODC-LacZ cells, the TetR-ODC-Luc cells also formed subcutaneous or liver tumors when directly inoculated at these sites. Without doxycycline treatment, mice bearing the TetR-ODC-Luc-derived liver tumors exhibited a very low background of bioluminescence. An increase of bioluminescence was clearly observed in tumor-bearing mice that were fed with doxycycline for 4 d (Fig. 3C). Quantification of bioluminescence before and after administration of doxycycline revealed a ∼30-fold increase of luminescent signals after doxycycline treatment. By contrast, tumor-bearing mice without doxycycline treatment only exhibited a marginal increase of bioluminescence over the same timeframe, suggesting that the increase of bioluminescence is due to the induction of luciferase expression instead of tumor growth during this period (Fig. 3D). A similar increase of luminescence was also observed in TetR-ODC-Luc-derived subcutaneous tumors (data not shown). Taken together, these results demonstrate that doxycycline can elicit a robust induction of reporter gene expression in tumors derived from both the TetR-ODC-LacZ and TetR-ODC-Luc cells. Therefore, an increase of reporter signal in the positive-readout tumors is anticipated upon functional delivery of the TetR siRNA.
Identifying promising delivery platforms for siRNA-based cancer therapy
Because the LacZ-based positive-readout model is expected to have better sensitivity than the luciferase-based model due to its ability to detect localized target knockdown in a few tumor cells, we examined a number of delivery methods to assess their ability to mediate siRNA delivery into tumors using the LacZ model. Delivery vehicles that required intratumor injections were tested in the TetR-ODC-LacZ-derived subcutaneous tumors, whereas delivery approaches that require systemic administration were tested in the TetR-ODC-LacZ-derived liver tumors because most delivery vehicles are expected to accumulate in the liver upon systemic administration. Among the delivery methods tested, some have been described in the literature or claimed by reagent vendors to have utility in tumor delivery (Verma et al. 2003; Takei et al. 2004; Urban-Klein et al. 2004; Takeshita et al. 2005; Zhang et al. 2008). Cationic lipids such as 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP), 1,2-dioleoyl-3-(dimethylamino) propane (DODAP), and 3β-[N-(N′,N′-dimethylaminoethane)-carbamoyl]cholesterol (DC-Chol) are effective transfection reagents for the delivery of nucleic acids in vitro (Templeton et al. 1997; Xing et al. 1997; Koltover et al. 1998), although the use of DC-Chol, DOTAP, and DODAP-based liposomes for tumor delivery of siRNA has not been reported. In addition, several methods that are known to effectively deliver siRNA to the liver, such as hydrodynamic injection of siRNA and adenovirus-mediated delivery of shRNAs (Lewis et al. 2002; Yang et al. 2007), were also included in the study to determine their potential utility in delivering siRNA into tumors.
Surprisingly, few of the delivery vehicles examined produced any detectable positive signal over the background in our model (Fig. 4A). One of the delivery systems that did exhibit consistent tumor delivery was a DODAP-based lipid nanoparticle formulation. The DODAP-based lipid nanoparticle contains DODAP/DSPC/Chol/PEG-DSPE at a 40/10/48/2 molar ratio. Intravenous injection of the DODAP-based lipid nanoparticle containing the TetR siRNA (2.5 mg siRNA/kg) resulted in an evident up-regulation of β-galactosidase in several areas within tumors in two independent experiments. As a control, the DODAP-based lipid nanoparticle that contains a nontargeted control siRNA (NTC) did not trigger any increase of β-galactosidase in tumors (Fig. 4B). However, the β-galactosidase expression induced by DODAP-based lipid nanoparticle is restricted to a few areas within the tumor (Fig. 4C). We estimated that DODAP-based lipid nanoparticle only delivered siRNA to ∼1/% of the tumor cells. No significant increase of bioluminescent signal was observed in the TetR-ODC-Luc-derived liver tumors following intravenous administration of the DODAP nanoparticle-encapsulated TetR siRNA (data not shown). The lack of significant response in the TetR-ODC-Luc-based model is expected for a delivery system that only delivers siRNA to a small fraction of tumor cells because the increase of luminescence in a few cells will likely be buried in the background signal during imaging analysis. Importantly, these results demonstrate the high sensitivity of our positive-readout system in detecting functional TetR knockdown in various regions across a tumor section.
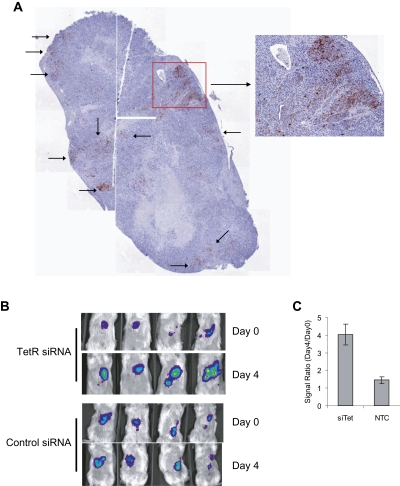
Compared with DODAP-based liposomes, the literature-described Stable Nucleic Acid Lipid Particle (SNALP) (Zimmermann et al. 2006) exhibited a more robust expression of β-galactosidase within the TetR-ODC-LacZ-derived tumors. When SNALP formulations were administered via tail-vein injection, significant increases of β-galactosidase were observed across the tumor sections (Fig. 5A). Consistent with these observations, SNALP mediated delivery of the TetR siRNA also led to a significant increase of luminescent signal (approximately fourfold) in the TetR-ODC-Luc-derived liver tumors, indicating functional siRNA delivery in these tumors (Fig. 5B,C). By contrast, a 30-fold increase of luminescence was observed in mice fed with doxycycline, a small molecule inducer that is expected to reach close to 100% of the tumor cells. Based on the differential induction of luminescence in SNALP- and doxycycline-treated mice, we estimated that the SNALP vehicle delivered sufficient amounts of the TetR siRNA to trigger reporter expression in 10%–15% of the tumor cells.
FIGURE 5.
SNALP-mediated delivery of a TetR siRNA triggers significant target knockdown in the positive-readout models. (A) Mice bearing the TetR-ODC-LacZ-derived liver tumors were administered with SNALP liposomes containing the TetR siRNA (i.v., 2.5 mg siRNA/kg on day 1 and day 4), and samples collected on day 7 for IHC analysis of β-galactosidase expression. n = 5 for each treatment group and representative pictures were shown here. (B) Mice bearing the TetR-ODC-Luc-derived liver tumors were imaged on day 0, administered with SNALP liposomes containing the control siRNA or the TetR siRNA on day 1 and day 2 (i.v., 2.5 mg siRNA/kg), and re-imaged on day 4 (48 h after the last dose). (C) The signal ratio of day 4/day 0 from each treatment group was plotted. N = 5 for each treatment group. Each bar in the graph represents the average signal ratio ± SEM for each group.
DISCUSSION
We report here on the development of a positive-readout system that elicits a rapid and robust response to siRNA delivery. Using this novel reporter system, we have created a panel of tumor models that serve as a versatile set of tools for evaluating siRNA delivery to tumors in vivo. The LacZ-based model has very high sensitivity and special resolution to detect siRNA delivery because a positive signal can be easily visualized at low levels in any region of the tumor in the presence of a negative background. As demonstrated in the example of using DODAP-liposome for siRNA delivery, the LacZ-based model allowed the detection of target knockdown in only 1%–2% of tumor cells, which is impossible to detect using most conventional methods. The enhanced sensitivity of the LacZ model allows us to distinguish between delivery systems that failed completely from those that successfully delivered siRNA to a certain percentage of tumor cells (e.g., 1%–20%). This degree of target knockdown is likely below the detection limit of most conventional assays. We believe that the moderately active delivery systems identified using the LacZ model could be a good starting point for further optimization to yield therapeutically viable siRNA delivery vehicles. It is also noteworthy that the LacZ model allowed us to detect significant heterogeneity in terms of susceptibility to siRNA delivery within tumors. Further understanding of the cause of this heterogeneity could have important ramifications on choosing the right tumor types or siRNA targets to match the activity profile of the delivery vehicle. For example, if a delivery system only gets access to cells adjacent to tumor vasculature, this delivery system may be better suited to deliver an antiangiogenic siRNA that targets the tumor vasculature compared to a cytotoxic siRNA, which targets the tumor cells. Alternatively, if a cytotoxic siRNA is to be used with this delivery vehicle, highly vascularized tumors need to be selected so that the delivery formulation can reach a sufficient number of tumor cells to produce a significant anti-tumor effect.
The Luciferase-based model is less sensitive than the LacZ-based model. Because the bioluminescence in the entire tumor, before and after siRNA delivery, is compared to determine whether there is an increase of reporter activity, the localized increase of reporter activity in a small fraction of tumor cells will likely be undistinguishable from the background signal. However, although the luciferase-based model is not as sensitive as the LacZ model, it is still much more sensitive than the conventional assays. As demonstrated in the example of SNALP-mediated siRNA delivery, target knockdown in 10%–15% tumor cells resulted in a readily detectable, three- to fourfold increase of luminescent signal, whereas, similar levels of delivery would be expected to result in a 10%–15% reduction of target at the protein or messenger levels in conventional assays, which would be very difficult to detect. Compared to the LacZ model, the luciferase-based model has the advantages of better quantification and higher throughput. Once a promising delivery vehicle is identified using the LacZ model, the higher throughput and better quantification of the Luciferase model would allow rapid comparative assessment of a large number of delivery systems, thus enabling an in vivo structure–activity relationship analysis of a particular delivery platform. In our experience, many formulations that are highly active in vitro are often inactive in vivo presumably due to poor PK properties. Considering the disconnection between in vitro and in vivo activities, the in vivo SAR capability is essential for the development of effective delivery vehicles for oncology applications.
In summary, we described here the creation and utilization of a positive-readout system for monitoring siRNA delivery to tumors in vivo. Results from our study suggest that various tumor models created using the positive-readout system could be extremely valuable assets to expedite the development of siRNA-based cancer therapeutics by serving as a reliable front-line screen tool to identify a promising delivery platform, enabling in vivo SAR analysis to support delivery system optimization, and providing in-depth information on a delivery system to guide the selection of tumor type and/or target for siRNA-based cancer therapy.
ACKNOWLEDGMENTS
This work is supported by Abbott Laboratories' internal R&D fund.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2546011.
REFERENCES
- Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ 2005. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing's sarcoma. Cancer Res 65: 8984–8992 [DOI] [PubMed] [Google Scholar]
- Koltover I, Salditt T, Rädler JO, Safinya CR 1998. An inverted hexagonal phase of cationic liposome–DNA complexes related to DNA release and delivery. Science 281: 78–81 [DOI] [PubMed] [Google Scholar]
- Landen CN Jr, Chavez-Reyes A, Bucana C, Schmandt R, Deavers MT, Lopez-Berestein G, Sood AK 2005. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res 65: 6910–6918 [DOI] [PubMed] [Google Scholar]
- Lewis DL, Hagstrom JE, Loomis AG, Wolff JA, Herweijer H 2002. Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nat Genet 32: 107–108 [DOI] [PubMed] [Google Scholar]
- Li L, Lin X, Staver M, Shoemaker A, Semizarov D, Fesik SW, Shen Y 2005. Evaluating hypoxia-inducible factor-1α as a cancer therapeutic target via inducible RNA interference in vivo. Cancer Res 65: 7249–7258 [DOI] [PubMed] [Google Scholar]
- Li L, Lin X, Khvorova A, Fesik SW, Shen Y 2007. Defining the optimal parameters for hairpin-based knockdown constructs. RNA 13: 1765–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirollo KF, Rait A, Zhou Q, Hwang SH, Dagata JA, Zon G, Hogrefe RI, Palchik G, Chang EH 2007. Materializing the potential of small interfering RNA via a tumor-targeting nanodelivery system. Cancer Res 67: 2938–2943 [DOI] [PubMed] [Google Scholar]
- Santel A, Aleku M, Keil O, Endruschat J, Esche V, Durieux B, Löffler K, Fechtner M, Röhl T, Fisch G, et al. 2006. RNA interference in the mouse vascular endothelium by systemic administration of siRNA-lipoplexes for cancer therapy. Gene Ther 13: 1360–1370 [DOI] [PubMed] [Google Scholar]
- Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, Wolfsberg TG, Umayam L, Lee JC, Hughes CM, Shanmugam KS, Bhattacharjee A, Meyerson M, et al. 2004. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci 101: 1892–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffelers RM, Ansari A, Xu J, Zhou Q, Tang Q, Storm G, Molema G, Lu PY, Scaria PV, Woodle MC 2004. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res 32: e149 doi: 10.1093/nar/gnh140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P 2005. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol 23: 709–717 [DOI] [PubMed] [Google Scholar]
- Takei Y, Kadomatsu K, Yuzawa Y, Matsuo S, Muramatsu T 2004. A small interfering RNA targeting vascular endothelial growth factor as cancer therapeutics. Cancer Res 64: 3365–3370 [DOI] [PubMed] [Google Scholar]
- Takeshita F, Minakuchi Y, Nagahara S, Honma K, Sasaki H, Hirai K, Teratani T, Namatame N, Yamamoto Y, Hanai K 2005. Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo. Proc Natl Acad Sci 102: 12177–12182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton NS, Lasic DD, Frederik PM, Strey HH, Roberts DD, Pavlakis GN 1997. Improved DNA: liposome complexes for increased systemic delivery and gene expression. Nat Biotechnol 15: 647–652 [DOI] [PubMed] [Google Scholar]
- Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A 2004. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther 12: 461–466 [DOI] [PubMed] [Google Scholar]
- Verma UN, Surabhi RM, Schmaltieg A, Becerra C, Gaynor RB 2003. Small interfering RNAs directed against beta-catenin inhibit the in vitro and in vivo growth of colon cancer cells. Clin Cancer Res 9: 1291–1300 [PubMed] [Google Scholar]
- Xing X, Liu V, Xia W, Stephens LC, Huang L, Lopez-Berestein G, Hung MC 1997. Safety studies of the intraperitoneal injection of E1A–liposome complex in mice. Gene Ther 4: 238–243 [DOI] [PubMed] [Google Scholar]
- Yang R, Wilcox DM, Haasch DL, Jung PM, Nguyen PT, Voorbach MJ, Doktor S, Brodjian S, Bush EN, Lin E et al. 2007. Liver-specific knockdown of JNK1 up-regulates proliferator-activated receptor γ coactivator 1β and increases plasma triglyceride despite reduced glucose and insulin levels in diet-induced obese mice. J Biol Chem 282: 22765–22774 [DOI] [PubMed] [Google Scholar]
- Zhang XJ, Ge YL, Hou L, Li Q 2008. Small interference RNAs directed against KDR gene inhibit the proliferation of breast cancer cells in vitro and in vivo. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi (Chin J Cell Mol Immunol) 24: 58–61 [PubMed] [Google Scholar]
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M 2006. RNAi-mediated gene silencing in non-human primates. Nature 441: 111–114 [DOI] [PubMed] [Google Scholar]