Abstract
Although generally regarded as functional in the cytoplasm, a number of microRNAs (miRNAs) have been found in the nucleus, possibly with a role in gene regulation. Here we report that, in fact, a substantial fraction of all human miRNAs are present in the nucleus of neural stem cells. Further, subsets of these miRNAs display consistently higher standardized rank in the nucleus than in the cytoplasm of these cells, as identified with an RT-qPCR technology and confirmed by microarray analysis. Likewise, other miRNAs display higher cytoplasmic standardized ranks. Three samples were partitioned into nuclear and cytoplasmic fractions in six assays for 373 miRNAs. From the 100 most highly expressed miRNAs, standard scores of nuclear and cytoplasmic concentrations were determined. Among those, 21 miRNAs had all three nuclear standard scores higher than all three cytoplasmic scores; likewise, 31 miRNAs had consistently higher cytoplasmic scores. Random concentrations would result in only five in each set. Remarkably, if one miRNA has a high standard score in a compartment, then other miRNAs having the same 5′ seeds and certain similar 3′ end patterns are also highly scored in the same way. That is, in addition to the seed sequence, 3′ sequence similarity criteria identify families of mature miRNAs with consistently high nuclear or cytoplasmic expression.
Keywords: miRNA, nucleus, nuclear localization, nuclear import
INTRODUCTION
Human miRNAs are a large group (>1000 members) of 16–27-nt, single-stranded RNA (ssRNA) molecules that are responsible principally for regulating mRNA translation and stability (Bartel 2004; Griffiths-Jones et al. 2008; Carthew and Sontheimer 2009). As presently understood, the biogenesis of mature miRNAs originates from large primary transcripts (pri-miRNAs). From the pri-miRNAs, the RNA binding protein Dgcr8 and the RNase III enzyme Rnasen recognize and excise precursor stem–loop structures with 44–180 nt creating pre-miRNAs that are then exported to the cytoplasm. Pre-miRNAs are further processed into the double-stranded (dsRNA) form of mature miRNAs by a second RNase III enzyme, Dicer. Cytoplasmic processing is followed by (or coupled to) assembly of one strand of the miRNA duplex into a number of multiprotein complexes, all of which contain a member of the miRNA-binding Argonaute protein superfamily. The assembled complexes bind via base pairing between the targeted mRNA (typically a subsequence of the 3′ UTR) and nucleotides from the 5′ end of the miRNA (often nucleotides 2–8, known as the seed sequence [Lewis et al. 2005; Grimson et al. 2007]). The Argonaute-containing complexes inhibit translation or stimulate degradation of mRNAs to varying degrees, so they are referred to as RNA-induced silencing complexes (RISCs). A typical mRNA may have multiple, putative targets for both the same and different miRNAs, and any particular miRNA may have as many as hundreds of putative mRNA targets. Hence, given the substantial number of miRNAs, there presumably exists a staggering combinatorial network of miRNA-mediated post-transcriptional gene regulation (Selbach et al. 2008). On top of the complexities of this network are questions about control of controllers, that is, control of biogenesis and processing of miRNAs, an area of much ongoing research (Slezak-Prochazka et al. 2010).
While miRNAs generally function in the cytoplasm, mature miRNAs have been found in the nucleus, and, in some cases, shown to alter promoter activity (Hwang et al. 2007a; Kim et al. 2008; Marcon et al. 2008; Ohrt et al. 2008; Place et al. 2008; Foldes-Papp et al. 2009). Other miRNAs have been localized to the nucleolus (Politz et al. 2009; Liao et al. 2010) and, notably, the nuclear and nucleolar abundances of some miRNAs have been reported to be greater than their cytoplasmic levels. As yet there is no complete molecular understanding for how miRNA subcellular localization is determined. However, siRNAs and piRNAs, molecular cousins to miRNAs that are produced by somewhat different processes, also function in the nucleus, e.g., in directing heterochromatin formation in plants, animals, and fungi, and specifying programmed DNA elimination in Tetrahymena species (Malone and Hannon 2009; Moazed 2009). Furthermore, exogenously supplied siRNAs can modify promoter activity in animal cells (Janowski et al. 2006; Li et al. 2006). Argonaute proteins are also found in the nucleus, and very recent evidence indicates that these proteins function therein in association with siRNAs. In an early report, Robb and colleagues (Robb et al. 2005) showed successful RNA interference (knockdown of the noncoding 7SK RNA) in the nucleus of human cells, thereby exciting interest in synthetic, nuclear control of gene expression. The means by which Argonaute proteins are imported into the nucleus is not known but, in some cases, their nuclear localization has been shown to require binding to siRNAs (Guang et al. 2008; Aravin et al. 2009). Thus, while Argonaute-dependent nuclear import of miRNAs has not been demonstrated directly, by analogy to siRNA localization, such a mechanism seems likely. In plants, however, miRNA biogenesis occurs completely within the nucleus, with the two sequential RNase III cleavages apparently carried out by a single Dicer-related enzyme (Voinnet 2009). This suggests the possibility that some mature miRNAs in animal cells may be generated in the nucleus, especially since a nuclear presence for Dicer has not been rigorously excluded (Emmerth et al. 2010).
In addition to questions as to how miRNAs may become located in nuclear compartments is the issue of specificity. In particular, why do some miRNAs show marked propensity to reside in the nucleus, especially since Argonaute proteins bind miRNAs and siRNAs independently of sequence. In this regard, Hwang and colleagues (Hwang et al. 2007a) first reported that a 6-nt sequence AGUGUU at the 3′ end of human miR-29b is responsible for it being preferentially localized in the nucleus (71%) compared to nuclear localization of miR-29a (42%), which lacks the sequence but is otherwise almost identical. In their supplement, 10 siRNAs similar to miR-29b were quantified as ∼30%–70% in the nucleus. Appending AGUGUU to the 3′ ends of other small ssRNAs was sufficient to cause their nuclear accumulation, and seven additional sequence motifs promoting the same have been described in a patent by the same researchers (Hwang et al. 2007b).
Most of the studies in which mature miRNAs were observed in animal cell nuclei have employed tumor cells that have undergone countless passages. Thus, in an effort to assess the generality of miRNA nuclear localization, the possible role of sequence dependence, and the influence of the tumor state on localization, we have analyzed miRNA distribution in cultured proliferating human neural progenitor cells in passages 4–6 after derivation from human embryonic stem cells (hESCs). We found that 188 of the 373 assayed mature miRNAs were detected in all experiments in both the nucleus and cytoplasm, a finding consistent with a recent report by Liao and colleagues (Liao et al. 2010). However, among the 100 most strongly detected miRNAs, a particular set of 21 miRNAs were found to have a consistently high concentration rank in the nucleus; similarly, another set of 31 miRNAs had a consistently high concentration rank in the cytoplasm (the number in each set expected to occur by chance is five). Further, compartmental preference appears to be sequence-specific. Thus, if one miRNA is preferentially localized, then the other miRNAs having the same 5′ seed sequence and certain similar 3′ end patterns are likewise preferentially localized to the same compartment. Finally, we have developed a particular mathematical method that yields sequence families sharing consistent miRNA localizations.
MATERIALS AND METHODS
Cell culture
Our interest in the potential role of miRNAs in schizophrenia (Perkins et al. 2007) prompted us to select human neural stem cell lines for in vitro experiments. Research into the etiology of schizophrenia has included neurodevelopmental factors (Lewis and Levitt 2002). Furthermore, many active research programs aim to induce pluripotency in differentiated cells, offering the possibility of deriving neuronal and glial cell cultures from, for example, somatic cells of schizophrenia patients. Success could lead to evaluations of diagnostic strategies, the cellular effects of pharmaceuticals, and indeed fundamental research into the basis of mental illnesses as complex diseases. These considerations motivated our choice of cell line.
Cells used in our experiments were neural stem cells derived from human embryonic WA09 cell lines (Aruna Biomedical) as described by Dhara and colleagues (Dhara et al. 2009). According to Dhara and Stice (2008) the goals of the provider are optimal differentiation of human embryonic stem cells (hESCs) to NP cells and their maintenance as a self-renewing population, available for further generation of pure populations of specified cell types in culture. Thus, ArunA has enabled differentiation of hESCs into NP cells expressing SOX2 and other neuroepithelial markers (NES, SOX1, SOX3, PSA-NCAM, and MUSASHI-1). Formation of “neural rosettes” was also reported by ArunA. Additional publications describe how the NP cells were derived from hESCs (Shin et al. 2006, Dhara et al. 2008). The provider certified the cells in the lot we used to have normal karyotype and consistent DNA fingerprints.
Cells were cultured according to the manufacturer's protocol, except cells were consistently cultured in an atmosphere of low oxygen (5% O2, 5% CO2, ∼90% N2) (Panchision 2009). Substrates were poly-L ornithine-coated and laminin-coated polystyrene plates in neurobasal medium supplemented with B27 and FGF2 (20 ng/μL). Cells were passaged at 90% confluence and harvested in linear growth. Assays were done with cells from passages 4–6.
Since miRNAs are key regulators of stem cell differentiation, precisely measured miRNA profiles can be expected to change as neural stem cells differentiate along neuronal or glial lines (Shi et al. 2010). Assurance that the cells used in this study remained a relatively homogenous population of neural stem cells across passages is thus given by analyses of miRNA profiles at each passage (for details, see Supplement 1).
As fully described in Supplement 1, we performed a nonparametric statistical test that indicated that passage number is not correlated with results. That is, the two experiments (one nuclear, one cytoplasmic) associated with any one batch yielded measurements that were not statistically distinguished from the other two batches. In dramatic contrast, the data of our six experiments were strongly distinguished by comparing nuclear and cytoplasmic fractions. These nonparametric tests imply that neither differentiation nor any other confounding factors generated any favorable or unfavorable bias for any of the three batches of cells in the sense of altered miRNA profiles.
Focusing further on differentiation, we note that Letzen et al. (2010) reported extensive miRNA profiles for eight stages of differentiation from human embryonic stem cells to terminally differentiated oligodendrocytes. Fortuitously, several miRNAs were assayed in both the Letzen work and in our work. Selected miRNAs show in Letzen data very dynamic concentration profiles stage-to-stage, but the same miRNAs in our measurements showed quantitatively much smaller changes batch-to-batch. Furthermore, our batch-to-batch changes were qualitatively inconsistent with stage-to-stage patterns of change. Thus, our batch-to-batch miRNA changes are both quantitatively and qualitatively inconsistent with stage-to-stage changes reported by Letzen et al. Full details are in Supplement 1.
Cell fractionation and RNA isolation
Cytoplasm and nuclear fractions were isolated using a Norgen Biotek (Thorold) kit following the manufacturer's instructions. Cells in a confluent monolayer were washed twice with PBS, and then ice-cold lysing solution was added. The cells were gently agitated on ice for 5 min. The lysate was then centrifuged for 10 min at 500 g. The cytoplasmic fraction was selected from the upper 250 μL and the nuclear fraction from the lower 45 μL, discarding an intermediate layer of ∼200 μL with the intention of controlling cross-contamination. This selection reflected our goal of obtaining fractions with minimal cross-contamination at the sacrifice of some quantities of the fractions. RNA was then isolated from each of the fractions using the supplied buffers and columns. RNA was vacuum desiccated and then resuspended in RNase free water to a concentration of ∼100 ng/μL. Quality and quantity of total RNA were determined using a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific). Using Agilent 2100 Bioanalyzer DNA and RNA chips, we verified that DNA concentration in the nuclear fraction was >100X that of the cytoplasmic fraction and that 28S ribosomal RNA peak fluorescence in the cytoplasmic fraction was >10X that of the nuclear fraction (Supplement 2). These two ratios for the same samples imply good fractionation.
miRNA RT-qPCR
We used ∼1000 ng total RNA for reverse transcriptions enabled by Megaplex RT Primers from Applied Biosystems, followed by real-time amplification using TaqMan Low Density Array (TaqMan) hydrolysis probes in v2.0 Pool A 384-well plates, according to the manufacturer's instructions. Megaplex RT Primers are highly multiplexed RT primers that convert miRNAs in a pool of 377 types (plus controls) to cDNA, after which the cDNA is amplified by a 40-cycle reaction. The TaqMan MicroRNA Arrays v2.0 and Megaplex Primer Pools enable amplification by a second 40-cycle reaction in an Applied Biosystems 7900HT qPCR instrument recording position-encoded dye concentration measurements. The output expression profile was keyed with entries in Sanger miRBase, updated to v14 (Griffiths-Jones et al. 2008). Supplement 3 correlates the TaqMan list to Sanger miRBase 14; four miRNA sequences have been recently delisted from previous Sanger releases, leaving 373 distinct probes for recognized miRNAs in TaqMan Pool A plates. (None of the delisted miRNA sequences previously known as miR-672, -674, -871, -872 was detected in any experiment.) Amplification and analysis were performed using SDS 2.2 software resulting in data displayed in Supplement 4. The SDS setting used was always “Automatic Baseline,” per the manufacturer's recommendation; however, the “Default” setting yielded similar final conclusions (data not shown). A complete account of our background program to validate the accuracy and reproducibility of TaqMan technology is in Supplement 5.
Data normalization with standard scores
A standard score is the dimensionless quantity defined by subtracting the mean from the raw score, then dividing by the standard deviation. If all raw scores were multiplied by a positive constant, the resulting standard scores would be unchanged. Depending upon cell type and state, the ratio of cytoplasm mass to nucleoplasm mass might vary substantially, so comparison of measurements of one set of miRNA levels across cell compartments, types, and states might best be performed with standard scores. With these facts in mind, we decided to rank miRNAs among multiple samples of nucleoplasm and cytoplasm with standard scores.
Three samples from confluent cell cultures were first partitioned into nuclear and cytoplasmic fractions, and then assayed for miRNA Cq values. Each of six experimental sets of Cq values was standardized by subtracting the mean of the 100 strongest miRNAs within the experiment and then dividing by the standard deviation. This standard score has the advantage of not requiring precise knowledge of the proportion of miRNA molecules successfully extracted from the nucleus or cytoplasm; that is, both mean and standard deviation are simple multiples of that proportion, so in the quotient, the standardized score is the same as would be that of 100% of the molecules. As will be shown in the following, this approach allows reproducible detection of small but consistent differences in miRNA populations. Of course care must be taken to control cross-contamination.
An miRNA was defined to have dominance in the nuclear fraction provided all three nuclear standard scores (derived from raw Cq values among 100 miRNAs) were greater than all three cytoplasmic standard scores; cytoplasmic dominance was defined analogously. Among the 100 most highly expressed miRNAs, 21 miRNAs displayed consistently higher standard scores in the nucleus than in the cytoplasm, and 31 miRNAs displayed consistently higher cytoplasmic standard scores. Both frequencies were much greater than expected by chance. In 100 simulations of 100 random assignments of six ranked labels into 20 such bins (three high and three low; see Supplement 6), the very greatest of all 10,000 bin occupancies was 13, attained five times. It can be shown that the probability of finding 21 or more nuclear dominant assignments from 100 sets of six random rankings is 2.08 × 10−8 and that for finding 31 or more cytoplasmic dominant assignments is 1.01 × 10−16 (Supplement 6). Hence, nonparametric analysis implies the above patterns of nuclear and cytoplasmic dominance cannot be credibly explained as chance events.
In each of the six tests, Cq values were recorded as well as the plate's average of its four RNU6 values. Undetected miRNAs had Cq = 40. Cq values of miRNA species in nuclear and cytoplasmic fractions were not directly comparable because the initial cellular volumes of the two fractions were not precisely known. However, in all six experiments (designated Nuci for nuclear fraction i and Cytj for cytoplasmic fraction j) the mean, raw Cq values were nearly the same (range = 32.13–34.30 for [in increasing order] Nuc2, Cyt2, Cyt1, Cyt3, Nuc1, Nuc3). We averaged the raw expressed values for each miRNA and chose the 100 miRNAs most strongly expressed (lowest Cq averages) for further analysis. The Cq values for the selected 100 miRNAs were all moderately or strongly detected with raw Cq values <28.
We then derived 100 miRNA standard scores for each of the six experiments. By definition, standard scores for one experiment have mean = 0 and standard deviation (SD) = 1. Standard scores across all six plates had a global range from −2.81 (mean of six lowest = −2.67, SD = 0.09) to +1.96 (mean = 1.80, SD = 0.15). Thus, all six plates had consistent distributions. The Grubb test (Barnett and Lewis 1994) was used to check for outliers, and none was detected.
The standard scores of 14 of the 100 strongest miRNAs had t-test P-values < 0.05, but only one survived correction for multiple testing (P < 0.05/100). Therefore quantitative evaluation of individual miRNAs could not be informative. Nonparametric methods were developed.
Rankings of concentrations of miRNAs in nucleus or in cytoplasm
Three samples from confluent cell cultures were first partitioned into nuclear and cytoplasmic fractions, and then assayed for miRNA Cq values. Using global, raw Cq values, the 100 strongest miRNAs were selected. Each of six experimental sets of Cq values was normalized by subtracting the mean within the experiment and then dividing by the standard deviation, yielding standard scores. A miRNA was defined to be preferentially ranked in the nuclear fraction provided all three nuclear standard scores were stronger than all three cytoplasmic standard scores; preferential cytoplasmic ranking was defined analogously. Among the 100 most highly expressed miRNAs, 21 miRNAs displayed consistently stronger (lower) standard scores in the nucleus, and 31 miRNAs displayed consistently stronger standard scores in the cytoplasm; both frequencies were much greater bin occupancies than expected by chance, as noted previously.
Of the 373 miRNAs, we detected 188 (Cq < 40) in all six experiments; 162 were strongly or moderately detected (Cq < 35) in all six experiments; and 260 were detected both in at least one nuclear sample and in at least one cytoplasmic sample (Supplement 4).
Regarding sensitivity and specificity of our experiments, we note results for six members of the let-7 family tested in the plate and designated let-7a, -b, -c, -d, -e, -g. As mature miRNA sequences, these are similar in that, mature let-7a, -b, -c sequences differ by only one or two bases, and the maximum number of different bases across all 15 pairs is four. None of these six let-7 miRNAs was strongly expressed in any of our samples, but all were detected in all samples with exactly one exception, let-7a in Nuc3. We found that the six-dimensional let-7 vectors had high Pearson correlations over the 15 possible pairs from the six samples; Pearson correlations ranged from 0.84 to 0.99 with mean = 0.93 and SD = 0.06 (Supplement 5 contains complete data). If each of the six measurements were random in the sense of some arbitrary base measurement plus some uniformly random value in an arbitrary range, then the Pearson correlations would be expected to populate the interval [−1, 1] with expected average zero. The observed Pearson correlations imply the let-7 probes are specific and, considering their consistent patterns of detection of weak expression, sensitive. These observations are in concordance with extensive tests of TaqMan hydrolysis probes reported by Chen and colleagues (Chen et al. 2005).
Verification of PCR results with a microarray
Cells from one additional batch were partitioned as in the above-mentioned experiments into nuclear and cytoplasmic fractions, and each fraction was subdivided into three technical replicates. All replicates were adjusted to a concentration of ∼28–30 ng/μL of small RNAs. A sample of 5 μL of each replicate was applied to six Agilent miRNA microarrays. The Agilent microarrays provided probes for 15 of the miRNAs in the TaqMan nuclear list (of 21); there were probes for 21 miRNAs in the TaqMan cytoplasmic list (of 31). Hence, there were 36 common miRNA probes in the TaqMan distinguished lists and in the inventory tested by the Agilent microarray.
Quality control parameters were satisfactory in all six Agilent microarrays (data not shown). Our statistical analysis used the log2 value of the “gTotalGeneSignal” from the Agilent text output (Supplement 7), called herein “log transformed values.” All 36 of the miRNAs assayed by the Agilent product and also distinguished in TaqMan experiments were detected in all six microarrays. The values for the 36 probes over all six microarrays ranged from 2.06 to 12.64 (Supplement 7).
Only one of the log transformed values of the 36 common miRNAs had a t-test P-value < 0.05, but none survived correction for multiple testing (P < 0.05/36). Therefore, quantitative evaluation of miRNAs individually could not be informative. Nonparametric methods were developed.
The global median of log transformed values for the 36 probes in nuclear samples was 0.36 less than the global median for the cytoplasmic samples. After adding that difference to nuclear sample median values, for each of the 36 miRNAs, a revised nuclear median was compared to a cytoplasmic median. We found that of the 15 TaqMan nuclear list probes represented in Agilent, 11 had higher nuclear medians. We found that of the 21 TaqMan cytoplasmic probes represented in Agilent, 14 had higher cytoplasmic medians. The probability of seeing the above nuclear result or better (11 or more of the 15 possible) by chance is 0.06. The probability of seeing the cytoplasmic result or better (14 or more of the 21 possible) by chance is 0.09. Thus, the chance probability of seeing both at least 11 of the TaqMan nuclear probes and at least 14 of the TaqMan cytoplasmic probes consistently distinguished by Agilent medians of log transformed values is 0.0056.
In all of the aforementioned Agilent assay analyses, the median can be replaced by the average. The analogous calculation shows that of the 15 TaqMan nuclear list probes represented in Agilent, nine had higher nuclear medians. Likewise, of the 21 TaqMan cytoplasmic probes represented in Agilent, 15 had higher cytoplasmic medians. The probability of such a chance alignment or better of TaqMan and Agilent results is 0.0119.
We conclude that, though microarray results are not as dramatic as the TaqMan results, microarray results do confirm that our TaqMan nuclear and cytoplasmic lists of dominant miRNAs were unlikely to have risen by chance.
Cytoplasm ClustalW2 (Larkin et al. 2007) employs a dynamic programming method that first scores alignments of small subsets of sequences, then merges larger and larger subsets with superior local scores to declare an overall alignment. ClustalW2 has numerous parameters that can be tuned, as befits a heuristic solution to a complex problem that can seldom be precisely defined by the user.
ClustalW2 yields a set of alignments that is a deterministic consequence of sequence input and parameter selection, identifying common bases in certain positions in aligned sequences. Importantly, there is further information in the ClustalW2 output, namely, in the order of sequences in the output. That is, if sequences A, B, and C appear in the order A, then B, then C, then A is at least as similar in the ClustalW2 sense to B as to C and possibly more so. This additional information is exploited in our miRNA Sequence Family Alignments.
There are numerous mathematical definitions of sequence families. Nonetheless we propose yet another algorithm because it is especially appropriate for emphasis of miRNA sequence similarity both in seed region and in the terminal 3′ bases. Of course other reasonable tests of exact or nearly exact seed identity and at least moderate 3′ similarity could be devised to organize the miRNA sequences in the context of the logic explained below (see Box 1). Indeed, enhancements that allow more complex tests might better define miRNA sequence families; the version below is merely a straightforward example.
BOX 1.
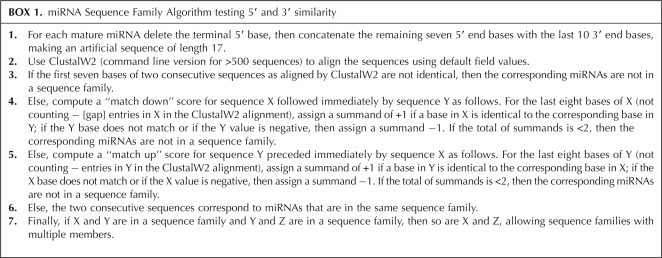
miRNA Sequence Family Algorithm testing 5′ and 3′ similarity
We have found the following algorithm to predict many cases of sequence families that are consistently in the nuclear or cytoplasmic concentration columns as shown in Table 1.
TABLE 1.
The miRNAs in human neural progenitor cells that are distinguished by nuclear or cytoplasmic concentration as in Figure 1
In other words, two miRNA sequences X and Y are in one sequence family if they have identical seed regions and if in the ClustalW2 alignment of the artificial 17-base sequences formed from them, they appear consecutively with very similar 3′ ends. Specifically, comparing the last consecutive eight bases of X down to Y and the last eight consecutive bases of Y up to X, they must exhibit at least five identical base matches.
Given our definition of sequence family, for the 373 distinct mature miRNA sequences in the intersection of Applied Biosystems TaqMan Pool A and Sanger release 16 of September 2010, there are 100 miRNAs in 43 sequence families of sizes 2–8. Sequence families include the familiar examples of the family miR-17-93 = {miR-17, -20a, -20b, -93, -106a, -106b} and a family of seven let-7 miRNAs together with miR-98.
RESULTS
miRNAs in neural stem cells
We chose to determine the subcellular distribution of miRNAs in human embryonic neural progenitor stem cells. The cells are commercially available with rigorous and well-described quality control procedures (Dhara et al. 2009). Cells were harvested while in linear growth, and lysates were quickly partitioned into nuclear and cytoplasmic fractions with a Norgen Biotek system. Good fractionation was confirmed by use of DNA chips and RNA chips in an Agilent 2100 Bioanalyzer (Supplement 2). Total RNA from both fractions was analyzed by an Applied Biosystems RT-qPCR assay configured to yield quantification cycle measurements (Cq values) for a set of 373 mature miRNAs. In total, the analysis was carried out using cells grown in three separate batches, that is, six assays of 373 mature miRNAs.
No biochemical protocol is likely to result in perfect nuclear/cytoplasmic fractionation, but perfect or even nearly perfect subcellular separation is not required in our nonparametric, bin occupancy analysis. That is, nonparametric analysis of our data leads to strong conclusions about relative rankings of mature miRNAs that would only be weakened by cross-contamination. The strongest 100 miRNA profiles were selected from the six experiments. Over each experiment, those Cq values were normalized by conversion to standard scores, thus having mean = 0 and SD = 1. This reduces the risk of bias from generally stronger signals in one experiment versus another.
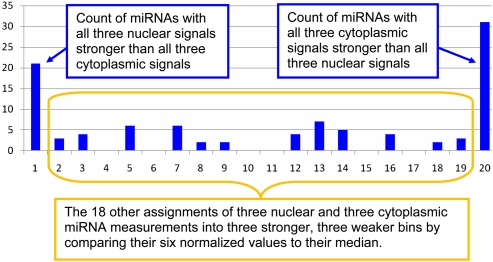
An miRNA was defined to have nuclear dominance if all its three nuclear standard scores were higher than all three cytoplasmic standard scores; in an analogous manner we defined instances cytoplasmic dominance. This is, in effect, a comparison of ranks of levels of an miRNA in nucleus versus ranks of the same miRNA levels in cytoplasm. There are 6!/(3!×3!) = 20 ways to assign three nuclear measurements and three cytoplasmic measurements into two bins of three below and above the median. Of the 20 possible assignments, one has all three nuclear standard scores less than (stronger than) all three cytoplasmic standard scores, and another has all three cytoplasmic standard scores less than all three nuclear standard scores. We refer to these two assignments as dominance of an miRNA in nucleus or cytoplasm. The null hypothesis would be consistent with expecting five of the 100 miRNAs in each of 20 bins including the two bins of nuclear or cytoplasmic dominance.
As shown in Figure 1, the assignments into bins from our data are quite different from this random expectation. The miRNAs with nuclear or cytoplasmic dominance are shown in Table 1 (complete data in Supplement 4). It is important to note that we expect five of the entries of each type to be present by chance. Allowing such chance membership is how nonparametric analysis of the collective informativeness of a set of miRNAs can be meaningful. By contrast, t-test P-values from the same miRNA data do not imply significance of any individual miRNA.
FIGURE 1.
A pattern of differential miRNA concentration from an assignment of our 100 most strongly expressed miRNAs into 20 bins. Each bin corresponds to an ordering by standard scores of three nuclear samples and three cytoplasmic samples, relative to measurements of all 100 miRNAs in each experiment. The far left bin has a count of 21 miRNAs with all three nuclear rankings stronger than all three cytoplasmic rankings. The far right bin has a count of 31 miRNAs with all three cytoplasmic rankings stronger than all three nuclear rankings. If miRNAs were not biased to preferential concentration in nuclear or cytoplasmic fractions, then the expected occupancy of each of the 20 bins would be five. Cross-contamination of sample types would generally weaken distinguishing bin allocations; the extraordinary allocations here are in spite of—not caused by—any imperfections of nuclear/cytoplasmic partitioning. It can be shown that the probability of finding 21 or more preferential assignments from 100 sets of six random rankings is 2.08×10−8 and that for finding 31 or more is 1.01×10−16 (Supplement 6).
Sequence relationships among top ranking miRNAs
Having identified particular miRNAs with compartmental dominance of standard scores, we next asked whether these miRNAs shared sequence similarity. Although there are numerous mathematical definitions for families of similar sequences, we proceeded to develop an algorithm that emphasizes miRNA sequence similarity based upon seed region and the terminal 3′ bases. For shared membership in a sequence family, the algorithm first requires that the seven-base seeds of two miRNA sequences be identical. Second, it requires that in a certain alignment of two sequences; of the last eight bases of one, at least five should match consecutive bases within the last 10 bases of the other. By focusing alignment on the two ends of a mature miRNA, this approach is a simple application of the principle of correspondence, that is, general alignment of RNAs with focus on blocks of residues (Brown et al. 2009). As described below, our experimental data imply that miRNAs having identical seeds and similar 3′ regions tend to have either entirely nuclear dominance or entirely cytoplasmic dominance (Table 2).
TABLE 2.
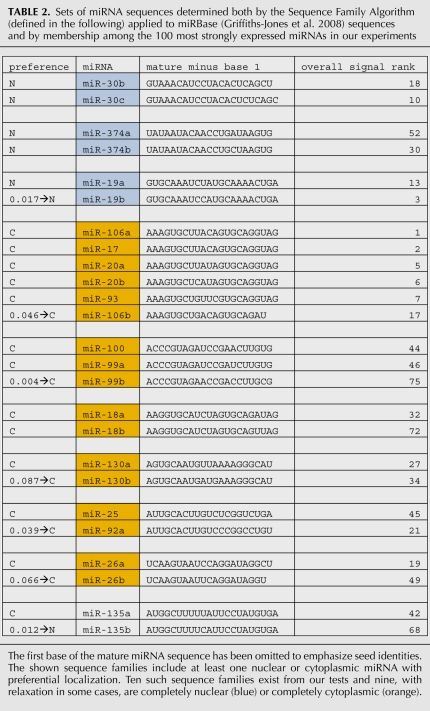
Sets of miRNA sequences determined both by the Sequence Family Algorithm (defined in the following) applied to miRBase (Griffiths-Jones et al. 2008) sequences and by membership among the 100 most strongly expressed miRNAs in our experiments
We also made use of a relaxation of the requirement that all three nuclear standard scores be strictly less than (or strictly greater than) all three cytoplasmic standard scores. That is, the two sets of values could have minimal overlap, so the lowest of the high bin is lower than the highest of the low bin by a small amount relative to the standard deviations = 1. Then an miRNA might be defined to be weakly nuclear if subtracting a small constant from all three of the nuclear standard scores leaves three nuclear values consistently below the adjusted median for all six values; relaxed cytoplasmic comparisons can be analogously defined. The gaps (minimum of upper minus maximum of lower) between the three high and the three low sets of standard scores for the 21 + 31 = 52 most highly ranked miRNAs have average ∼ 0.141 and SD ∼ 0.122. In that context, we can select a typical overlap for relaxed normalized values of miRNAs to be something less than 0.100 = one-tenth of 1 SD for the sample. For example, in Table 2 the notation “0.046→C” for miR-106b means that the consequence of subtracting 0.046 from the three cytoplasmic standard scores leaves consistently lower (stronger) cytoplasmic standard scores. Interestingly, miR-106b is the only miRNA in the conventional miR-17-93 sequence family (as defined by Mendell [2008]) that does not display cytoplasmic dominance without relaxation and also is the only one with a significantly different 3′ end that precludes membership in the same sequence family as defined herein. Regarding verification, we assayed three technical replicates of the nuclear fraction of one sample versus three technical replicates of the cytoplasmic fraction of the same sample using six miRNA microarrays on one Agilent slide. Focus was on the 36 miRNAs that are both included as Agilent assays and included among the 52 miRNAs distinguished by the TaqMan experiments. Pearson correlations among the 36 nuclear and cytoplasmic replicates averaged ∼0.983 and ∼0.991, respectively (Supplement 7). However, the nine cross-compartment Pearson correlations averaged ∼0.988, that is, slightly better than correlations among the three nuclear technical replicates; this reflects the close similarities of the nuclear and cytoplasmic signatures relative to the accuracy available with the Agilent microarrays. The implication is that parametric analysis of measurements for individual miRNAs cannot be informative. Nonparametric tests that identify collectively informative sets of miRNAs are essential. As noted previouisly, nonparametric methods provided validation using median standard score data (P = 0.0056 for chance consistency) or average data (P = 0.0119 for chance consistency). Other miRNAs are not in the top 100, but otherwise do conform to the rule that if one in a sequence family has nuclear or cytoplasmic dominance, then all in the sequence family have the same dominance (Supplement 8).
It should be noted that declaring sequence families is always heuristic and that experimentation with various settings of tunable parameters can lead to different alignments and so to different sequence families. Furthermore, even with the same settings, selecting subsets of a pool can revise pairwise alignments. However, our experiments establish the general principle that a combination of 5′ and 3′ alignments can yield sequence families that are correctly predicted to be mostly or entirely miRNAs with nuclear dominance or mostly or entirely miRNAs with cytoplasmic dominance. In contrast, two mature miRNAs with the same seed but very different 3′ ends do not typically exhibit correlated rankings or, in experiments of others, correlations of other types.
In contrast with sequence families, we may consider physical clusters. Of the 52 listed miRNAs, miR-342-3p and miR-345 are in a physical cluster and both exhibit nuclear dominance. Likewise the six-member miR-363-106a physical cluster includes three miRNAs (miR-106a, -18b, -20b) with cytoplasmic dominance and the others with no dominance. However, in the six-member miR-17-92-1 physical cluster are three with high cytoplasmic ranking, but one (miR19a) with a high nuclear ranking. Strangely, although miR-28-3p and miR-219-5p have higher than average nuclear ranking, their partners, miR-28-5p and miR-219-2-3p, show higher cytoplasmic ranking. And although miR-16-2 and miR-15b are in a physical cluster, miR-16 shows nuclear preference, whereas miR-15b shows cytoplasmic. Thus, there appear to be no simple rules that bias miRNAs in a physical cluster to share nuclear or cytoplasmic dominance.
Sequences that have identical seeds but are not members of one sequence family
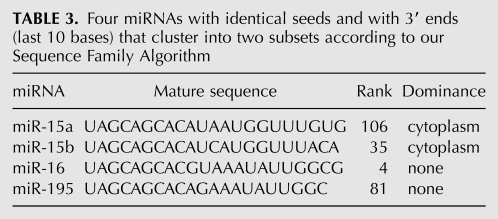
Aside from the sequence families that are consistently highly ranked in the nucleus or cytoplasm in Table 2, we note two other pairs. Some miRNAs have identical seeds but fail to match strongly by the two-ended test of our miRNA Sequence Family Algorithm (Box 1). Consider the four miRNAs in Table 3.
TABLE 3.
Four miRNAs with identical seeds and with 3′ ends (last 10 bases) that cluster into two subsets according to our Sequence Family Algorithm
Here miR-15a, -15b, -16, and -195 all share the same seed AGCAGCA, but the first two are in one sequence family and the last two are in another (Supplement 8). The first two have stronger cytoplasmic standard scores but the last two do not (Supplement 4). Likewise miR-21 and miR-590-5p have identical seeds but do not belong to one sequence family and do not display the same compartment dominance (see Table 4).
TABLE 4.
Two miRNAs with identical seeds that do not belong to one sequence family and do not display the same compartment dominance
Thus, seed identity in itself is not sufficient to predict membership in a sequence family of miRNAs with shared nuclear or cytoplasmic dominance.
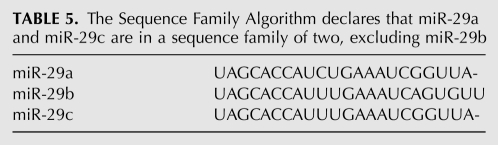
Regarding the three human miRNAs with seed AGCACCA, Hwang and colleagues (Hwang et al. 2007a) found that miR-29b in HeLa cells was preferentially located in the nucleus (71% nuclear), miR-29a was predominantly cytoplasmic (58%), and miR-29c was not detectable. Over base positions 2–17 from the 5′ end these miRNAs are nearly identical, but in the 3′ ends, miR-29b is markedly different. Interestingly, the above-mentioned miRNA Sequence Family Algorithm (Box 1) declares that miR-29a and miR-29c are in a sequence family of two, excluding miR-29b (Table 5; Supplement 8).
TABLE 5.
The Sequence Family Algorithm declares that miR-29a and miR-29c are in a sequence family of two, excluding miR-29b
We, too, have studied the apparent role of …AGUGUU in miR-29b in HeLa cells (Jeffries et al. 2009), finding that indeed that AGUGUU associates with nuclear import. However, in our present experiments using human neural progenitors, miR-29b was only weakly detected, and miR-29a, -c were not clearly biased to nuclear or cytoplasmic dominance.
Motif ASUS in 3′ ends of miRNAs
As noted in the Introduction, Hwang, Wentzel, and Mendell further found that other small ssRNAs, when modified to include, like miR-29b, the sequence …AGUGUU at their 3′ termini, could also accumulate in the nucleus. This was reflected in a patent listing UGUGUU, ACUGUU, AGAGUU, AGUCUU, AGUGAU, AGUGUA, AGNGUN (Hwang et al. 2007b); notably ASUS (S = C or G) appeared in six of the eight (underlined) motifs. This observation prompted us to look for ASUS in the 3′ regions of the 21 miRNAs that displayed stronger nuclear standard scores and in the 31 with stronger cytoplasmic standard scores. Of the 21 in the nuclear list, seven included ASUS in the last nine bases (recall five of the 21 were expected to be in the list by chance). From the 3′ ends of the seven with stronger nuclear ranking in our experiments, we found instances of ASUS as underlined in Table 6.
TABLE 6.
The pattern ASUS appears in 3′ ends of seven miRNAs with stronger nuclear ranking in our experiments
Although official tables usually show one miRNA sequence for one miRNA label, often there are actually multiple mature miRNA subtypes with different ends, as can be detected with deep sequencing (Liao et al. 2010). The 3′ end is particularly variable, possibly relevant to the variable positions of ASUS in these examples. Among the 31 miRNAs that consistently display stronger cytoplasmic standard scores, no mature sequences include ASUS in the last nine bases. Thus, we speculate some role for ASUS in bringing about higher than average levels of miRNA nuclear localization.
Additional relevance to earlier work
Regarding other work in the same area, we note a report by Lakshmipathy and colleagues (Lakshmipathy et al. 2007) that compared miRNA expression in undifferentiated versus differentiated hESCs. Testing hESCs, differentiated cells, and terminally differentiated adult cells, they found some miRNAs that decrease sharply with hESC differentiation (e.g., miR-371, -372, -302a, -302d) and others that increase (let-7a, miR-143, -29a). This pioneering work suggested miRNAs and also their mRNA targets transcribed from pluripotency genes could serve as signatures of stages of stem cell differentiation.
Laurent and colleagues (Laurent et al. 2008) reported miRNA signatures for a variety of stem cell and differentiated cell types. Among miRNAs distinguished by high expression levels in hESCs were many with seeds AAAGAGC or AAGGUGC, such as some members of the miR-17-92 physical cluster (physically close miRNAs on chromosome 13). Wilson and colleagues (Wilson et al. 2009) used miRNA microarrays to compare miRNA signatures of human-induced pluripotent stem cells (hiPSCs) versus hESCs. They found statistically significant differences between the two cell types, evidenced in expression of some physical clusters such as miR-17-92 and the miR-302 cluster.
Chin and colleagues (Chin et al. 2009) compared 15 hiPSC lines, 12 hESC lines, and three fibroblast lines. They reported that gene expression profiles of hiPSCs showed a recurrent signature distinguished from that of hESCs, regardless of somatic source. The hiPSC expression signatures became, upon extended passaging, progressively similar to but always distinct from those of hESCs. As evident in early passages, hiPSCs seemed to be incompletely reset to hESC expression patterns. Regarding gene expression, hiPSCs relative to hESCs generally favored genes associated with differentiation at the expense of genes associated with basic processes, such as energy production, nucleic acid processing, and mitosis. Using an miRNA bioarray (Zhang et al. 2008) they showed that several miRNAs serve as signatures of types and stages of stem cell differentiation.
DISCUSSION
A number of reports have described the occurrence of particular miRNAs in animal cell nuclei and nucleoli (Hwang et al. 2007a; Kim et al. 2008; Marcon et al. 2008; Ohrt et al. 2008; Place et al. 2008; Foldes-Papp et al. 2009; Politz et al. 2009; Liao et al. 2010). In this article, we wished to evaluate the generality of miRNA nuclear localization by assaying a nontumor, minimally passaged, progenitor cell line.
Our experiments have established that in a human neural progenitor cell line, several miRNAs have consistently stronger standard scores in nuclear fractions than in cytoplasmic fractions. Likewise, other miRNAs have stronger cytoplasmic standard scores, and about half of all strongly expressed miRNAs have no clear preference. Nonparametric statistical analyses convincingly established that the observations cannot be ascribed to chance. The mature sequences of miRNAs with the first base omitted can be organized according to their identity among their seeds and a certain similarity among bases at the 3′ ends. Interestingly, if one member of a sequence family displays consistently stronger standard scores among the 100 most strongly expressed miRNAs in nucleus or cytoplasm, then the others show the same compartmental ranking.
In addition to a higher than average nuclear or cytoplasmic ranking for particular miRNAs, our study also confirms with an alternative technology the finding of Liao and colleagues (Liao et al. 2010) that the majority of all miRNAs are present not only in the cytoplasm, as expected, but in the nucleus as well. However, there are important differences between our results and those of Liao et al. First, the miRNAs, identified by Liao and colleagues, as being over-represented in the nucleus were different from those that we identified. However, Liao et al. examined a nasopharyngeal carcinoma cell line, whereas we used neural progenitor cells. Perhaps differences in distinguished lists may be related to cell type or to the untransformed versus tumor state of cells; indeed, such differences may be useful biomarkers. Second, Liao and colleagues used one sequencing experiment, each, for nucleus and cytoplasm; we have used three biological replicates. Third, Liao and colleagues use Illumina sequencing validated with a microarray, while we used TaqMan assays, also validated with a microarray. Importantly, Willenbrock and colleagues (Willenbrock et al. 2009) reported that Exiqon and Agilent miRNA microarray expression measures correlate better with synthetic miRNA-like spikes than expression measures obtained from sequencing data and that the two technologies perform equivalently in terms of reproducibility and relative ratio quantification. The same performance would likely be expected of the miRNA microarray used by Liao et al. (Tang et al. 2007). Notably, our microarray experience with two triplets of technical replicates resulted in data much noisier than that from TaqMan assays of triple biological replicates. Fourth, we have explored a motif associated with miRNAs displaying a high nuclear ranking preference, i.e., ASUS in the 3′ end of mature miRNAs; this motif includes the sequence AGUGUU previously found to be associated with nuclear localization of miR-29b (Hwang et al. 2007a, Jeffries et al. 2009) and is allowed in six of the eight patterns in a patent (Hwang et al. 2007b).
Although the cytoplasmic functions of miRNAs are well established, what might be the role for miRNAs in the nucleus, especially the subsets showing a higher than average nuclear prevalence? Two human miRNAs, miR-373 and miR-320, have been reported to regulate the activities of specific promoters, the former inducing and the latter repressing transcription (Kim et al. 2008; Place et al. 2008). These findings establish a potential nuclear role for miRNAs, and, although bioinformatic analyses suggest that many more miRNAs might target promoter regions (Kim et al. 2008), the generality of miRNA-mediated transcriptional regulation is yet to be established directly. Interestingly, our present study revealed a nuclear preference for the aforementioned miR-320.
Alternatively, return of some miRNAs to the nucleus might be simply a way to sequester them from the cytoplasm and fine-tune their impact on translational regulation and mRNA stability. If so, the particular distribution of miRNAs across the nuclear envelope could be a useful signature of, e.g., the state of cell division, differentiation, or response to stress. Failure to regulate miRNA nuclear versus cytoplasmic distribution could conceivably contribute to cellular pathology.
Other connections to gene regulation are suggested by findings reported by Smalheiser and colleagues (Smalheiser and Torvik 2006). The sequence GCACUU in the Alu consensus is predicted to be a hairpin loop, so it might be available for targeting by several miRNAs that include AAGUGC in their seeds, such as the miR-17-93 sequence family, miR-302a(b,c,d,e), miR-372, miR-373, miR-519a, b-3p, c-3p, d, miR-520a-3p, b, c-3p, d-3p, and e. This list strongly intersects lists of miRNAs distinguishing stem cells by other researchers (Laurent et al. 2008; Chin et al. 2009; Wilson et al. 2009). However, what mechanistic role the cytoplasm-preferring miR-17-93 physical sequence family or the other families play in stem cell phenomena remains to be discovered.
Similarly, one might ask what genes are mutually targeted by the nucleus-preferring miR-30b, c, miR-19a, b, and miR-374a, b sequence families (Table 2). Given mRNA targets for these miRNAs as suggested by TargetScan 5.1 (Lewis et al. 2005; Friedman et al. 2009), the search engine DAVID (Dennis et al. 2003; Huang da et al. 2009) declares 33 genes potentially targeted by all of them. The list includes genes for proteins known to be active in the nucleus, in neurodevelopment, and in DNA-dependent transcription regulation including ELL2, HLF, MECP2, MEF2D, NEUROD1, NFIB, NRK, SATB1, and SOX4. These intriguing findings point to potential avenues of research.
Complementing the above findings for miR-373 and miR-320, Younger and Corey (2009) reported that siRNAs in human cells can target noncoding RNAs (ncRNAs) that overlap promoters and coding sequences, often affecting expression of the coding gene and suggesting a similar regulatory role for endogenous miRNAs. At least in the case of miRNAs displaying a nuclear preference, their corresponding targets may not be annotated mRNAs, but as yet unannotated ncRNAs for which miRNA base-pairing rules may differ from those that apply to miRNA–mRNA interactions in the cytoplasm. The conservation of 3′ motifs we find associated with miRNAs showing a nuclear preference may be a reflection of such alternative rules.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Ms. Vandana Turaga for skillful management of cell lines. We also thank Dr. Patrick Sullivan and the members of his lab for their technical support and the staff at ArunA Biomedical, Inc., for their assistance with cell culture methods. Also, we are very grateful to the referees who made many valuable suggestions about how the present work is positioned within a broad research landscape. The research reflected in this paper was supported in part by an anonymous donor through the San Francisco Foundation, Stanley Medical Research Foundation grant no. 08R-1978, National Institutes of Health NIEHS grant no. 5P01ES014635-02, and National Institutes of Health NIGMS grant no. 2R01GM066940-05A1. The contents of the paper are solely the responsibility of the authors and do not necessarily represent the official views of any supporting agency.
Footnotes
Abbreviations: Cq, quantification cycle; Cytj, cytoplasmic fraction j; dsRNA, double strand RNA; hESC, human embryonic stem cell; hiPSC, human induced pluripotency stem cell; miRNA, microRNA; ncRNA, noncoding RNA; nt, nucleotide; Nuci, nuclear fraction i; pri-miRNA, primary miRNA; RISC, RNA induced silencing complex; S, C or G; SD, standard deviation; ssRNA, single-stranded RNA; TaqMan, Applied Biosystems TaqMan low density array.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2006511.
REFERENCES
- Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A 2009. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet 5: e1000764 doi: 10.1371/journal.pgen.1000764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett V, Lewis T 1994. Outliers in statistical data, 3rd ed Wiley, Chichester, UK [Google Scholar]
- Bartel DP 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Brown JW, Birmingham A, Griffiths PE, Jossinet F, Kachouri-Lafond R, Knight R, Lang BF, Leontis N, Steger G, Stombaugh J, et al. 2009. The RNA structure alignment ontology. RNA 15: 1623–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. 2005. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33: e179 doi: 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, et al. 2009. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 5: 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: 3. [PubMed] [Google Scholar]
- Dhara SK, Stice SL 2008. Neural differentiation of human embryonic stem cells. J Cell Biochem 105: 633–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhara SK, Hasneen K, Machacek DW, Boyd NL, Rao RR, Stice SL 2008. Human neural progenitor cells derived from embryonic stem cells in feeder-free cultures. Differentiation 76: 454–464 [DOI] [PubMed] [Google Scholar]
- Dhara SK, Gerwe BA, Majumder A, Dodla MC, Boyd NL, Machacek DW, Hasneen K, Stice SL 2009. Genetic manipulation of neural progenitors derived from human embryonic stem cells. Tissue Eng Part A 15: 3621–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerth S, Schober H, Gaidatzis D, Roloff T, Jacobeit K, Buhler M 2010. Nuclear retention of fission yeast dicer is a prerequisite for RNAi-mediated heterochromatin assembly. Dev Cell 18: 102–113 [DOI] [PubMed] [Google Scholar]
- Foldes-Papp Z, Konig K, Studier H, Buckle R, Breunig HG, Uchugonova A, Kostner GM 2009. Trafficking of mature miRNA-122 into the nucleus of live liver cells. Curr Pharm Biotechnol 10: 569–578 [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S 2008. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science 321: 537–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT 2007a. A hexanucleotide element directs microRNA nuclear import. Science 315: 97–100 [DOI] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT 2007b. Patent No. WO 2007/149521 A2: Nucleotide motifs providing localization elements and methods of use. World Intellectual Property Organization, Geneva, Switzerland [Google Scholar]
- Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, Minna JD, Corey DR 2006. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol 13: 787–792 [DOI] [PubMed] [Google Scholar]
- Jeffries CD, Fried HM, Perkins DO 2009. Additional layers of gene regulatory complexity from recently discovered microRNA mechanisms. Int J Biochem Cell Biol 42: 1236–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Saetrom P, Snove O Jr, Rossi JJ 2008. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci 105: 16230–16235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmipathy U, Love B, Goff LA, Jornsten R, Graichen R, Hart RP, Chesnut JD 2007. MicroRNA expression pattern of undifferentiated and differentiated human embryonic stem cells. Stem Cells Dev 16: 1003–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Laurent LC, Chen J, Ulitsky I, Mueller FJ, Lu C, Shamir R, Fan JB, Loring JF 2008. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells 26: 1506–1516 [DOI] [PubMed] [Google Scholar]
- Letzen BS, Liu C, Thakor NV, Gearhart JD, All AH, Kerr CL 2010. MicroRNA expression profiling of oligodendrocyte differentiation from human embryonic stem cells. PLoS ONE 5: e10480 doi: 10.1371/journal.pone.0010480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Levitt P 2002. Schizophrenia as a disorder of neurodevelopment. Annu Rev Neurosci 25: 409–432 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R 2006. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci 103: 17337–17342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H, Shao P, Chen YQ, Qu LH 2010. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS One 5: e10563 doi: 10.1371/journal.pone.0010563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ 2009. Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon E, Babak T, Chua G, Hughes T, Moens PB 2008. miRNA and piRNA localization in the male mammalian meiotic nucleus. Chromosome Res 16: 243–260 [DOI] [PubMed] [Google Scholar]
- Mendell JT 2008. miRiad roles for the miR-17-92 cluster in development and disease. Cell 133: 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D 2009. Small RNAs in transcriptional gene silencing and genome defence. Nature 457: 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohrt T, Mutze J, Staroske W, Weinmann L, Hock J, Crell K, Meister G, Schwille P 2008. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res 36: 6439–6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchision DM 2009. The role of oxygen in regulating neural stem cells in development and disease. J Cell Physiol 220: 562–568 [DOI] [PubMed] [Google Scholar]
- Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM 2007. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol 8: R27 doi: 10.1186/gb-2007-8-2-r27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R 2008. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci 105: 1608–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Hogan EM, Pederson T 2009. MicroRNAs with a nucleolar location. RNA 15: 1705–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb GB, Brown KM, Khurana J, Rana TM 2005. Specific and potent RNAi in the nucleus of human cells. Nat Struct Mol Biol 12: 33–37 [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Shi Y, Zhao X, Hsieh J, Wichterle H, Impey S, Banerjee S, Neveu P, Kosik KS 2010. MicroRNA regulation of neural stem cells and neurogenesis. J Neurosci 30: 14931–14936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Mitalipova M, Noggle S, Tibbitts D, Venable A, Rao R, Stice SL 2006. Long-term proliferation of human embryonic stem cell-derived neuroepithelial cells using defined adherent culture conditions. Stem Cells 24: 125–138 [DOI] [PubMed] [Google Scholar]
- Slezak-Prochazka I, Durmus S, Kroesen BJ, van den Berg A 2010. MicroRNAs, macrocontrol: regulation of miRNA processing. RNA 16: 1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Torvik VI 2006. Alu elements within human mRNAs are probable microRNA targets. Trends Genet 22: 532–536 [DOI] [PubMed] [Google Scholar]
- Tang X, Gal J, Zhuang X, Wang W, Zhu H, Tang G 2007. A simple array platform for microRNA analysis and its application in mouse tissues. RNA 13: 1803–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Willenbrock H, Salomon J, Søkilde R, Barken KB, Hansen TN, Nielsen FC, Møller S, Litman T 2009. Quantitative miRNA expression analysis: comparing microarrays with next-generation sequencing. RNA 15: 2028–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KD, Venkatasubrahmanyam S, Jia F, Sun N, Butte AJ, Wu JC 2009. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev 18: 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger ST, Corey DR 2009. The puzzle of RNAs that target gene promoters. ChemBioChem 10: 1135–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K, et al. 2008. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci 105: 7004–7009 [DOI] [PMC free article] [PubMed] [Google Scholar]