Abstract
Genes with sequence similarity to the yeast tRNAHis guanylyltransferase (Thg1) gene have been identified in all three domains of life, and Thg1 family enzymes are implicated in diverse processes, ranging from tRNAHis maturation to 5′-end repair of tRNAs. All of these activities take advantage of the ability of Thg1 family enzymes to catalyze 3′-5′ nucleotide addition reactions. Although many Thg1-containing organisms have a single Thg1-related gene, certain eukaryotic microbes possess multiple genes with sequence similarity to Thg1. Here we investigate the activities of four Thg1-like proteins (TLPs) encoded by the genome of the slime mold, Dictyostelium discoideum (a member of the eukaryotic supergroup Amoebozoa). We show that one of the four TLPs is a bona fide Thg1 ortholog, a cytoplasmic G−1 addition enzyme likely to be responsible for tRNAHis maturation in D. discoideum. Two other D. discoideum TLPs exhibit biochemical activities consistent with a role for these enzymes in mitochondrial 5′-tRNA editing, based on their ability to efficiently repair the 5′ ends of mitochondrial tRNA editing substrates. Although 5′-tRNA editing was discovered nearly two decades ago, the identity of the protein(s) that catalyze this activity has remained elusive. This article provides the first identification of any purified protein that appears to play a role in the 5′-tRNA editing reaction. Moreover, the presence of multiple Thg1 family members in D. discoideum suggests that gene duplication and divergence during evolution has resulted in paralogous proteins that use 3′-5′ nucleotide addition reactions for diverse biological functions in the same organism.
Keywords: 3′-5′ polymerase, G−1 addition, RNA editing, tRNA repair, tRNAHis
INTRODUCTION
The enzyme Thg1 (tRNAHis guanylyltransferase), whose gene was originally identified in yeast (Gu et al. 2003), is part of a family of enzymes found in all three domains of life and united by the ability to catalyze the 3′-5′ addition of nucleotides to nucleic acid substrates. Thg1 is widely distributed throughout eukaryotes (domain Eucarya), where it uses 3′-5′ nucleotide addition to attach a single essential guanylate residue (G−1) to the 5′ end of tRNAHis species (Cooley et al. 1982; Gu et al. 2005; Preston and Phizicky 2010). The presence of G−1 is unique to tRNAHis and acts as a necessary recognition element for efficient histidylation, both in vitro and in vivo in yeast (Rudinger et al. 1994, 1997; Nameki et al. 1995; Rosen and Musier-Forsyth 2004). The biological functions of Thg1-like proteins (TLPs) found in the domains Bacteria and Archaea are less well understood. In some archaeons, such as Methanobacterium thermoautotrophicus and Methanopyrus kandleri, TLPs likely add the required G−1 to tRNAHis, similar to their eukaryotic counterparts (Abad et al. 2010; Heinemann et al. 2010). However, in other archaeons, such as Methanosarcina acetivorans and Methanosarcina barkeri, and in nearly all bacterial species, tRNAHis genes already contain an encoded G−1 residue. The genomically encoded G−1 is presumably incorporated into the precursor tRNA during transcription, and can be retained in the mature tRNAHis due to an unusual 5′-end processing reaction catalyzed by RNase P, as occurs in Escherichia coli (Orellana et al. 1986). The lack of an obligate requirement for post-transcriptional G−1 addition in these Thg1-containing species suggests that some Thg1 family members may use 3′-5′ addition for alternative functions, beyond a simple role in tRNAHis maturation.
Thg1/TLP family members from Archaea and Bacteria exhibit at least two biochemical differences from eukaryotic family members studied so far (Abad et al. 2010; Rao et al. 2010). First, unlike yeast Thg1, bacterial/archaeal TLPs only add Watson–Crick base-paired nucleotides in the 3′-5′ addition reaction (Abad et al. 2010). Template-dependent 3′-5′ addition activity appears to be an ancestral property, as Thg1/TLP enzymes from all domains of life, including eukaryotes, catalyze this reaction (Jackman and Phizicky 2006b; Heinemann et al. 2009, 2010; Abad et al. 2010; Rao et al. 2010). Second, TLPs from Bacteria and Archaea exhibit a strong kinetic preference for the templated addition of nucleotides to the 5′ end of truncated tRNA substrates over the addition of −1 nucleotides to full-length tRNAs, and they add the missing 5′-nucleotides to tRNAs other than tRNAHis (Rao et al. 2010). Neither the addition to 5′-truncated tRNAs nor the addition to other tRNAs is catalyzed efficiently by yeast Thg1. The ability of bacterial/archaeal TLPs to restore a fully base-paired aminoacyl stem suggests a role for TLPs in previously unidentified pathways of tRNA 5′-end repair. Interestingly, TLPs from Archaea that require post-transcriptional G−1 addition to tRNAHis also catalyze the 5′-end repair reaction, suggesting that some Thg1 family members may participate in both tRNAHis maturation and general tRNA repair activities (Rao et al. 2010).
5′-End repair of tRNA is also an essential component of a 5′-tRNA editing activity that occurs in the mitochondria of a number of distantly related eukaryotic microbes (Lonergan and Gray 1993a, b; Laforest et al. 1997, 2004; Price and Gray 1999a; Gott et al. 2010). In these protists, such as the amoebozoon Acanthamoeba castellanii, where 5′-tRNA editing was first discovered, certain mitochondrially encoded tRNAs are predicted to contain up to three mismatched nucleotides at the first three positions in their aminoacyl-acceptor stem. The 5′-tRNA editing activity is thought to comprise at least two components: first, a nucleolytic activity that removes the incorrectly paired nucleotides from the 5′ end of the tRNA, and second, a 3′-5′ nucleotide addition activity that restores the missing nucleotides, using the 3′ end of the tRNA as the template to generate a fully paired aminoacyl-acceptor stem (Price and Gray 1999b; Bullerwell and Gray 2005). Although 5′-tRNA editing was the first of a number of tRNA editing activities to be described, the identity of the protein(s) that catalyze this reaction has remained unknown. However, the striking similarity of the 3′-5′ addition component of the 5′-tRNA editing reaction to the 5′-end repair activity recently associated with bacterial and archaeal TLPs suggested that enzymes from the Thg1 superfamily could play a role in 5′-tRNA editing.
In support of this hypothesis, multiple Thg1-related sequences have been identified in the genomes of several protists predicted to require 5′-tRNA editing to produce functional mitochondrial tRNAs. BLAST searches using yeast Thg1 have revealed the presence of two Thg1-related sequences in A. castellanii and in the chytid fungus, Spizellomyces punctatus (unpublished results), as well as four different genes with sequence similarity to yeast Thg1 in another amoebozoon, the slime mold Dictyostelium discoideum (Altschul et al. 1997; Fey et al. 2009). We chose the D. discoideum enzymes for further investigation based on the availability of a well-characterized genomic assembly for this organism and because of its biochemical tractability. Interestingly, one of the four D. discoideum sequences (initially called DdiTLP1 and here renamed DdiThg1) shares more sequence similarity with eukaryal Thg1 enzymes, such as yeast Thg1 (54% identity/72% similarity between DdiTLP1 and yeast Thg1), than with prokaryal TLPs, such as M. acetivorans TLP (30% identity/47% similarity between DdiTLP1 and archaeal MaTLP), consistent with a function for DdiTLP1 in cytoplasmic tRNAHis maturation (Fig. 1). The other three DdiTLPs (DdiTLP2–4) share more sequence identity with bacterial and archaeal family members (Fig. 1), suggesting that one or more of these could be components of the 5′-tRNA editing enzyme. Two of the DdiTLPs (TLP2 and TLP3) are predicted to contain mitochondrial targeting sequences, which would also be consistent with a role for these enzymes in mitochondrial 5′-tRNA editing.
FIGURE 1.
Sequence alignment of D. discoideum TLPs with Thg1 family members. Sequences of the four D. discoideum TLPs (DdiTLP1–4) were aligned with representative Thg1/TLP sequences from Eucarya (human; HsThg1 and Saccharomyces cerevisiae; ScThg1), Archaea (M. acetivorans; MaTLP and M. thermoautotrophicus; MtTLP) and Bacteria (Bacillus. thuringiensis; BtTLP) using Multalin (Corpet 1988). Dark and light shading indicate >90% and >50% sequence identity, respectively. Numbers in parentheses at the end of the sequence are the number of additional C-terminal residues not shown in the alignment. Residues highlighted in red are putative N-terminal mitochondrial targeting sequences predicted by MitoProt (Claros 1995; Claros and Vincens 1996). The Q/N-rich domain found in DdiTLP3 is indicated by underlining; M107 is indicated by a black star. Red stars indicate universally conserved residues that are critical for Thg1/TLP function (Jackman and Phizicky 2008). Residues indicated with blue stars are catalytically important for the activity of yeast Thg1, but are conserved only in eukaryotic Thg1 family members (including DdiTLP1). This includes the highly conserved HINNLYN motif (underlined in blue) that is catalytically critical for yeast Thg1 activity but is not found in bacterial or archaeal Thg1 enzymes sequences.
To test these hypotheses, we characterized the biochemical activities of each of the four Thg1-related sequences found in D. discoideum. Here we demonstrate that the bona fide Thg1 ortholog from Ddi (DdiThg1) adds G−1 to tRNAHis, both in vitro and in vivo consistent with its predicted function from sequence analysis and predicted cytoplasmic localization. Using an assay for 3′-5′ nucleotide addition to 5′-truncated mitochondrial-tRNA (mt-tRNA) substrates from D. discoideum, we also show that two of the TLPs (DdiTLP3 and DdiTLP4) exhibit biochemical activities consistent with their participation in editing. These results constitute the first identification of any purified protein that could play a role in 5′-tRNA editing in eukaryotes. As 5′-tRNA editing reactions require the addition of any of four nucleotides to complete the acceptor stem of substrate tRNAs, and occur with many different mt-tRNA species, this observation suggests more generalized roles for 3′-5′ nucleotide addition than the simple addition of a single nucleotide to tRNAHis first associated with members of this enzyme family in yeast and multicellular eukaryotes.
RESULTS
Purification of D. discoideum TLP enzymes
The four D. discoideum Thg1-related sequences identified by BLAST search (Fig. 1) were amplified by polymerase chain reaction (PCR) from a D. discoideum cDNA library and cloned with an N-terminal His6 tag into a vector for overexpression in and subsequent purification from E. coli (Fig. 2A). Based on the BLAST results, one of the four sequences (Q54E29) appeared to be more closely related than the others to yeast Thg1 and was consequently the strongest candidate to be the Thg1 ortholog in D. discoideum. Nonetheless, so as not to bias our approach, we initially chose to call the four genes DdiTLPs 1–4 (Q54E29 is designated DdiTLP1, Q54HW0 is DdiTLP2, Q54WD4 is DdiTLP3, and Q86IE7 is DdiTLP4).
FIGURE 2.
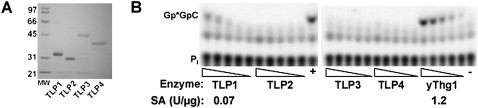
DdiTLP1 exhibits Thg1 activity in vitro. (A) SDS-PAGE analysis of purified TLPs from D. discoideum. Purified proteins (3 μg each) were resolved by 15% SDS-PAGE; lane MW, molecular weight markers. Predicted molecular weights: TLP1, 32 kDa; TLP2 (without mitochondrial peptide), 32 kDa; TLP3, 50 kDa, and TLP4, 36 kDa. (B) G−1 addition activity tested with 5′-32P-labeled yeast tRNAHis using each of the purified D. discoideum TLPs and yeast Thg1. Lane +, G−1 addition product generated by yeast Thg1; −, no enzyme control. The phosphatase protection assay was performed on reactions containing decreasing amounts of each purified protein (fivefold serial dilutions, see Materials and Methods for exact concentrations of each protein). The trimeric G−1 addition product (Gp*GpC) was resolved from inorganic phosphate (P*i) derived from remaining substrate using silica TLC. Specific activities for DdiTLP1 and yeast Thg1 were calculated by quantification of the amount of purified protein that catalyzed 10% conversion of substrate to G−1-containing product in 2 h at room temperature (defined as 1 Unit of enzyme activity); specific activities (SA) were expressed as Units per microgram of purified protein and indicated below each panel.
All four genes contain a 3′-5′ nucleotidyltransferase domain marked by the presence of several highly conserved residues that are critical for catalysis by yeast and human Thg1 (Fig. 1) (Jackman and Phizicky 2008; Hyde et al. 2010). DdiTLP2 and DdiTLP3 both contain a predicted mitochondrial targeting peptide (Fig. 1, red sequence). For DdiTLP2, this sequence was removed during cloning so that the final protein product expressed in E. coli lacked the N-terminal mitochondrial peptide. For DdiTLP3, the predicted sequence also contains an additional unusual Q/N-rich domain (underlined in Fig. 1) between the predicted N-terminal mitochondrial targeting sequence and the 3′-5′ nucleotidyltransferase domain. The Q/N-rich coding sequence is present in the PCR product derived from the cDNA expression library, indicating that this sequence is not attributable to an incorrect gene model (e.g., due to retention of an in-frame intron) but is contained in the DdiTLP3 mRNA. We cannot exclude the possibility that a translational start site downstream of the Q/N-rich domain (such as at M107, the beginning of the sequence that aligns with yeast Thg1) is actually utilized in this case, although an unusually high number (34%) of predicted protein-coding sequences in D. discoideum contain poly Q/N sequences (Eichinger et al. 2005). Due to the uncertainties regarding translation start site and the role of the Q/N domain, the full-length DdiTLP3 sequence (including mitochondrial-targeting peptide and Q/N domain) was cloned for expression and purification in this study. We note that the Q/N domain-containing DdiTLP3 product was purified in similar yields to the other TLPs (Fig. 2A) and was active in our assays (see below).
DdiTLP1 catalyzes the addition of G−1 to tRNAHis in vitro
We first tested the activity of all four purified DdiTLPs with a 5′-32P-labeled yeast tRNAHis substrate, using a phosphatase protection assay described previously (Jackman and Phizicky 2006a). Briefly, the addition of a −1 nucleotide results in protection of the 5′-labeled phosphate from dephosphorylation by alkaline phosphatase, and, after RNase A digestion, the resulting phosphatase-treated oligomeric reaction product (Gp*GpC) can be resolved from inorganic phosphate (P*i, generated from unreacted substrate) using thin-layer chromatography (TLC). In these assays, only DdiTLP1 catalyzed any detectable amount of G−1 addition to the yeast tRNAHis substrate, although the overall specific activity of the D. discoideum protein was ∼20-fold lower than that observed with yeast Thg1 (Fig. 2B). To test whether the reduced level of activity was due to the use of the nonnative yeast tRNA substrate instead of D. discoideum cytoplasmic tRNAHis, we used in vitro transcription to generate a tRNA with the sequence of Ddi-tRNAHis and tested G−1 addition activity using the same assay. However, although specific activity improved slightly, DdiTLP1 was still ∼10-fold less active than yeast Thg1 in this assay, even with a D. discoideum tRNAHis (data not shown).
DdiTLP1 catalyzes the addition of G−1 to tRNAHis in vivo in yeast
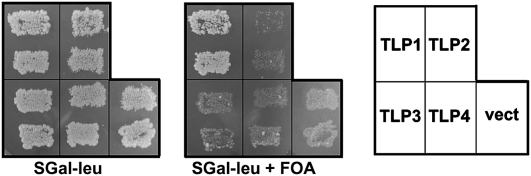
We used a yeast genetic assay to determine whether the relatively weaker G−1 addition activity catalyzed by DdiTLP1 is robust enough to support the growth of the yeast thg1Δ strain when DdiTLP1 is expressed as the sole source of Thg1 in the cells. A plasmid shuffle assay was performed, using a previously described yeast thg1Δ strain that contains a wild-type copy of yeast THG1 on a plasmid [CEN URA3 PTHG1-THG1] to maintain viability of the strain. Plasmids containing each of the four DdiTLPs under control of a galactose-inducible promoter [CEN LEU2 PGAL-TLPx] were transformed into the thg1Δ strain. For these constructs, the putative mitochondrial targeting sequences for DdiTLP2 and DdiTLP3 were removed, as G−1 addition occurs with cytoplasmic tRNAHis in yeast. Positive transformants were tested for their ability to support growth on media containing 5-fluoroorotic acid, which causes loss of the URA3 yeast THG1 plasmid. If the TLP supports growth of the thg1Δ strain, this result indicates that it is able to catalyze the essential G−1 addition reaction with tRNAHis in vivo. Of the four TLP-containing plasmids tested, complementation was only observed in the DdiTLP1-containing strain, consistent with the in vitro assay results (Fig. 3). The in vivo G−1-addition activity of DdiTLP1 in yeast is sufficient to fully substitute for the deletion of yeast THG1, as the DdiTLP1-complemented thg1Δ strain exhibits no growth defect relative to the wild-type strain on either rich (YPGal) or minimal media (SGal-leu) at a range of growth temperatures (18°C, 30°C, and 37°C) (data not shown). As both in vitro and in vivo data support a function for DdiTLP1 in G−1 addition to cytoplasmic tRNAHis, similar to yeast Thg1, we have renamed this gene product DdiThg1.
FIGURE 3.
DdiTLP1 catalyzes Thg1 activity in vivo in yeast. Complementation of the yeast thg1Δ growth defect by each of the D. discoideum TLPs was tested using a plasmid shuffle assay, as previously described (Abad et al. 2010). DdiTLPs, as indicated, expressed under control of a galactose-inducible promoter on LEU2 plasmids, were transformed into the yeast thg1Δ strain and two independent positive transformants were tested by replica plating onto the indicated media. Clones of TLP2 and TLP3 lacked their respective mitochondrial targeting sequences and the Q/N-rich sequence for TLP3 (see Fig. 1). Empty vector control strains (vect) were also tested. Photographs were taken after 3–4 d growth at 30°C.
DdiTLP3 and DdiTLP4 add a missing G+1 to mt-tRNAIleCAU
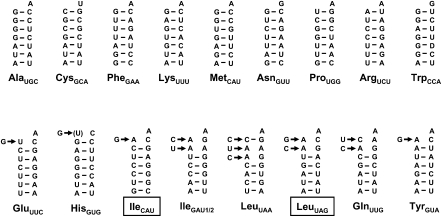
In D. discoideum, nine of 18 mtDNA-encoded tRNA genes contain predicted mismatched nucleotides at their 5′ ends (Fig. 4; Ogawa et al. 2000). Although 5′ editing has not been directly demonstrated in D. discoideum through comparison of tRNA gene sequences with the sequences of isolated mt-tRNA, the observation of nucleotide mismatches at any of the first three positions of the tRNA aminoacyl-acceptor stem is strongly predictive of the occurrence of 5′-tRNA editing in other protists. Moreover, mitochondrial 5′-tRNA editing has been directly demonstrated in a close relative of D. discoideum, the amoebozoon Polysphondylium pallidum (Schindel 2004). The two-step 5′-tRNA editing reaction requires first, removal of the mismatched nucleotides from the 5′ end of the transcript, and second, the 3′-5′ addition of the correct residues to the resulting 5′-truncated tRNA to generate a fully paired aminoacyl-acceptor stem (Price and Gray 1999b).
FIGURE 4.
Mitochondrial tRNA sequences from D. discoideum indicate a need for 5′-tRNA editing. Aminoacyl-acceptor stems of the 18 mt-tRNAs encoded in the D. discoideum mitochondrial genome are shown; the remainder of each tRNA sequence is omitted for clarity. Two genes for mt-tRNAIleGAU that differ by only two nucleotides are indicated as IleGAU1/2 as they are identical in their aminoacyl-acceptor stems. The need for mt-tRNA editing is reflected by the presence of non-Watson–Crick unpaired residues in the acceptor stem for the nine tRNAs (including the two IleGAU isoforms) shown on the bottom row. Residues expected to be added by 5′ editing are indicated by arrows to the left of each incorrect nucleotide.
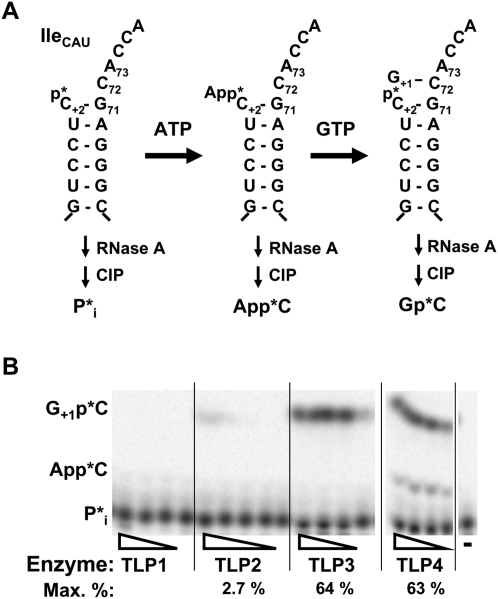
We tested the ability of each of the four TLPs to catalyze the second (nucleotide incorporation) step of the editing reaction, and added back in the missing 5′-end nucleotides to mt-tRNAIleCAU from D. discoideum. A 5′-truncated mt-tRNAIleCAU substrate was designed to mimic the tRNA species after removal of the original mismatched nucleotides by the as-yet-unidentified nuclease activity (Fig. 5). In this case, deletion of the encoded A+1 that forms a mismatch with C72 yields a C+2-initiating tRNA coding sequence, which is not a preferred template for in vitro transcription using T7 RNA polymerase. Therefore, the tRNA sequence was cloned with a 5′-hammerhead ribozyme sequence that catalyzes cotranscriptional cleavage, yielding a 5′-hydroxylated C+2-initiating tRNA. The tRNA was 5′-32P-labeled and tested for the addition of the missing G+1 nucleotide using the phosphatase protection assay (Fig. 5). The nucleotide incorporation activity previously observed in partially purified mitochondrial extracts with 5′-monophosphorylated tRNA transcripts required ATP (presumably for 5′-end activation) (Price and Gray 1999b), so ATP was included in the reactions.
FIGURE 5.
DdiTLP3 and DdiTLP4 repair 5′-truncated mt-tRNAIleCAU. (A) Schematic of phosphatase protection assay used with 5′-32P-labeled tRNA to detect G+1 addition catalyzed by TLPs in the presence of ATP (0.1 mM) and GTP (1 mM). Only aminoacyl-acceptor stem sequences of the tRNAs are shown for clarity; lines indicate connection to the tRNA body. Expected products of RNase A/CIP treatment are shown beneath each tRNA. (B) The addition of G+1 assayed as shown in A with decreasing concentrations of each purified TLP (fivefold serial dilutions of each). Identities of the labeled species are indicated to the left of the figure. Lane −, no-enzyme control reaction. The maximum percent reaction products (sum of G+1 and AppG reaction products, if observed) formed by TLP2, -3, and -4 is indicated below each panel.
The addition of G+1 yields a Gp*C product following treatment of the reaction products with RNase A and phosphatase (Fig. 5A). DdiTLP3 and DdiTLP4 both catalyzed robust formation of a major reaction product in the presence of GTP and ATP (Fig. 5B); this product contains a G residue linked to the labeled phosphate as revealed by release of Gp* following RNase T2 digestion of the TLC-purified reaction product (data not shown). In reactions with DdiTLP4, a small amount of the activated precursor for G+1 addition (App*C+2, also confirmed by nuclease digestion of the TLC-purified reaction product) was also observed (Fig. 5B). DdiThg1 (DdiTLP1 above) did not exhibit detectable 5′-end repair of mt-tRNAIle, consistent with a distinct role for this enzyme in cytoplasmic tRNAHis maturation. Barely detectable levels of G+1 addition were observed with DdiTLP2 (2.7% G+1-containing product), suggesting that this is not a preferred activity for this enzyme.
Steady-state kinetic parameters for the addition of the missing G+1 residue were determined for DdiTLP3 and DdiTLP4. The kcat for G+1 addition catalyzed by DdiTLP4 is approximately fivefold greater than kcat measured for DdiTLP3 (Table 1). Precise determination of KM exhibited by DdiTLP3 was somewhat difficult due to the slow rates of reaction, but saturation of the initial rates at tRNA substrate concentrations >0.2 μM, observed repeatedly in the steady-state assays, suggests that KM is lower for DdiTLP3 than for DdiTLP4, and therefore the overall catalytic efficiencies of both enzymes are similar (Table 1). Notably, the measured kcat/KM for 5′-end repair reactions catalyzed by TLPs are in close agreement with kcat/KM values measured previously for the physiological G−1 addition reaction catalyzed by yeast Thg1 with monophosphorylated tRNA substrate (shown in Table 1 for comparison) (Jackman and Phizicky 2006a). As with G−1 addition to tRNAHis, the measured in vitro rates for 5′-repair of mt-tRNAs are lower than might be expected for a typical enzyme-catalyzed reaction and may indicate participation of other factors in enhancing activity in vivo.
TABLE 1.
Steady-state kinetic parameters for 5′-end repair catalyzed by DdiTLPs
DdiTLP3 and DdiTLP4 catalyze 5′-end repair of mt-tRNALeuUAG
For 5′-tRNA editing in vivo, the addition of the missing nucleotides is not limited to the addition of G, and requires 3′-5′ polymerization of more than one nucleotide for some substrates (Fig. 4). Therefore, we tested addition to 5′-truncated mt-tRNALeuUAG missing both C+2 and G+1 residues (Fig. 6A). The 5′-32P-labeled U+3-initiating transcript (also generated using a hammerhead ribozyme transcript) was first used to test for the addition of the missing C+2 nucleotide in the presence of ATP and CTP. Phosphatase protection assays were performed as above, except that RNase T1 was used instead of RNase A, yielding labeled products terminating with G+4. As with the mt-tRNAIle substrate, prominent reaction products were observed in reactions containing DdiTLP3 and DdiTLP4, but not with the other two DdiTLPs (Fig. 6B).
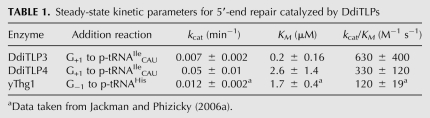
FIGURE 6.
DdiTLP3 and DdiTLP4 add a missing C+2 to 5′-truncated mt-tRNALeuUAG. (A) Schematic of phosphatase protection assay used with 5′-32P-labeled tRNA to detect addition catalyzed by TLPs in the presence of ATP (0.1 mM) and CTP (1 mM). Only aminoacyl-acceptor stem sequences are shown for clarity and reaction products of RNase T1/CIP treatment are shown below each tRNA sequence. (B) 3′-5′ Addition activity was tested using the assay shown in A with fivefold serial dilutions of each of the four purified TLPs. Lane Y, yeast Thg1 (1 μg), lane −, no-enzyme control. The major reaction products produced by TLP3 and TLP4 are indicated as P1 and P2; the identities of these products were assigned as shown in parentheses based on further nuclease digestion. (C) Analysis of P1 and P2 reaction products of DdiTLP4 reaction with 5′-truncated mt-tRNALeuUAG. The TLC-purified P1 and P2 products (shown in the first lane of each panel, labeled −) were tested by further nuclease digestion using RNAse T1 (T1), RNase T2 (T2), nuclease P1 (P1), and RNase A (A), as indicated. The expected reaction products for each digestion are shown below each panel. Products were resolved by PEI-cellulose TLC; positions of cold NMP standards (C, A, U, and G) were visualized by UV shadowing and are indicated by circles to the left of the figure.
In the absence of oligonucleotide standards, the identities of the two major products isolated from the TLC plates (indicated P1 and P2 in Fig. 6B) were determined by further nuclease digestion. Both P1 and P2 are C+2-containing products, as evidenced by the production of Cp* and p*U by RNase T2 and nuclease P1, respectively (Fig. 6C). However, the different behavior of P1 and P2 upon RNase A treatment suggests that P2 corresponds to C+2p*UpG, whereas P1 corresponds to A+1pC+2p*UpG. This assignment is based on the fact that, unlike for P2, treatment of P1 with RNase A did not yield Cp*, indicating that another RNase A-resistant nucleotide (either A or G linked by a standard phosphodiester bond, based on the RNase T2 result) has been added at the +1 position. As ATP was the only other nucleotide included in the reactions, we conclude that the product results from the addition of a non-Watson–Crick-pairing A+1 nucleotide, after C+2 is added. C+2 addition by yeast Thg1 was barely detectable, consistent with the inefficient 5′-end repair activities observed previously with yeast Thg1 using other 5′-truncated tRNA substrates (Jackman and Phizicky 2006b; Rao et al. 2010).
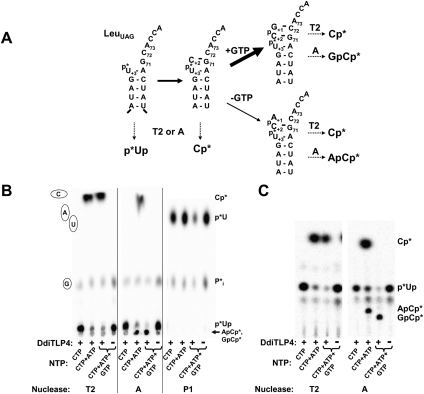
We then tested the ability of TLPs to add both missing nucleotides to U+3-tRNALeu. As complete repair of the p*U+3-initiating tRNA would generate labeled products that are sensitive to both RNase A (C+2-containing) and RNase T1 (G+1-containing), a different assay was used to detect product formation. Following incubation of labeled substrate with each enzyme and the indicated NTPs, reactions were first treated with phosphatase, then purified by phenol extraction and ethanol precipitation, and treated with various nucleases (Fig. 7A). For clarity, the results shown in Figure 7 are only those for reactions containing DdiTLP4; however, DdiTLP3 generated identical reaction products in parallel assays. The labeled 5′-phosphate was not completely removed from unreacted substrate by the phosphatase treatment, as evidenced by the labeled p*Up product generated by RNase A and T2 digestion and p*U generated by nuclease P1 digestion in control reactions that lack enzyme (see Fig. 7B, fourth lane for each nuclease). Nonetheless, pre-treatment with phosphatase was important to remove unlabeled 5′-phosphates from the various nucleotide addition products. As expected, C addition does not occur in the absence of ATP for 5′ end activation (Fig. 7B, first lane for each nuclease). As only p*U was observed after nuclease P1 digestion, the activated (App*U+3) intermediate for C+2 addition does not accumulate in any of the reactions. Consistent with the results above, reactions that contained only ATP and CTP generated predominantly Cp* after RNase T2 or RNase A digestion, and a second minor reaction product that remains at the origin (likely ApCp* based on the analysis in Fig. 6) (Fig. 7B). However, in reactions that contained all three NTPs, no Cp* product was detected after RNase A digestion, indicating that the C+2-containing intermediate species (Fig. 7A) does not accumulate. Instead, the major RNase A reaction product is a spot that remains at the origin, and the observation of Cp* by RNase T2 digest reveals that this major product contains an RNase A-resistant +1 nucleotide, linked by standard phosphodiester bond to C+2, as expected for the G+1-containing product (Fig. 7B). The two RNase A-generated products remaining at the origin can be further resolved by silica TLC (Fig. 7C), which reveals that (1) the dinucleotide reaction products formed in the presence and absence of GTP are not the same and (2) the presence of GTP drives the addition reaction to completion, as there is no evidence for accumulation of products that contain only C+2. Thus, although nontemplated A+1 addition can occur when only ATP is present in the assays, this reaction is less efficient than the addition of the correct Watson–Crick base-pairing G+1 nucleotide. This result is consistent with the preference for formation of Watson–Crick base pairs exhibited by Thg1/TLP family enzymes.
FIGURE 7.
DdiTLP4 (and DdiTLP3) fully repair 5′-truncated mt-tRNALeuUAG using 3′-5′ polymerization. (A) Schematic of nuclease digestion assay used with 5′-32P-labeled tRNA to detect addition catalyzed by TLPs in the presence of various nucleotides, as indicated. The relative thicknesses of the reaction arrows reflect the experimentally observed (see B, C below) preference for the addition of templated G+1 over the addition of nontemplated A+1, observed when GTP was omitted from the assays. For this assay, reactions were treated with phosphatase and purified by phenol extraction and ethanol precipitation prior to further nuclease digestion; expected reaction products for each tRNA are shown by dashed arrows. (B) Assays contained DdiTLP4 (30 μM), or no enzyme (−) as indicated, and either CTP only (1 mM), CTP (1 mM) + ATP (0.1 mM), or all three NTPs (1 mM each CTP and GTP, and 0.1 mM ATP), and were digested with the indicated nuclease. Reaction products were resolved using PEI-cellulose TLC; positions of unlabeled NMP standards (C, A, U, and G) were visualized by UV shadowing and are indicated to the left of the figure. Identical results were observed in reactions with DdiTLP3, not shown here for clarity. (C) Reaction products from panel B were resolved by silica TLC to further separate products that remained at the origin following RNase A digestion. Identities of labeled species are indicated to the right of the figure.
DISCUSSION
Despite identification of mitochondrial 5′-tRNA editing in certain protists nearly 20 years ago, the identities of any of the protein components of the 5′-tRNA editing machinery has remained unknown. By studying the activities of four Thg1-like proteins from the slime mold D. discoideum, we have identified the first purified proteins that are likely to participate in the 5′-tRNA editing process. Two of the four TLPs encoded by D. discoideum (DdiTLP3 and DdiTLP4) catalyze 5′-tRNA repair of mt-tRNA substrates derived from D. discoideum in vitro (Figs. 5–7), and thus are well-suited to participate in the second step of the 5′-editing reaction, forming a fully base-paired aminoacyl-acceptor stem for each tRNA. During the course of our investigation we also demonstrated that one of the TLPs (DdiThg1) is the likely cytoplasmic G−1 addition enzyme in D. discoideum, based on its ability to catalyze G−1 addition to tRNAHis both in vitro and in vivo in yeast (Figs. 2, 3). These results are the first demonstration of multiple uses for 3′-5′ nucleotide addition reactions catalyzed by Thg1/TLP family enzymes in the same organism.
The potential for an evolutionary relationship between the 5′-editing enzymes and the enzyme that catalyzes tRNAHis maturation in eukaryotes, the identity of which was also unknown at that time, was suggested soon after the discovery of the 5′-editing activity (Price and Gray 1999b). This hypothesis was based on the fact that both activities required nucleotide addition at the 5′ end of tRNA, potentially through the involvement of a 3′-5′ nucleotide polymerase. The unexpected discovery of Watson–Crick-templated 3′-5′ polymerase activity catalyzed by yeast Thg1 (Jackman and Phizicky 2006b) further supported this hypothesis. However, attempts to demonstrate an “editing-like” activity of yeast Thg1 were not successful, as yeast Thg1 does not efficiently add missing nucleotides to 5′-truncated tRNAs (Jackman and Phizicky 2006b). Instead, the connection between 5′ editing and the Thg1 enzyme family became evident only recently upon characterization of TLPs found in Bacteria and Archaea. Like DdiTLP3 and DdiTLP4, bacterial and archaeal TLPs catalyze efficient repair of 5′-truncated tRNA substrates (Rao et al. 2010). Moreover, they exhibit a strong kinetic preference for 5′-end repair over nucleotide additions at the −1 position of tRNA substrates, suggesting that this is the preferred activity for bacterial and archaeal TLPs in vivo. Endogenous substrates for tRNA 5′-end repair in Bacteria and Archaea have not yet been identified, as genomic tRNA sequences from TLP-containing species do not contain aminoacyl-acceptor stem mismatches. Thus, the occurrence of 5′ editing in Bacteria and Archaea seems unlikely. Instead, the 5′-end repair activity in these species is proposed to play a more general role in repair of tRNA that has been damaged or misprocessed (Rao et al. 2010). Nonetheless, the results presented here are the first demonstrated involvement of tRNA 5′-end repair in a known physiological activity, and further support the proposed repair function of TLPs in Bacteria and Archaea.
The lack of apparent 5′-end repair activity catalyzed by bona fide Thg1 enzymes such as yeast Thg1 and DdiThg1 is intriguing with respect to the evolution of the Thg1/TLP enzyme family. The universal ability of members of the Thg1/TLP enzyme family from all domains of life to catalyze 3′-5′ Watson–Crick polymerization suggests that this is an ancestral activity of Thg1 enzymes (Abad et al. 2010). Evolutionary pressure on eukaryotic Thg1 enzymes to catalyze the nontemplated addition of G−1 opposite A73 with strong selectivity for tRNAHis may have resulted in a concurrent reduction in their ability to add, in a templated fashion, different nucleotides in repairing a diverse set of tRNA substrates. However, the possibility remains that even eukaryotic Thg1 enzymes may similarly be able to catalyze 5′-end repair, perhaps for other substrates or under certain conditions. The demarcation between enzymes that are involved in G−1 addition and those that appear to be involved in 5′-end repair is not always clear. In some archaeons, such as M. thermoautotrophicus, the single encoded TLP (MtTLP) exhibits a biochemical and kinetic preference for 5′-end repair of 5′-truncated tRNA similar to that shown by other archaeal TLPs. Yet the lack of a genomically encoded G−1 residue on tRNAHis in M. thermoautotrophicus suggests that MtTLP predictably functions in post-transcriptional G−1 addition (Heinemann et al. 2010; Rao et al. 2010). Further investigation of mt-tRNA editing in certain protists appears likely to reveal a similar situation. In A. castellanii, we have identified only two TLPs in the virtually complete draft genome sequence, but unlike in D. discoideum, neither of these TLPs appears to be an authentic ortholog to the canonical “Thg1” enzymes from yeast and humans. However, cytoplasmic G−1 addition to tRNAHis is presumably also required in A. castellanii, so that one of the A. castellanii TLPs may catalyze this reaction, despite its lack of similarity to yeast Thg1. Investigation of the functions of these members of the Thg1/TLP enzyme family are likely to be important for understanding the evolution of the distinct activities catalyzed by Thg1/TLP enzymes.
Steady-state kinetic analysis of 5′-end repair catalyzed by DdiTLP3 and DdiTLP4 compared with G−1 addition catalyzed by yeast Thg1 revealed similar in vitro activity for each enzyme with respect to its expected substrates (Table 1). However, questions remain about the nature of the editing enzymes in vivo. In particular, the reason for the presence of two enzymes in D. discoideum with apparently similar abilities to catalyze the same reaction is intriguing. DdiTLP3 and DdiTLP4 may exhibit distinct tRNA substrate recognition properties not yet observed with the limited number of substrates tested so far, and thus both enzymes could be required in vivo to repair the full complement of mt-tRNAs to be edited. It is also possible that the in vivo editing enzyme contains both DdiTLP3 and DdiTLP4, and that the activity of the in vivo enzyme is enhanced over that of each enzyme in isolation, a possibility that could also explain the relatively low rates observed in vitro. The kcat for the 5′-repair activity is not enhanced above that observed with either enzyme alone when purified DdiTLP3 and DdiTLP4 are mixed in the reactions. Nonetheless, the recently observed tetrameric structure of human Thg1 (Hyde et al. 2010) suggests the possibility of formation of heteromultimeric complexes in organisms that encode more than one Thg1/TLP gene, as is the case in D. discoideum. Finally, the possibility that only one of the enzymes catalyzes 5′-tRNA editing in vivo cannot be ruled out; in this case, the other enzyme may be required for another biological function, perhaps related to other tRNA repair activities. With respect to this possibility, it is interesting to note that only DdiTLP3 possesses an identifiable targeting signal for mitochondrial localization (Fig. 1). Although a nonstandard targeting signal in DdiTLP4 may yet be identified, or interactions with other proteins may bring DdiTLP4 into the mitochondria, the possibility of a cytosolic function for DdiTLP4 (e.g., in the repair of 5′-truncated cytosolic tRNAs) remains intriguing.
Of the four TLPs investigated in this work, DdiTLP2 remains the only enzyme so far not associated with any of the typical biochemical activities of Thg1/TLP family enzymes. As the gene was cloned from a D. discoideum cDNA library, the mRNA is expressed, although DdiTLP2 protein expression in vivo has not been tested. Possibilities for this protein include noncatalytic roles in G−1 addition and/or 5′-editing activities, perhaps by association with other TLPs, or the possibility that DdiTLP2 exhibits activity with other as-yet-unidentified substrates. In vitro mixing experiments again revealed no stimulation of G−1 addition or 5′-editing activities by inclusion of purified DdiTLP2 in the assays (data not shown). Nonetheless, DdiTLP2 does not lack any of the highly conserved residues that function in 3′-5′ nucleotide addition, including critically important metal-binding residues (Jackman and Phizicky 2008; Hyde et al. 2010), an absence of which could readily explain the lack of in vitro activities observed in these assays (Fig. 1). The strongly predicted mitochondrial localization of DdiTLP2 suggests a likely function for the protein in the organelle, but further studies regarding this function will be necessary to elucidate the role of DdiTLP2 in D. discoideum.
Although this investigation establishes the foundation for the detailed biochemical analysis of 5′-tRNA editing and suggests that at least some of the protein components of the in vivo editing enzyme have been identified, many questions remain about the complete form of the editing holoenzyme, including identification of the postulated nuclease needed to remove the initial 5′-mismatched nucleotides from the transcript. Further investigations of the functions of the multiple Thg1/TLP family enzymes in protists will likely yield important insights into the evolution and mechanism of 3′-5′ nucleotide addition reactions in biology.
MATERIALS AND METHODS
Cloning and purification of DdiTLPs
The four Ddi TLP sequences were identified by BLAST search and amplified by PCR from purified DNA recovered from a D. discoideum cDNA library. For ligation-independent cloning (LIC) into a pET vector, appropriate sequences for expression with an N-terminal His6 tag (AVA421) were introduced with the PCR primers. The gel-purified PCR products for each DdiTLP were inserted into AVA421 vector using LIC with T4 DNA polymerase (Invitrogen) according to the manufacturer's instructions; clones were verified by sequencing. Plasmids encoding the four TLPs were transformed into E. coli strain (Rosetta pLysS) for expression. Cultures were grown in LB media with 100 μg/mL ampicillin at 37°C and induced with isopropyl-β-thiogalactose (1 mM final concentration) upon reaching OD600 = 0.4. After incubation for 4–5 h at 37°C, cells were harvested by centrifugation and lysed by French press, followed by centrifugation to obtain a cell-free crude extract. Purification was performed using immobilized metal-ion affinity chromatography with TALON resin (Clontech), as previously described (Abad et al. 2010). Following dialysis into buffer containing 50% glycerol, concentrations of the purified proteins (stored at −20°C) were determined by BioRad protein assay. Each of the purified TLPs was estimated to be >90% pure by SDS-PAGE, as judged by intensity of the Coomassie-stained protein bands (Fig. 2A).
Cloning and preparation of D. discoideum mt-tRNA constructs
Plasmids for in vitro transcription of mt-tRNAs (mt-tRNAIleCAU and mt-tRNALeuUAG) were constructed using pairs of overlapping oligonucleotides (50 nucleotides each) and successive rounds of fill-in polymerization to yield DNA fragments with the final sequence: [T7 RNA polymerase promoter]–[hammerhead ribozyme]–[tRNA], as described in Morl et al. (2005). A final round of PCR was performed to introduce a BstNI restriction site for run-off transcription, and BamHI/PstI restriction sites for cloning by ligation into a digested pUC19 vector. Final constructs were verified by sequencing. tRNAs were produced from the BstNI-digested plasmid by in vitro transcription using T7 RNA polymerase under standard conditions, except that an additional cycling step (60°C × 3 min followed by 25°C × 3 min, repeated 10 times) was added following the initial 1 h transcription reaction at 37°C. The cycling step ensures maximal formation of ribozyme-cleaved tRNA initiating at the desired position. The gel-purified 5′-hydroxylated tRNA that results from transcription/hammerhead cleavage was subsequently 5′-monophosphorylated using T4 polynucleotide kinase and either unlabeled or [γ-32P]-labeled ATP, according to standard protocols. Unreacted ATP was removed by spin column chromatography using BioGel P6 columns (BioRad).
3′-5′ Nucleotide addition assay
Phosphatase protection assays were performed using 5′-32P-labeled tRNA substrates, as previously described (Jackman and Phizicky 2006a). Reactions were initiated by the addition of fivefold serial dilutions of purified enzyme. Concentration ranges spanned for each enzyme varied depending on the concentration of each purified protein preparation and were: DdiTLP1, 6–0.01 μM; DdiTLP2, 100–0.2 μM; DdiTLP3, 3–0.005 μM; and DdiTLP4, 34–0.05 μM. Reactions were allowed to proceed for 2 h at room temperature, stopped by the addition of nuclease (either RNase A or RNase T1, as indicated for each substrate) and subsequently treated with phosphatase. The resulting reaction mixtures were resolved by silica TLC in a 55:35:10 (v:v:v, 1-propanol:NH4OH:H2O) solvent system. Plates were dried and visualized using a Typhoon Trio imager. For product identification experiments, labeled products were eluted from silica TLC plates with water, dried, and resuspended in ddH2O for further digestion. Nuclease digestion reactions were performed in 20 mM sodium acetate, pH 5.2; nuclease P1 digestion reactions also contained 200 μM ZnCl2, whereas RNase A, T2 and T1 digestions additionally contained 100 μM EDTA. Steady-state kinetic parameters for addition to monophosphorylated tRNAs were determined by measurement of initial rates for TLP-catalyzed G+1 addition at varied tRNA substrate concentrations (0.1–10 μM tRNAIle and 0.05–2 μM tRNALeu) using the above-mentioned assay, also as previously described (Jackman and Phizicky 2006a). For C+2/G+1 addition to mt-tRNALeu, reactions were first treated with phosphatase (0.5 U/10 μL reaction) and subsequently purified by phenol extraction and ethanol precipitation. The labeled products were then subjected to digestion with various nucleases, under the same conditions described above.
Yeast complementation assay
The DdiTLP genes were cloned to a shuttle plasmid containing a galactose-inducible promoter [CEN LEU2 PGAL-TLPx]. Four DdiTLP gene inserts, TLP1, TLP2-mito (lacking the predicted mitochondrial-targeting sequence), TLP3-M107 (lacking the predicted mitochondrial-targeting sequence and Q/N-rich domain, and starting from M107) and TLP4, were generated by PCR using pairs of primers that introduce BamHI or PstI restriction sites for cloning. BamHI/PstI-digested PCR products and vector (treated with calf intestinal phosphatase, CIP) were ligated using T4 DNA ligase (USB, Affymetrix, Inc.) according to the manufacturer's protocol. Each construct was verified by DNA sequencing. Five plasmids (the four TLP constructs and the empty vector) were transformed to a yeast thg1Δ strain containing a yeast THG1 gene on a URA3 plasmid [CEN URA3 PTHG1-THG1], which is needed to maintain viability. Positive (Leu+) transformants were selected at 30°C on synthetic dextrose (SD) media, and tested by replica plating at 30°C to synthetic galactose (SGal) media containing 5-fluoroorotic acid, to induce expression of the DdiTLPs and select against the URA3 plasmid-containing wild-type yeast THG1.
ACKNOWLEDGMENTS
Funding for this work was provided by NIH grant nos. GM087543 (to J.E.J.) and GM054663 (to J.M.G.). A.W. was supported by the NSF-REU in Molecular Genetics and Biochemistry at The Ohio State University. The authors thank Ralf Bundschuh for valuable discussions and assistance with TLP sequence identification.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2517111
REFERENCES
- Abad MG, Rao BS, Jackman JE 2010. Template-dependent 3'-5' nucleotide addition is a shared feature of tRNAHis guanylyltransferase enzymes from multiple domains of life. Proc Natl Acad Sci 107: 674–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullerwell CE, Gray MW 2005. In vitro characterization of a tRNA editing activity in the mitochondria of Spizellomyces punctatus, a Chytridiomycete fungus. J Biol Chem 280: 2463–2470 [DOI] [PubMed] [Google Scholar]
- Claros MG 1995. MitoProt, a Macintosh application for studying mitochondrial proteins. Comput Appl Biosci 11: 441–447 [DOI] [PubMed] [Google Scholar]
- Claros MG, Vincens P 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241: 779–786 [DOI] [PubMed] [Google Scholar]
- Cooley L, Appel B, Soll D 1982. Post-transcriptional nucleotide addition is responsible for the formation of the 5' terminus of histidine tRNA. Proc Natl Acad Sci 79: 6475–6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435: 43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey P, Gaudet P, Curk T, Zupan B, Just EM, Basu S, Merchant SN, Bushmanova YA, Shaulsky G, Kibbe WA, et al. 2009. dictyBase—a Dictyostelium bioinformatics resource update. Nucleic Acids Res 37: D515–D519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gott JM, Somerlot BH, Gray MW 2010. Two forms of RNA editing are required for tRNA maturation in Physarum mitochondria. RNA 16: 482–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Jackman JE, Lohan AJ, Gray MW, Phizicky EM 2003. tRNAHis maturation: an essential yeast protein catalyzes addition of a guanine nucleotide to the 5' end of tRNAHis. Genes Dev 17: 2889–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Hurto RL, Hopper AK, Grayhack EJ, Phizicky EM 2005. Depletion of Saccharomyces cerevisiae tRNA(His) guanylyltransferase Thg1p leads to uncharged tRNAHis with additional m(5)C. Mol Cell Biol 25: 8191–8201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann IU, O'Donoghue P, Madinger C, Benner J, Randau L, Noren CJ, Soll D 2009. The appearance of pyrrolysine in tRNAHis guanylyltransferase by neutral evolution. Proc Natl Acad Sci 106: 21103–21108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann IU, Randau L, Tomko RJ Jr, Soll D 2010. 3'-5' tRNAHis guanylyltransferase in bacteria. FEBS Lett 584: 3567–3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde SJ, Eckenroth BE, Smith BA, Eberley WA, Heintz NH, Jackman JE, Doublie S 2010. tRNAHis guanylyltransferase (THG1), a unique 3'-5' nucleotidyl transferase, shares unexpected structural homology with canonical 5'-3' DNA polymerases. Proc Natl Acad Sci 107: 20305–20310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman JE, Phizicky EM 2006a. tRNAHis guanylyltransferase adds G−1 to the 5' end of tRNAHis by recognition of the anticodon, one of several features unexpectedly shared with tRNA synthetases. RNA 12: 1007–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman JE, Phizicky EM 2006b. tRNAHis guanylyltransferase catalyzes a 3'-5' polymerization reaction that is distinct from G−1 addition. Proc Natl Acad Sci 103: 8640–8645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman JE, Phizicky EM 2008. Identification of critical residues for G−1 addition and substrate recognition by tRNA(His) guanylyltransferase. Biochemistry 47: 4817–4825 [DOI] [PubMed] [Google Scholar]
- Laforest MJ, Roewer I, Lang BF 1997. Mitochondrial tRNAs in the lower fungus Spizellomyces punctatus: tRNA editing and UAG ‘stop’ codons recognized as leucine. Nucleic Acids Res 25: 626–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforest MJ, Bullerwell CE, Forget L, Lang BF 2004. Origin, evolution, and mechanism of 5' tRNA editing in chytridiomycete fungi. RNA 10: 1191–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonergan KM, Gray MW 1993a. Editing of transfer RNAs in Acanthamoeba castellanii mitochondria. Science 259: 812–816 [DOI] [PubMed] [Google Scholar]
- Lonergan KM, Gray MW 1993b. Predicted editing of additional transfer RNAs in Acanthamoeba castellanii mitochondria. Nucleic Acids Res 21: 4402 doi: 10.1093/nar/21.18.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morl M, Lizano E, Willkomm DK, Hartmann RK 2005. Production of RNAs with homogeneous 5' and 3' ends. In Handbook of RNA biochemistry (ed. Hartmann RK, Schon A, Westhof E), pp. 22–35 Wiley-VCH, Wenheim, Germany [Google Scholar]
- Nameki N, Asahara H, Shimizu M, Okada N, Himeno H 1995. Identity elements of Saccharomyces cerevisiae tRNA(His). Nucleic Acids Res 23: 389–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Yoshino R, Angata K, Iwamoto M, Pi M, Kuroe K, Matsuo K, Morio T, Urushihara H, Yanagisawa K, et al. 2000. The mitochondrial DNA of Dictyostelium discoideum: complete sequence, gene content and genome organization. Mol Gen Genet 263: 514–519 [DOI] [PubMed] [Google Scholar]
- Orellana O, Cooley L, Soll D 1986. The additional guanylate at the 5' terminus of Escherichia coli tRNAHis is the result of unusual processing by RNase P. Mol Cell Biol 6: 525–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston MA, Phizicky EM 2010. The requirement for the highly conserved G−1 residue of Saccharomyces cerevisiae tRNAHis can be circumvented by overexpression of tRNAHis and its synthetase. RNA 16: 1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DH, Gray MW 1999a. Confirmation of predicted edits and demonstration of unpredicted edits in Acanthamoeba castellanii mitochondrial tRNAs. Curr Genet 35: 23–29 [DOI] [PubMed] [Google Scholar]
- Price DH, Gray MW 1999b. A novel nucleotide incorporation activity implicated in the editing of mitochondrial transfer RNAs in Acanthamoeba castellanii. RNA 5: 302–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao BS, Maris EL, Jackman JE 2010. tRNA 5'-end repair activities of tRNAHis guanylyltransferase (Thg1)-like proteins from Bacteria and Archaea. Nucl Acids Res doi: 10.1093/nar/gkq976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen AE, Musier-Forsyth K 2004. Recognition of G−1:C73 atomic groups by Escherichia coli histidyl-tRNA synthetase. J Am Chem Soc 126: 64–65 [DOI] [PubMed] [Google Scholar]
- Rudinger J, Florentz C, Giege R 1994. Histidylation by yeast HisRS of tRNA or tRNA-like structure relies on residues −1 and 73 but is dependent on the RNA context. Nucleic Acids Res 22: 5031–5037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudinger J, Felden B, Florentz C, Giege R 1997. Strategy for RNA recognition by yeast histidyl-tRNA synthetase. Bioorg Med Chem 5: 1001–1009 [DOI] [PubMed] [Google Scholar]
- Schindel E 2004. Editing of mitochondrial tRNAs in Polysphondylium pallidum. In Biochemistry and molecular biology. Dalhousie University, Halifax, Nova Scotia, Canada [Google Scholar]