Abstract
Animals have evolved mechanisms to ensure the robustness of developmental outcomes to changing environments. MicroRNA expression may contribute to developmental robustness because microRNAs are key post-transcriptional regulators of developmental gene expression and can affect the expression of multiple target genes. Caenorhabditis elegans provides an excellent model to study developmental responses to environmental conditions. In favorable environments, C. elegans larvae develop rapidly and continuously through four larval stages. In contrast, in unfavorable conditions, larval development may be interrupted at either of two diapause stages: The L1 diapause occurs when embryos hatch in the absence of food, and the dauer diapause occurs after the second larval stage in response to environmental stimuli encountered during the first two larval stages. Dauer larvae are stress resistant and long lived, permitting survival in harsh conditions. When environmental conditions improve, dauer larvae re-enter development, and progress through two post-dauer larval stages to adulthood. Strikingly, all of these life history options (whether continuous or interrupted) involve an identical pattern and sequence of cell division and cell fates. To identify microRNAs with potential functions in buffering development in the context of C. elegans life history options, we used multiplex real-time PCR to assess the expression of 107 microRNAs throughout development in both continuous and interrupted life histories. We identified 17 microRNAs whose developmental profile of expression is affected by dauer life history and/or L1 diapause, compared to continuous development. Hence these microRNAs could function to regulate gene expression programs appropriate for different life history options in the developing worm.
Keywords: microRNA, dauer, development
INTRODUCTION
Animal development is a complex, highly regulated process, whose outcome is vital to the survival of the individual and the species. Animals in their natural environments can develop normally despite varying and harsh environmental conditions. Hence, animals have evolved mechanisms to ensure that developmental outcomes occur robustly, regardless of changing conditions. MicroRNAs (miRNAs) may play an important role in developmental robustness to changing environments. These small, noncoding RNAs bind to partially complementary sequences in target mRNAs and regulate translation and mRNA stability through a variety of mechanisms, depending on cellular context and environmental conditions (Hammell 2008; Holtz and Pasquinelli 2009). Because a given miRNA can regulate the expression of multiple target genes, miRNAs can potentially provide coordinated rapid changes in gene expression in response to developmental, environmental, and physiological cues (Bartel 2004, 2009). Indeed, the expression of many miRNAs is developmentally regulated, consistent with roles for miRNAs in the regulation of gene expression in conjunction with developmental processes (Bartel 2004; Kato and Slack 2008). Furthermore, roles for miRNAs in providing robustness of developmental outcomes to various perturbations have been demonstrated (Li et al. 2009; Hilgers et al. 2010; Rasmussen et al. 2010).
Caenorhabditis elegans is an excellent system for the study of developmental robustness to environmental perturbations because this animal possesses mechanisms to respond to fluctuations in environmental conditions and to adjust developmental progression accordingly, without compromising the integrity of cell fate specification events (Liu and Ambros 1991; Euling and Ambros 1996; Riddle and Albert 1997; Braendle and Félix 2008). In order to identify miRNAs that could mediate interactions between physiological and developmental pathways, we assayed the expression level of 107 miRNAs during development through alternative life histories: the continuous development that occurs in favorable environments, and the interrupted development that can occur in unfavorable environments.
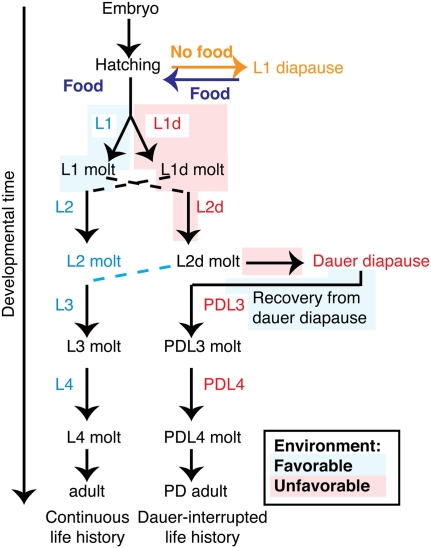
C. elegans development consists of embryogenesis followed by four larval stages (L1–L4) punctuated by molts (see Fig. 1). In favorable environments, larval development proceeds rapidly and continuously over ∼40–50 h at 20°C (Sulston and Horvitz 1977). Unfavorable environmental conditions sensed during L1 cause the larva to progress to the L2d stage, which is ∼50% greater in duration than the rapid L2 stage (Golden and Riddle 1984). L2d larvae prepare for future harsh conditions, while continuing to assess environmental cues. If conditions remain poor, larvae will enter the dauer diapause, a developmentally arrested, long-lived, and highly stress-resistant stage (Riddle and Albert 1997). All cells remain quiescent during dauer diapause, and the dauer larvae remain at a developmental stage equivalent to the L2-to-L3 molt (L2m) (Cassada and Russell 1975). When environmental conditions improve, dauer larvae restart their cell division cycles and re-enter development, progressing through post-dauer L3 and post-dauer L4 (PDL3, PDL4) stages (Riddle and Albert 1997). We refer to these two alternative developmental paths—of either continuous or dauer-interrupted development—as “life histories” (Fig. 1; Fielenbach and Antebi 2008).
FIGURE 1.
Schematic representation of continuous and interrupted life history options. C. elegans larvae develop continuously through four larval stages in favorable environmental conditions, but can interrupt their development by entry to the stress-resistant L1 diapause or dauer diapause in unfavorable environmental conditions. (Red shading) Unfavorable or (blue shading) favorable environmental conditions sensed by larvae. These environmental conditions drive larvae to either continuous or diapause-interrupted life histories. (Dashed lines) Junctures where larvae may switch between diapause-interrupted and continuous life history depending on the severity of the conditions encountered. Developmental stages at which miRNA levels were assessed in each life history: (blue) stages are within the continuous life history; (red) stages are within the dauer-interrupted life history. Developmentally equivalent stages were compared (Table 1); these are the red and blue stages at the same point along the vertical axis. (Orange) L1 diapause, which can lead to either continuous or dauer life histories, was compared to both embryos and continuously developing L1 larvae (Table 2).
Besides the dauer diapause, which occurs after the L2d stage, C. elegans larvae can undergo another, distinct diapause at the beginning of L1. The L1 diapause occurs prior to larval development, in situations in which embryos hatch in the absence of food (Fig. 1). Larvae in L1 diapause share some features with larvae in dauer diapause: They are developmentally arrested and nonfeeding. Furthermore, larvae in L1 diapause display some stress resistance and lengthened life span, although to a lesser extent than dauer larvae (Johnson et al. 1984). Upon exposure to food, these arrested larvae exit L1 diapause and proceed with larval development. Post-diapause L1 animals are not necessarily committed to L2d or dauer arrest; they can choose either continuous or dauer-interrupted life histories depending on the environmental conditions encountered as they progress through the first two larval stages (Golden and Riddle 1984; Johnson et al. 1984).
Since the pace and/or continuity of larval development are profoundly affected by larval life history, while the patterns of cell divisions and the associated sequences of cell lineage fates are unaffected, there must be mechanisms to integrate the progression of cell lineage development with the environmental and physiological signals that govern the dauer versus continuous life history choices. Genes involved in enabling animals to successfully traverse these distinct alternative life histories could include genes that display life history–dependent differences in expression.
There is evidence that life history affects the expression of protein-coding genes. The majority of previously published studies have compared mRNA levels or protein levels in dauer larvae to non-dauer animals of various developmental stages (Dalley and Golomb 1992; Jones et al. 2001; Holt and Riddle 2003; Wang and Kim 2003; Ruzanov et al. 2007; Jeong et al. 2009b; Ruzanov and Riddle 2010). An additional study monitored gene expression changes during entry to dauer (Jeong et al. 2009a). These studies focused on identifying genes that could contribute to the stress resistance, altered metabolism, and/or long life span characteristic of dauer larvae, rather than on the developmental effects of dauer diapause. However, two previous studies did compare mRNA levels between developmentally equivalent stages in different life histories. Liu et al. (2004) compared late L2d/early dauer TGFβ mutants to late L2/early L3 staged wild-type larvae, and Harvey et al. (2009) compared wild-type early-staged L2d larvae to wild-type continuously developing L2 larvae. Liu et al. (2004) found changes in the expression of many genes that regulate dauer formation, and also in several developmental regulators such as Notch pathway genes, Hedgehog pathway genes, and genes that regulate stage-specific cell fate decisions. Some of these changes could be part of the coordination between pathways that regulate development and pathways that regulate life history choice.
Less is known about gene expression changes in post-dauer larval stages. However, a recent study compared wild-type young adult hermaphrodites that had developed either continuously or whose development was interrupted by dauer (Hall et al. 2010). The investigators found not only numerous mRNAs whose levels were altered, but also global chromatin-remodeling signatures in post-dauer adults. Furthermore, they found that the post-dauer chromatin state was associated with increased brood size, suggesting that the epigenetic changes that occur in the dauer life history can affect the phenotype of wild-type individuals. It is not known whether transcription of miRNA-encoding genes is similarly affected by these chromatin modifications.
Previous work has identified certain miRNAs whose developmental expression is affected by life history. We and others have shown that let-7-family miRNA expression is reduced in L2d larvae and in dauer larvae by the repressive activity of the DAF-12 nuclear hormone receptor (Bethke et al. 2009; Hammell et al. 2009). This repression is part of a feedback loop that functions to integrate stage-specific cell fate decisions with life history choice (Hammell et al. 2009). In our previous study (Hammell et al. 2009), we compared the expression of 107 miRNAs during continuous L2 and L2d. We found four let-7-family miRNAs and one additional miRNA, miR-788, to be down-regulated in L2d. In a separate report, miR-788 was shown to be down-regulated during dauer diapause, likely by direct action of the DAF-3 transcription factor (Martinez et al. 2008a). In addition, both miR-248 and miR-234 were found to be up-regulated during dauer diapause by unknown mechanisms (Lim et al. 2003).
In the present study, we expand on those previous studies and present a comprehensive profiling of 107 worm miRNAs across the entirety of larval development—including animals in L1 diapause, animals at all stages of continuous development, and animals at all stages of dauer-associated life histories. We find 14 miRNAs whose expression is affected by dauer life history during at least one larval stage. In addition, we identify five miRNAs that are up-regulated during L1 diapause, two of which are also up-regulated in the dauer life history. We discuss these effects of life history on miRNA expression in the context of overall development by comparing the trajectory of their expression across all larval stages in both life histories. These findings serve as the basis for forming hypotheses about the function of miRNA regulatory mechanisms in life history choices and stress resistance.
RESULTS
The expression of 14 miRNAs is affected by dauer life history
To ascertain the expression of most (107) C. elegans miRNAs during development through both continuous and dauer-associated life histories, we isolated RNA from animals grown at 20°C in either favorable or dauer-promoting environmental conditions. To collect staged larvae developing continuously in favorable environmental conditions, embryos were isolated by hypochlorite treatment of eggs and gravid adults. These embryos were added to plates containing plenty of food and space. Larvae grew at sparse population density until the appropriate stage (L1, L2, L2m, L3, or L4), as determined developmentally (see Supplemental Methods). To collect staged L1 and L2 larvae that would have been likely to enter dauer diapause had they not been harvested, embryos were added to growth medium containing dauer pheromone, a potent environmental cue for eliciting L1d, L2d, and dauer formation (Golden and Riddle 1982; Butcher et al. 2007). Larvae were grown in parallel with or without pheromone, and larvae from each culture were harvested for RNA extraction at equivalent developmental stages. Dauer larvae were isolated by SDS selection from plates that had a high population density and on which animals had exhausted their food supply. For comparison to dauer larvae, animals that had developed continuously in favorable environmental conditions were harvested at the L2 molt and early L3 (see Fig. 1). Similarly, post-dauer L3 (PDL3) and post-dauer L4 (PDL4) larvae were obtained by selecting dauer larvae as above, then adding the dauer larvae to plates with abundant food to support the resumption of development. PDL3 and PDL4 larvae were harvested when they reached a stage of development equivalent to the continuously developing L3 and L4 larvae, respectively. For each stage and life history, three independent biological replicates were obtained in this manner.
There are numerous methods for quantifying miRNA levels including mir-Taqman real-time PCR, Northern blot, deep sequencing, microarrays, and reporter transgenes. All have some benefits and some caveats associated with them (Git et al. 2010). We chose the multiplex miR-Taqman assay to quantify miRNA levels because it is highly quantitative, can discriminate between miRNA family members, can measure levels of multiple miRNAs simultaneously, and measures the endogenous, mature miRNA species. Furthermore, a very small amount of RNA is required, permitting us to grow relatively small numbers of animals in specific environmental conditions. To verify the specificity of this assay, we tested strains lacking eight different miRNAs within the let-7 and miR-35 families. None of the miRNAs that were genetically deleted were detectable in this assay, whereas wild-type miRNAs were readily detectable (Supplemental Table S1).
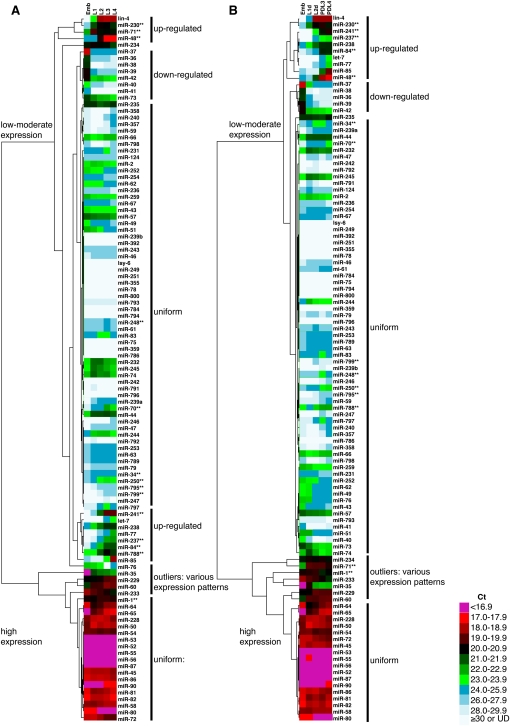
To ascertain which miRNAs differed significantly between the life histories, we obtained the mean Ct values for each stage and life history across all three biological replicates. Mean Ct values were normalized to the 25 microRNAs that were least variant across all stages and conditions. The complete list of these normalized mean Ct values and the standard deviations associated with them are listed in Supplemental Table S2. We observed a range of expression levels for the microRNAs that we assayed, and a range of developmental profiles (Fig. 2).
FIGURE 2.
Hierarchical clustering of miRNA expression profiles during developmental progression. Hierarchical clustering was carried out using Cluster 3.0 and visualized using Java TreeView programs (Materials and Methods) on (A) embryo (Emb), L1, L2, L3, and L4 staged continuously developing larvae or (B) embryo (Emb), L1d, L2d, PDL3, and PDL4 staged larvae progressing through the dauer life history. Note that a 1-Ct difference corresponds to a twofold change in expression level. (**) An miRNA whose expression changes significantly and substantially between life histories (see Fig. 3; Table 1).
The normalized mean Ct values and their standard deviations were used to calculate fold-changes, error, and statistical significance between developmentally equivalent stages of continuous versus dauer-associated development (see Fig. 1). These calculations were carried out by standard methods (see Materials and Methods; AppliedBiosystems 2008). Fold-changes were deemed significant if they met all of the following criteria: (1) The fold-change had a P-value of ≤0.05 as assessed by t-test (Materials and Methods); (2) the fold-change was ≥twofold; and (3) the miRNA was expressed highly enough to be well measured by the assay (Supplemental Methods). The complete list of fold-changes of all 107 miRNAs at each larval stage is provided in Supplemental Table S3. To assess the reproducibility of these results, particularly during dauer diapause when gene expression in general is substantially affected, we performed completely independent experiments, assaying all miRNA levels in samples of staged populations of dauer and L3 larvae obtained independently of those in Supplemental Tables S2 and S3. The results of these experiments were essentially in agreement with those in Supplemental Table S3. There were a few cases of discordant results, in which an miRNA displayed a significant change in dauer larvae in one set of experiments, but not the other set. The data for these are listed in Supplemental Table S4. Only those miRNAs for which the results of both sets of experiments were in agreement are reported in Table 1 and Figure 3.
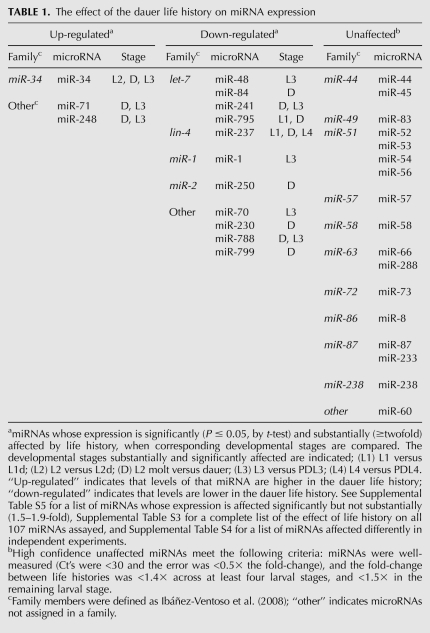
TABLE 1.
The effect of the dauer life history on miRNA expression
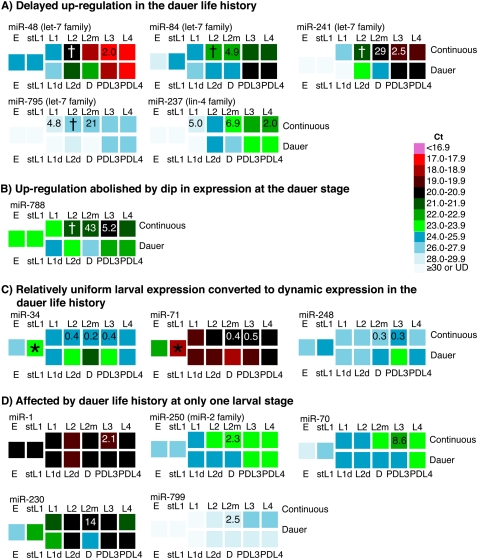
FIGURE 3.
Developmental trajectory of miRNAs whose expression is significantly different between life histories. Four different effects on developmental trajectory were observed (A–D). See text for details. The expression of miRNAs whose expression is substantially and significantly affected at one or more larval stages (Table 1) was represented by the same color-coding scheme as in Figure 2. To directly compare progression through continuous and dauer-interrupted life histories, the corresponding developmental stages are shown in parallel, with the continuous life history on the top and the dauer life history on the bottom. Developmental diapauses are also shown, including L1 diapause (stL1) and dauer diapause (D). Note that embryo and stL1 stages can lead to either continuous or dauer life history. Numbers within squares indicate the fold-change in expression between continuous and dauer life histories (continuous/dauer). Note that in some cases, an approximately twofold change in expression does not cause a change in the color, or conversely that a small change in Ct can cause a color change if the Ct was near the boundary defined. Also note that a 1-Ct difference is a twofold change in miRNA level. See Supplemental Table S3 for a complete list of the fold-change in miRNA expression between different life histories at all larval stages for all 107 miRNAs, including the error, and P-value, Supplemental Table S5 for a list of miRNAs that change significantly but not substantially (1.5–1.9-fold), and Supplemental Table S4 for a list of miRNAs that are affected differently in independent experiments. (*) A statistically significant fold-change is observed when the miRNA level in stL1 is compared to the miRNA level in both embryo and continuously developing L1 stages (see Table 2). (†) The substantial fold-change observed at this stage is not statistically significant in this analysis (Supplemental Table S3). These miRNAs are dramatically up-regulated in L2 staged larvae (Supplemental Table S2; Abbott et al. 2005), causing any small variation in the stage of each sample to lead to a large variation in the amount of miRNA present in different biological replicates. However, when these same samples were compared using methods to assess fold-change for cultures grown side by side, we did find a statistically significant difference at this stage (Hammell et al. 2009; Pradervand et al. 2009).
Using the above criteria, we identified 14 miRNAs that were significantly different in level when corresponding stages of the continuous and dauer life histories are compared (for at least one stage) (Fig. 3; Table 1). In Supplemental Table S5, we also provide a list of 15 miRNAs whose expression changes are less substantial (1.5–1.9-fold), yet still statistically significant by t-test (Materials and Methods). In our analysis, however, we focus on miRNAs whose levels were changed at least twofold, because we have greater confidence in the biological relevance of expression changes of this magnitude.
In contrast to the miRNAs that we found to be significantly affected by life history, we also identified a set of miRNAs that, based on our data, appear to be unaffected by life history (Table 1). These miRNAs are well expressed, as defined by having Ct values <30; their mean fold-change across the three biological replicates was ≤1.4–1.5, and the error was ≤0.5× the fold-change. miRNAs that do not appear in Table 1 are either poorly expressed (Supplemental Table S2), variable between experiments (Supplemental Table S4), or more ambiguous with respect to fold-change between life histories, such that they do not meet the criteria we define above (Supplemental Table S3).
We noticed that miRNAs whose expression changes dramatically during a particular larval stage displayed a high standard deviation at that stage. For example, lin-4 expression is very low in newly hatched L1s but high by the end of L1 (Feinbaum and Ambros 1999). We find a high standard deviation for the lin-4 Ct values of the three biological replicates during the L1 stage, but not other stages when lin-4 levels are more stable (Supplemental Table S2). This is likely due to the fact that each biological replicate captures an imperfectly synchronized population of larvae (see Supplemental Methods).
Five miRNAs are significantly up-regulated during L1 diapause
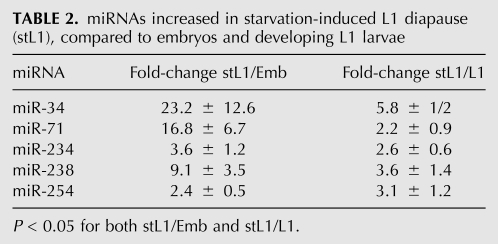
In order to identify miRNAs whose expression is affected by L1 diapause, we compared starved L1 (stL1) larvae (starvation of newly hatched L1 larvae causes them to arrest in L1 diapause) to embryos and to continuously developing L1 larvae. We define miRNAs whose expression responds to L1 diapause to be those whose expression is higher (or lower) in stL1 animals compared to both embryo and L1 stages. From this comparison, we identified five miRNAs that were significantly up-regulated in stL1 larvae (Table 2), and no miRNAs to be significantly down-regulated in stL1 larvae (Supplemental Table S6).
TABLE 2.
miRNAs increased in starvation-induced L1 diapause (stL1), compared to embryos and developing L1 larvae
StL1 larvae share some features with dauer larvae: They are developmentally arrested, quiescent, and nonfeeding. However, unlike dauer larvae, stL1 larvae are active in pharyngeal pumping, do not adopt the dauer-characteristic cuticle morphology, and lack the extreme longevity and stress resistance of dauer larvae (Johnson et al. 1984). The five miRNAs we find to be significantly up-regulated in stL1 larvae show a range of behaviors in the dauer larva life history. miR-238 is unaffected by dauer life history at any stage (Table 1), suggesting that its up-regulation in stL1 larvae is a specific response to L1 diapause. In contrast, miR-34 and miR-71 are also up-regulated in L2d, dauer, and PDL3 stages (Fig. 3; Table 1), suggesting that up-regulation of these miRNAs may be a more general response to environmental stress. miR-234 and miR-254 are both up-regulated in dauer diapause (P ≤ 0.05), but the fold-change is <2, such that we have less confidence in the biological significance of this effect (Supplemental Table S5).
Developmental profiles of miRNA expression
To characterize how the developmental trajectories of miRNA expression are affected by life history, we performed hierarchical clustering on the Ct values of all 107 miRNAs in dauer versus continuous life histories. In order to focus on progression through development, we used only stages that were not developmentally arrested (i.e., L1 diapause and dauer diapause were omitted) (Fig. 2). The clusters we observed were generally similar between life histories (Fig. 2). In each case, hierarchical clustering produced two major groups of miRNAs: those with low to moderate expression and those with high expression. As subclasses within these groups we find (1) developmentally regulated clusters of miRNAs—those whose expression dramatically increases or decreases during progression through development; (2) developmentally uniform clusters of miRNAs; and (3) a small set of outliers whose expression changes in different directions across development (Fig. 2).
The developmental profiles of miRNA expression that we observe for continuous development are generally consistent with previously published developmental profiles. For example, we find lin-4-family and let-7-family miRNAs to be up-regulated during development, whereas the levels of miR-35 family miRNAs are highest in embryos and decline thereafter (Abbott et al. 2005; Esquela-Kerscher et al. 2005; Kato et al. 2009a). To better quantify how our data for miRNA expression correspond to previously published data, we compared our Ct values to the number of sequence reads in a deep-sequencing analysis performed in embryo, L1, L2, L3, and L4 stages (Kato et al. 2009a). At each of these developmental stages, we find that our data correspond well to the deep-sequencing data for ∼75% of the miRNAs (Supplemental Table S7). The 25% of miRNAs with poor agreement between the two data sets could reflect various factors including (1) variability in miRNA expression during a stage when it is being up- or down-regulated, (2) different sequence biases inherent to the mirTaqman and deep sequencing assays (Willenbrock et al. 2009; Git et al. 2010), and (3) differences in growth conditions in the two studies (Ruzanov and Riddle 2010; for more details, see Supplemental Methods and Supplemental Table S8).
Developmental trajectory of miRNAs significantly affected by life history
To ascertain how developmental profiles of miRNA expression may be affected by life history, we conducted a side-by-side comparison of corresponding stages of dauer versus continuous life histories for the 14 miRNAs whose expression is significantly affected by life history at one or more larval stage (Fig. 3). In this comparison, we included expression levels for the L1 diapause and dauer larva stages. We find four patterns of effect on the trajectory of miRNA expression. First, the developmental up-regulation of lin-4 and let-7-family microRNAs is delayed in the dauer life history (Fig. 3A). The dramatic up-regulation of miR-237, miR-48, miR-84, and miR-241 observed during continuous development is consistent with previous reports (Abbott et al. 2005; Esquela-Kerscher et al. 2005). The delay in up-regulation of let-7-family microRNAs that we observe throughout the dauer life history (Fig. 3A) is consistent with the model that unfavorable environmental conditions cause the repressive form of DAF-12 to dampen the intrinsic up-regulation of let-7-family miRNAs as part of a feedback loop that coordinates developmental timing with life history choice (Hammell et al. 2009).
Second, the developmental up-regulation of miR-788 that occurs during continuous development was abolished in the dauer life history (Fig. 3B). Indeed, miR-788 clusters with developmentally up-regulated miRNAs in continuous development, but with developmentally uniform miRNAs in the dauer life history (Fig. 2).
Third, the trajectory of three miRNAs—miR-34, miR-71, and miR-248—is shifted from relatively uniform during continuous larval development to dynamic expression during development through the dauer life history (Fig. 3C). These three miRNAs are all up-regulated in the dauer life history (Table 1). Interestingly, the levels of miR-34, miR-71, and miR-248 are also increased in stL1 larvae relative to embryos and L1 larvae (Fig. 3C), suggesting that these miRNAs are generally activated during periods of environmental stress. This increase is substantial (twofold to 23-fold) for all three miRNAs, but statistically significant for only miR-34 and miR-71 (Table 2; Supplemental Table S6). The substantial change in the developmental trajectory of these miRNAs suggests that their targets might need to be down-regulated during a large portion of progression through the dauer life history.
Finally, for five additional miRNAs, the developmental trajectories are similar in continuous and dauer life histories, with the exception of a single stage at which they are significantly affected in the dauer life history (Fig. 3D).
Life history can affect the transcription of miRNAs
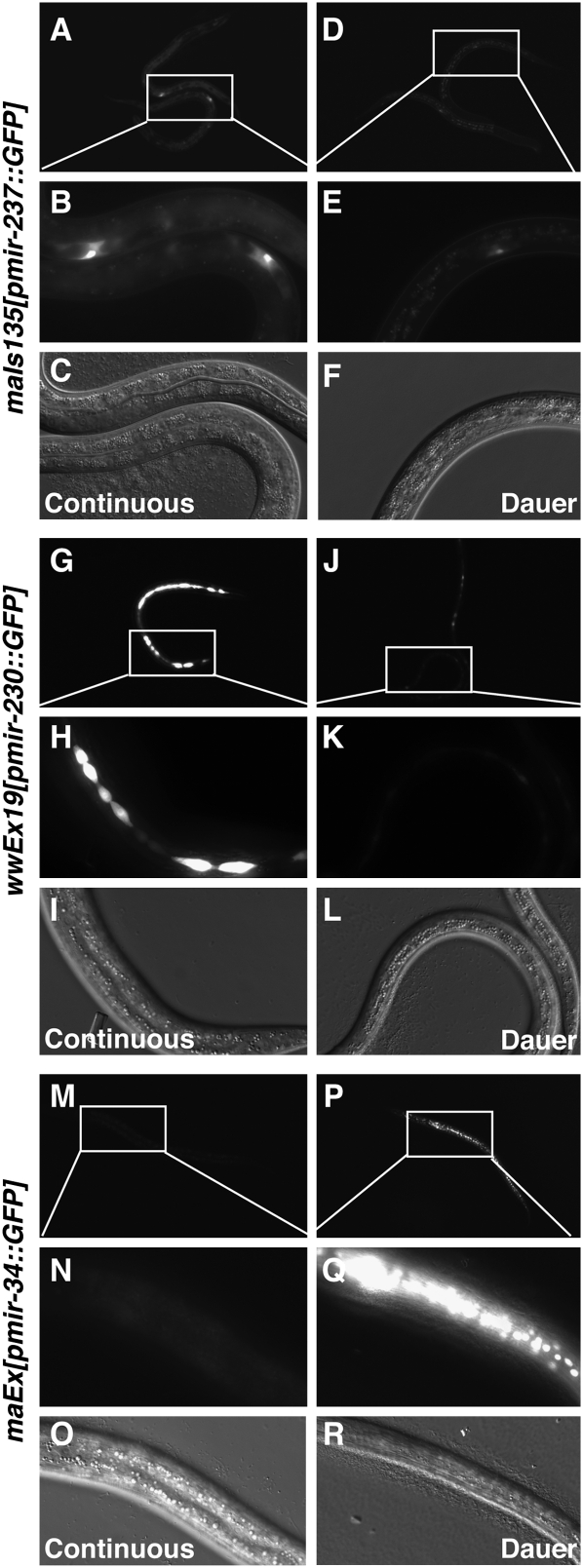
Previously published reports have demonstrated transcriptional down-regulation of several miRNAs during dauer diapause. These include the let-7-family miRNAs that are direct targets of the DAF-12 nuclear hormone receptor, and miR-788, which is a direct target of DAF-3/Smad (Martinez et al. 2008a; Bethke et al. 2009). We therefore wondered whether the effect of life history on the levels of other miRNAs might be mediated transcriptionally. Since dauer diapause is the stage at which life history appears to exert the greatest effect on the levels of mature miRNAs (Fig. 3), we focused on the comparison between dauer diapause and the corresponding stage of continuous development (approximately the L2 molt and early L3 stage). We examined the expression of transcriptional reporters for the three miRNAs whose levels are most affected by dauer diapause, aside from the above-mentioned let-7 family and miR-788 (Fig. 3; Supplemental Table S3). These miRNAs are miR-34 (sixfold up-regulated), miR-237 (sevenfold down-regulated), and miR-230 (14-fold down-regulated). All three of these reporters exhibited changes in expression consistent with our observations of the mature levels (Fig. 4). Specifically, levels of mir-237∷GFP and mir-230∷GFP were reduced in dauer larvae, whereas levels of mir-34∷GFP were increased in dauer larvae (Fig. 4). Therefore, the effect of life history on the levels of these particular miRNAs is mediated in large part transcriptionally.
FIGURE 4.
Transcriptional regulation of miRNA expression in dauer diapause. Expression of transcriptional reporters of miRNA expression during continuous development (L2 molt or early L3 stage, prior to VPC division) or during dauer diapause. (A–F) Expression of a mir-237∷GFP reporter (Martinez et al. 2008b). (A,D) GFP fluorescence observed at a 200-msec exposure time using a 10× objective. (B,E) Magnification of the boxed region in A and D. Fluorescence observed at a 19-msec exposure time using a 40× objective. (C,F) DIC images corresponding to B and E, respectively. (G–L) Expression of a mir-230∷GFP reporter (Martinez et al. 2008b). (G,J) GFP fluorescence observed at a 100-msec exposure time using a 10× objective. (H,K) Magnification of the boxed region in G and J, respectively. Fluorescence observed at a 3-msec exposure time using a 40× objective. (I,L) DIC images corresponding to H and K, respectively. (M–R) Expression of an mir-34∷GFP reporter containing 5 kb of promoter sequence. (M,P) GFP fluorescence observed at a 200-msec exposure time using a 10× objective. (N,Q) Magnification of the boxed region in M and P, respectively. Fluorescence observed at a 50-msec exposure time using a 40× objective. (O,R) DIC images corresponding to N and Q, respectively.
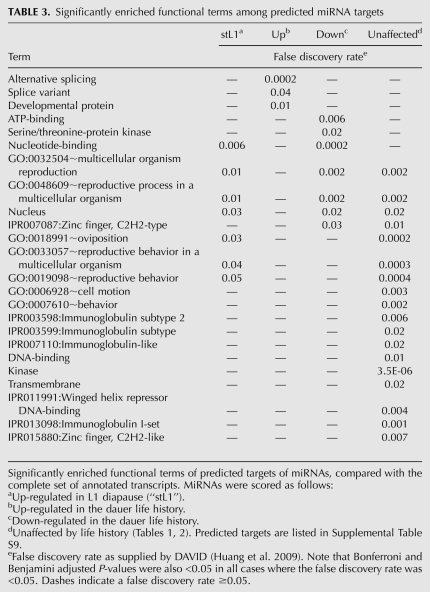
Functional terms enriched among predicted targets of miRNAs whose expression is affected by life history
As genetic studies of miRNA mutants are only beginning to reveal the function of most miRNAs in C. elegans (Miska et al. 2007; Alvarez-Saavedra and Horvitz 2010; Brenner et al. 2010), we estimated the types of biological processes that might be affected by life history as a result of changes in miRNA levels. First, miRNAs were placed into one of four groups: up-regulated during L1 diapause, up-regulated in the dauer life history, down-regulated in the dauer life history, or unaffected by dauer life history (Tables 1, 2). Next, predicted targets of miRNA families within each group were identified using mirWIP (Supplemental Table S9; Hammell et al. 2008). Finally, for each group of predicted targets, significantly enriched functional terms were identified using DAVID (Huang et al. 2009).
We find that predicted targets of miRNAs up-regulated in the dauer life history are enriched for developmental proteins and alternative splicing-related functions (Table 3). Since alternative splicing is an important aspect of the developmental control of gene expression in C. elegans, the predicted down-regulation of these genes is consistent with the interruption of development within the dauer life history (Zahler 2005; Rukov et al. 2007). These terms were not enriched within other groups (Table 3). In contrast, predicted targets of the other three groups show a high degree of overlap of terms with significant enrichment (Table 3). Exceptions include kinase-related functions enriched among predicted targets of miRNAs down-regulated in the dauer life history, and several primarily molecular categories enriched among predicted targets of miRNAs that are unaffected by life history (Table 3). Interestingly, we did not find dauer-related terms to be enriched in any group. We did find three important genes that regulate dauer formation—daf-5, daf-12, and daf-16—to be predicted targets of several miRNAs identified in this study (Supplemental Table S9). However, because duplicate entries are not included in the analysis, in order for a functional term to be enriched there would need to be many different predicted targets that have been assigned that term, rather than a few key genes appearing multiple times.
TABLE 3.
Significantly enriched functional terms among predicted miRNA targets
DISCUSSION
The stress-resistant dauer diapause permits C. elegans larvae to withstand the harsh environmental conditions that occur in their natural environment. The importance of the ability of C. elegans to choose the dauer life history is underscored by the observation that samples of C. elegans isolated from the wild are most often found in the dauer stage (Barriere and Felix 2005). This choice between continuous and dauer-interrupted life histories involves dramatic effects on developmental progression, including a lengthening of pre-dauer stages and the postponement of post-dauer stages (Golden and Riddle 1984). These processes certainly involve widespread and coordinated regulation of gene expression, on the transcriptional and post-transcriptional levels. Changes in gene transcript levels have been documented that accompany the increased stress resistance, reduced metabolism, and lengthened life span characteristic of dauer larvae (Dalley and Golomb 1992; Jones et al. 2001; Holt and Riddle 2003; Wang and Kim 2003; Ruzanov et al. 2007; Jeong et al. 2009b; Ruzanov and Riddle 2010). Similarly, transcriptional changes have been described associated with L1 diapause (Baugh et al. 2009). Less is known about the contribution of post-transcriptional regulation of gene expression in the choice between continuous and interrupted life histories. microRNAs are candidates for acting post-transcriptionally to coordinate programs of differential gene expression, as individual microRNAs have the potential to regulate multiple target genes (Bartel 2004, 2009). We present a quantitative assessment of miRNA expression throughout continuous and diapause-interrupted life histories that provides a valuable resource to researchers studying any aspect of development that involves L1 diapause, dauer diapause, or other aspects of dauer-associated life history.
Biological processes that may be affected by altered miRNA levels in different life histories
We hypothesize that miRNAs may be involved in integrating the developmental and physiological effects of life history choice. Consistent with this hypothesis, we find that miRNAs that are affected by life history are also likely to fall into developmentally regulated clusters in the continuous life history (seven of 14 miRNAs) (Fig. 2A), whereas miRNAs that are unaffected by life history are likely to fall into developmentally uniform clusters (13 of 17 miRNAs) (Fig. 2A). This general trend suggests a relationship between developmental progression and the physiological changes that accompany life history choice, such that the targets of miRNAs that are present at relatively uniform levels throughout development may remain at those same levels in either life history.
A relationship between gene expression changes in dauer larvae and gene expression changes that occur in aging adults has been described (for review, see Golden and Melov 2007). Indeed, many of the same genes that regulate dauer formation also regulate life span (see Golden and Melov 2007; Hu 2007). Two previous studies characterized the expression of miRNAs during aging in wild-type adult C. elegans hermaphrodites (Ibáñez-Ventoso et al. 2006; de Lencastre et al. 2010). While many of the miRNAs whose expression is affected by aging are distinct from those whose expression is affected by life history, it is interesting to note that miR-34 and miR-71, two of the three miRNAs that are up-regulated in the dauer life history (Table 1), are also up-regulated in older adults (Ibáñez-Ventoso et al. 2006; de Lencastre et al. 2010). Intriguingly, miR-71 promotes longevity, as well as resistance to both heat and oxidative stress in adulthood, whereas miR-34 is required for a robust DNA damage response (Kato et al. 2009b; de Lencastre et al. 2010). These roles are consistent with the up-regulation of these miRNAs during L1 diapause and in the dauer life history, in response to environmental stress (Fig. 3).
Transcriptional regulation of miRNA expression
The data presented in Figure 4, combined with previous reports, demonstrate that the effect of dauer diapause on miRNA expression can be mediated transcriptionally (Martinez et al. 2008a; Bethke et al. 2009). The previously characterized transcriptional regulation was assessed by the association of core dauer pathway transcriptional regulators with miRNA promoters. These core components include the DAF-12 nuclear hormone receptor, which is absolutely required for dauer formation, and the DAF-3 Smad, which promotes dauer formation downstream from TGF-β signaling (Thomas et al. 1993; Patterson et al. 1997; Antebi et al. 2000). Yeast one-hybrid assays have been performed for the promoters of most miRNAs (Martinez et al. 2008a). Interestingly, DAF-3 binds to seven of the nine tested promoters of miRNAs whose mature levels are affected by life history. In contrast, DAF-3 binds to only one of the 10 tested promoters of miRNAs whose expression is unaffected by life history (Supplemental Table S10; Martinez et al. 2008a). This raises the possibility that the effect of life history on miRNA expression may be mediated directly by the factors that regulate the life history choice itself, at least in some cases.
Additionally, information about the expression of transcriptional reporters is available for 10 of the promoters of miRNAs whose expression is affected by life history (Esquela-Kerscher et al. 2005; Martinez et al. 2008b; Kato et al. 2009b; Alvarez-Saavedra and Horvitz 2010). These miRNA gene transcriptional reporters showed various expression patterns, including broad temporal and anatomical expression and more specific expression patterns. The three tissues where reporter expression was most often seen were the hypodermis, the vulva, and the nervous system (Supplemental Table S10). The hypodermis is an endocrine tissue where signaling regulating the dauer formation decision occurs (Gerisch and Antebi 2004). Furthermore, the timing of stage-specific cell fate decisions within the hypodermis is regulated by lin-4 and let-7-family miRNAs. This regulation can be altered by dauer formation (Liu and Ambros 1991). The vulva is an ectodermal tissue that is lineally related to some hypodermal cells (Sulston and Horvitz 1977). It has been noted that mutations in genes affecting vulval development can produce altered phenotypes when these animals progress through dauer life history (Ferguson and Horvitz 1985; Euling and Ambros 1996; Braendle and Félix 2008). Finally, certain sensory neurons are the source of signals regulating the dauer formation decision. Furthermore, the morphology of some neurons is altered in dauer diapause (for review, see Riddle and Albert 1997; Hu 2007). Therefore, many of the miRNAs whose levels are affected by life history are apparently transcribed in tissues that are also affected by life history and/or that participate in life history choices.
In conclusion, we have identified a core set of miRNAs whose expression is altered by diapause-associated life history. These microRNA include four let-7-family microRNAs. The let-7 family had been previously implicated in a mechanism that coordinates stage-specific cell fate decisions with life history choice (Hammell et al. 2009). This coordination is necessary for the robustness of cell fate specification to environmental changes and life history that is observed in wild-type C. elegans animals (Liu and Ambros 1991; Euling and Ambros 1996; Braendle and Félix 2008). We have also identified 10 other microRNAs whose expression is altered by life history choice. Future genetic analysis of the functions of these additional miRNAs should reveal whether and how they may also participate in the coordination of developmental and physiological gene expression in the context of diapause-associated life history choices.
MATERIALS AND METHODS
Strains and growth conditions
The wild-type reference strain N2 was grown on the OP50 bacterial strain at 20°C in all cases. To synchronize larvae, we isolated embryos by hypochlorite treatment and then added them directly to either standard NGM plates to prepare continuously developing larvae, or small (2 mL) NGM plates lacking peptone and with the addition of 30 μg/mL streptomycin and 50-μL crude dauer pheromone (Vowels and Thomas 1994) to isolate L1d and L2d larvae. Pheromone-containing plates were seeded with 20 μL of 6× OP50. To prepare dauer larvae and post-dauer stages, a mixed-stage population of animals was grown on standard NGM plates until the food supply was exhausted and the population density was sufficient to induce dauer larvae formation. Dauer larvae were then selected by washing with 1% SDS and either harvested immediately (for dauer larvae) or added to fresh NGM plates seeded with OP50 (for post-dauer stages). All stages were confirmed by microscopic examination of stage-specific anatomical characteristics, with the exception of the dauer stage, which was defined by SDS resistance. For more details, see the Supplemental Methods.
Strains containing transcriptional reporters of miRNAs include VT1113 unc-119(ed3)III; maIs135[pmir-237∷GFP, unc-119+], VL347 unc-119(ed3)III; wwEx19[pmir-230∷GFP, unc-119+] (Martinez et al. 2008b), and VT2514 unc-119(ed3)III; maEx221[pmir-345kb∷GFP, unc-119+]. These strains were grown on 60-mm NGM plates seeded with OP50 at 20°C. Dauer larvae were isolated from crowded and starved plates and examined in parallel with continuously grown larvae at the L2m or early L3 stage. Dauer larvae were picked from starved plates and examined directly (Fig. 4). The presence of dauer alae was verified in these larvae. Images were obtained on a Zeiss Axio Imager D1 with an AxioCam MRm camera, and a X-Cite 120Q light source (EXFO Photonic Solutions, Inc.).
RNA preparation and mir-Taqman assay
Harvested larvae were snap-frozen in liquid nitrogen. RNA was extracted using the Trizol reagent (Invitrogen). Dauer larvae (which have very tough cuticles) were subjected to three or more rounds of freeze-cracking by alternating between liquid nitrogen and a 37°C water bath. Two microliters of a 3 ng/μL RNA dilution was used for multiplex miR-Taqman reactions according to the manufacturer, with an ABI 7900HT Fast-Real Time PCR System (Applied Biosystems).
mir-Taqman analysis
To identify the 25 least variant miRNAs across all conditions, we calculated the variance among the mean Ct's of three biological replicates across all stages and conditions (embryo, stL1, L1, L2, L2m, L3, L4, L1d, L2d, dauer, PDL3, PDL4). The miRNAs identified are listed in Supplemental Table S2. For each larval stage and life history, the mean of the 25 least variant miRNAs was calculated and then used to calculate ΔCt, ΔΔCt, and fold-change values as per the “Relative Quantitation Method” outlined in materials provided by Applied Biosystems (AppliedBiosystems 2008). See the Supplemental Methods for more details. Statistical significance was assessed by performing a t-test (two-tailed, heteroscedastic) on the normalized Ct values of each miRNA at each developmental stage, comparing the three biological replicates obtained for one life history with the three biological replicates obtained for the other life history.
Functional terms
Predicted targets of the miRNA families in Tables 1 and 2 were identified using mirWIP (Hammell et al. 2008). The smallest number of predicted targets of any of these miRNAs was 34. We therefore used the top 34 predicted targets of each of the miRNA families of the indicated group (up-regulated in L1 diapause, up-regulated in the dauer life history, down-regulated in the dauer life history, or unaffected by dauer life history) to compare to the entire C. elegans genome using functional annotation clustering within DAVID (Huang et al. 2009) in order to ask for significantly enriched functional terms.
Hierarchical clustering
To compare normalized expression levels of miRNAs during developmental progression, we used Cluster 3.0 to analyze −ΔCt values using an uncentered correlation and average linkage (de Hoon et al. 2004). The ΔCt is the mean Ct of the three biological replicates, minus the mean Ct of the 25 least variant miRNAs (see the Supplemental Methods). We used the negative of the ΔCt so that higher Ct values would be visualized as lower expression. The mean Ct of the 25 least variant miRNAs was 20.4 when normalized; therefore miRNAs with Ct values close to 20.4 had a −ΔCt of 0. The clustering was visualized with Java Tree View 1.1.3. Because the range of −ΔCt values displayed by this program was limited to −3 to +3 (corresponding to normalized Ct values of 17.4 to 23.4), we added additional colors using Adobe Illustrator to better represent miRNA expression outside this range. For clarity, the key shown in Figures 1 to 3 is in terms of normalized Ct values, instead of −ΔCt values.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article, including a Supplemental Methods section (with additional experimental details) and 10 Supplemental Tables of data, as referred to in the Results and Discussion sections.
ACKNOWLEDGMENTS
We thank Caifu Chen of Applied Biosystems Inc. for development and use of the C. elegans–specific miR-TaqMan assays. We also thank Iva Greenwald for the use of equipment and reagents. We are grateful to James Chen for advice about enrichment of functional terms, and to Maria Sallee for helpful discussions and critical reading of the manuscript. This work was supported by National Institutes of Health Grants GM30428 (to V.A.), F32 GM73307 (to X.K.), F32 GM087039 (to M.H.), and F32 GM070118 (to M.C.O.). Core resources supported by the Diabetes Endocrinology Research Center grant (DK32520) were also used. V.A. is a member of the UMass DERC (DK32520).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2310111.
REFERENCES
- Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V 2005. The let-7 microRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell 9: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Saavedra E, Horvitz HR 2010. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol 20: 367–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi A, Yeh WH, Tait D, Hedgecock EM, Riddle DL 2000. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev 14: 1512–1527 [PMC free article] [PubMed] [Google Scholar]
- AppliedBiosystems 2008. Guide to performing relative quantitation of gene expression using real-time quantitative PCR. http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_042380.pdf
- Barriere A, Felix MA 2005. High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Curr Biol 15: 1176–1184 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2004. MicroRNAs genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Demodena J, Sternberg PW 2009. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science 324: 92–94 [DOI] [PubMed] [Google Scholar]
- Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A 2009. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science 324: 95–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braendle C, Félix MA 2008. Plasticity and errors of a robust developmental system in different environments. Dev Cell 15: 714–724 [DOI] [PubMed] [Google Scholar]
- Brenner JL, Jasiewicz KL, Fahley AF, Kemp BJ, Abbott AL 2010. Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr Biol 20: 1321–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher RA, Fujita M, Schroeder FC, Clardy J 2007. Small-molecule pheromones that control dauer development in Caenorhabditis elegans. Nat Chem Biol 3: 420–422 [DOI] [PubMed] [Google Scholar]
- Cassada RC, Russell RL 1975. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev Biol 46: 326–342 [DOI] [PubMed] [Google Scholar]
- Dalley BK, Golomb M 1992. Gene expression in the Caenorhabditis elegans dauer larva: developmental regulation of Hsp90 and other genes. Dev Biol 151: 80–90 [DOI] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S 2004. Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- de Lencastre A, Pincus Z, Zhou K, Kato M, Lee SS, Slack FJ 2010. MicroRNAs both promote and antagonize longevity in C. elegans. Curr Biol 20: 2159–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Johnson SM, Bai L, Saito K, Partridge J, Reinert KL, Slack FJ 2005. Post-embryonic expression of C. elegans microRNAs belonging to the lin-4 and let-7 families in the hypodermis and the reproductive system. Dev Dyn 234: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euling S, Ambros V 1996. Reversal of cell fate determination in Caenorhabditis elegans vulval development. Development 122: 2507–2515 [DOI] [PubMed] [Google Scholar]
- Feinbaum R, Ambros V 1999. The timing of lin-4 RNA accumulation controls the timing of postembryonic developmental events in Caenorhabditis elegans. Dev Biol 210: 87–95 [DOI] [PubMed] [Google Scholar]
- Ferguson EL, Horvitz HR 1985. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics 110: 17–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N, Antebi A 2008. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev 22: 2149–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch B, Antebi A 2004. Hormonal signals produced by DAF-9/cytochrome P450 regulate C. elegans dauer diapause in response to environmental cues. Development 131: 1765–1776 [DOI] [PubMed] [Google Scholar]
- Git A, Dvinge H, Salmon-Divon M, Osborne M, Kutter C, Hadfield J, Bertone P, Caldas C 2010. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA 16: 991–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden TR, Melov S 2007. Gene expression changes associated with aging in C. elegans. WormBook 2007: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden JW, Riddle DL 1982. A pheromone influences larval development in the nematode Caenorhabditis elegans. Science 218: 578–580 [DOI] [PubMed] [Google Scholar]
- Golden JW, Riddle DL 1984. The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev Biol 102: 368–378 [DOI] [PubMed] [Google Scholar]
- Hall SE, Beverly M, Russ C, Nusbaum C, Sengupta P 2010. A cellular memory of developmental history generates phenotypic diversity in C. elegans. Curr Biol 20: 149–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell CM 2008. The microRNA-argonaute complex: A platform for mRNA modulation. RNA Biol 5: 123–127 [DOI] [PubMed] [Google Scholar]
- Hammell M, Long D, Zhang L, Lee A, Carmack CS, Han M, Ding Y, Ambros V 2008. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat Methods 5: 813–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell CM, Karp X, Ambros V 2009. A feedback circuit involving let-7-family miRNAs and DAF-12 integrates environmental signals and developmental timing in Caenorhabditis elegans. Proc Natl Acad Sci 106: 18668–18673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey SC, Barker GL, Shorto A, Viney ME 2009. Natural variation in gene expression in the early development of dauer larvae of Caenorhabditis elegans. BMC Genomics 10: 325 doi: 10.1186/1471-2164-10-325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V, Bushati N, Cohen SM 2010. Drosophila microRNAs 263a/b confer robustness during development by protecting nascent sense organs from apoptosis. PLoS Biol 8: e1000396 doi: 10.1371/journal.pbio.1000396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SJ, Riddle DL 2003. SAGE surveys C. elegans carbohydrate metabolism: evidence for an anaerobic shift in the long-lived dauer larva. Mech Ageing Dev 124: 779–800 [DOI] [PubMed] [Google Scholar]
- Holtz J, Pasquinelli AE 2009. Uncoupling of lin-14 mRNA and protein repression by nutrient deprivation in Caenorhabditis elegans. RNA 15: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu PJ 2007. Dauer. WormBook 2007: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Ibáñez-Ventoso C, Yang M, Guo S, Robins H, Padgett RW, Driscoll M 2006. Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell 5: 235–246 [DOI] [PubMed] [Google Scholar]
- Ibáñez-Ventoso C, Vora M, Driscoll M 2008. Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PLoS ONE 3: e2818. doi: 10.1371/journal.pone.0002818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong PY, Kwon MS, Joo HJ, Paik YK 2009a. Molecular time-course and the metabolic basis of entry into dauer in Caenorhabditis elegans. PLoS ONE 4: e4162 doi: 10.1371/journal.pone.0004162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong PY, Na K, Jeong MJ, Chitwood D, Shim YH, Paik YK 2009b. Proteomic analysis of Caenorhabditis elegans. Methods Mol Biol 519: 145–169 [DOI] [PubMed] [Google Scholar]
- Johnson TE, Mitchell DH, Kline S, Kemal R, Foy J 1984. Arresting development arrests aging in the nematode Caenorhabditis elegans. Mech Ageing Dev 28: 23–40 [DOI] [PubMed] [Google Scholar]
- Jones SJ, Riddle DL, Pouzyrev AT, Velculescu VE, Hillier L, Eddy SR, Stricklin SL, Baillie DL, Waterston R, Marra MA 2001. Changes in gene expression associated with developmental arrest and longevity in Caenorhabditis elegans. Genome Res 11: 1346–1352 [DOI] [PubMed] [Google Scholar]
- Kato M, Slack FJ 2008. microRNAs: small molecules with big roles—C. elegans to human cancer. Biol Cell 100: 71–81 [DOI] [PubMed] [Google Scholar]
- Kato M, de Lencastre A, Pincus Z, Slack FJ 2009a. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol 10: R54 doi: 10.1186/gb-2009-10-5-r54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Paranjape T, Müller RU, Ullrich R, Nallur S, Gillespie E, Keane K, Esquela-Kerscher A, Weidhaas JB, Slack FJ 2009b. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene 28: 2419–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW 2009. A microRNA imparts robustness against environmental fluctuation during development. Cell 137: 273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP 2003. The microRNAs of Caenorhabditis elegans. Genes Dev 17: 991–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Ambros V 1991. Alternative temporal control systems for hypodermal cell differentiation in Caenorhabditis elegans. Nature 350: 162–165 [DOI] [PubMed] [Google Scholar]
- Liu T, Zimmerman KK, Patterson GI 2004. Regulation of signaling genes by TGFβ during entry into dauer diapause in C. elegans. BMC Dev Biol 4: 11 doi: 10.1186/1471-213X-4-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, Roth FP, Ambros VR, Walhout AJ 2008a. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev 22: 2535–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Reece-Hoyes JS, Barrasa MI, Ambros VR, Walhout AJ 2008b. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res 18: 2005–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR 2007. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet 3: e215 doi: 10.1371/journal.pgen.0030215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GI, Koweek A, Wong A, Liu Y, Ruvkun G 1997. The DAF-3 Smad protein antagonizes TGF-β-related receptor signaling in the Caenorhabditis elegans dauer pathway. Genes Dev 11: 2679–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradervand S, Weber J, Thomas J, Bueno M, Wirapati P, Lefort K, Dotto GP, Harshman K 2009. Impact of normalization on miRNA microarray expression profiling. RNA 15: 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen KD, Simmini S, Abreu-Goodger C, Bartonicek N, Di Giacomo M, Bilbao-Cortes D, Horos R, Von Lindern M, Enright AJ, O'Carroll D 2010. The miR-144/451 locus is required for erythroid homeostasis. J Exp Med 207: 1351–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle DL, Albert PS 1997. Genetic and environmental regulation of dauer larva development. In C. elegans II (ed. Riddle DL et al. ), pp. 739–768 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- Rukov JL, Irimia M, Mork S, Lund VK, Vinther J, Arctander P 2007. High qualitative and quantitative conservation of alternative splicing in Caenorhabditis elegans and Caenorhabditis briggsae. Mol Biol Evol 24: 909–917 [DOI] [PubMed] [Google Scholar]
- Ruzanov P, Riddle DL 2010. Deep SAGE analysis of the Caenorhabditis elegans transcriptome. Nucleic Acids Res 38: 3252–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzanov P, Riddle DL, Marra MA, McKay SJ, Jones SM 2007. Genes that may modulate longevity in C. elegans in both dauer larvae and long-lived daf-2 adults. Exp Gerontol 42: 825–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56: 110–156 [DOI] [PubMed] [Google Scholar]
- Thomas JH, Birnby DA, Vowels JJ 1993. Evidence for parallel processing of sensory information controlling dauer formation in Caenorhabditis elegans. Genetics 134: 1105–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels JJ, Thomas JH 1994. Multiple chemosensory defects in daf-11 and daf-21 mutants of Caenorhabditis elegans. Genetics 138: 303–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Kim SK 2003. Global analysis of dauer gene expression in Caenorhabditis elegans. Development 130: 1621–1634 [DOI] [PubMed] [Google Scholar]
- Willenbrock H, Salomon J, Søkilde R, Barken KB, Hansen TN, Nielsen FC, Møller S, Litman T 2009. Quantitative miRNA expression analysis: Comparing microarrays with next-generation sequencing. RNA 15: 2028–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM 2005. Alternative splicing in C. elegans. WormBook 2005: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]