Abstract
Translation initiation factor eIF5B promotes GTP-dependent ribosomal subunit joining in the final step of the translation initiation pathway. The protein resembles a chalice with the α-helix H12 forming the stem connecting the GTP-binding domain cup to the domain IV base. Helix H12 has been proposed to function as a rigid lever arm governing domain IV movements in response to nucleotide binding and as a molecular ruler fixing the distance between domain IV and the G domain of the factor. To investigate its function, helix H12 was lengthened or shortened by one or two turns. In addition, six consecutive residues in the helix were substituted by Gly to alter the helical rigidity. Whereas the mutations had minimal impacts on the factor's binding to the ribosome and its GTP binding and hydrolysis activities, shortening the helix by six residues impaired the rate of subunit joining in vitro and both this mutation and the Gly substitution mutation lowered the yield of Met-tRNAiMet bound to 80S complexes formed in the presence of nonhydrolyzable GTP. Thus, these two mutations, which impair yeast cell growth and enhance ribosome leaky scanning in vivo, impair the rate of formation and stability of the 80S product of subunit joining. These data support the notion that helix H12 functions as a ruler connecting the GTPase center of the ribosome to the P site where Met-tRNAiMet is bound and that helix H12 rigidity is required to stabilize Met-tRNAiMet binding.
Keywords: IF2, eIF5B, subunit joining, translation initiation
INTRODUCTION
Eukaryotic translation initiation involves the stepwise assembly of a ribosome–Met-tRNAiMet–mRNA complex in which the specific initiator Met-tRNAiMet in the ribosomal P site is base-paired to the AUG start codon of the mRNA (for review, see Sonenberg and Hinnebusch 2009). Assembly of this 80S complex starts with the binding of the Met-tRNAiMet in complex with the factor eIF2 and GTP to the small (40S) ribosomal subunit. This 43S preinitiation complex also contains the factors eIF1 and eIF1A, bound near the ribosomal E and A sites, respectively. The 43S complex binds to an mRNA near the 5′ cap and then scans in a 3′ direction in search of a start codon. During scanning, a fraction of the GTP bound to eIF2 is hydrolyzed to GDP + Pi under the influence of eIF5. Upon start site recognition eIF1 dissociates and Pi is released. Subsequent release of eIF2 leaves Met-tRNAiMet in the P site bound to the start codon and eIF1A in the ribosomal A site. The factor eIF5B, another GTPase, promotes large (60S) ribosomal subunit joining. Formation of the 80S ribosome triggers GTP hydrolysis by eIF5B and release of both eIF5B and eIF1A from the ribosome, which is now poised to enter the elongation phase of protein synthesis.
The factor eIF5B is the eukaryotic ortholog of the prokaryotic translation initiation factor IF2. Whereas IF2 plays roles both in binding Met-tRNAiMet to the small ribosomal subunit and in subunit joining, eIF5B function appears to be mainly restricted to ribosomal subunit joining with the exception of a few viral mRNAs (Pestova et al. 2000, 2007, 2008; Roll-Mecak et al. 2001; Terenin et al. 2008). Like yeast and human eIF5B, Escherichia coli IF2 contains a centrally located GTP-binding (G) domain. The N-terminal portions of these proteins display little sequence conservation, and consistently this region is not required for yeast eIF5B function either in vitro or in vivo (Choi et al. 2000; Shin et al. 2002). The archaeal aIF5B, which naturally lacks the N-terminal region, functionally substitutes for yeast eIF5B both in vitro and in vivo (Lee et al. 1999). The X-ray crystal structure of aIF5B from Methanobacterium thermoautotrophicum (M. thermoautotrophicum) revealed that the protein consists of four domains: an N-terminal G domain, a β-barrel domain II, an α-β-α sandwich domain III, and another β-barrel domain IV (Roll-Mecak et al. 2000). Overall aIF5B resembles a chalice with domains I–III forming the cup and domain IV forming the base. The cup and base are connected by α-helix H12, the stem of the chalice. Comparison of the structures of aIF5B bound to GDP and to GDPNP revealed that modest reorientation of the switch elements, that contact the phosphates of the nucleotide in the G domain, triggers conformational changes in the cup portion of the protein that are amplified through helix H12 acting as a lever arm and result in a more significant ∼5 Å swing of domain IV (Fig. 1A; Roll-Mecak et al. 2000). Based on cryo-EM images of the bacterial initiation complex with IF2, the G domain of IF2 binds near the GTPase-activating center of the 50S subunit and domain IV contacts the fMet portion of fMet-tRNAiMet bound in the ribosomal P site (Allen et al. 2005; Myasnikov et al. 2005; Simonetti et al. 2008). Hydroxyl radical cleavage studies indicated that eIF5B binds to a similar position on mammalian 80S ribosomes (Unbehaun et al. 2007). Moreover, consistent with these mapping studies, eIF5B and rRNA suppressor studies placed domain II of eIF5B near 18S rRNA helix h5 in the 40S subunit (Shin et al. 2009). Taken together these studies indicate that helix H12 might function as a ruler to properly position the G domain and domain IV when the factor is bound to the ribosome or as a rigid lever arm to enable proper movement of domain IV in response GTP binding or hydrolysis.
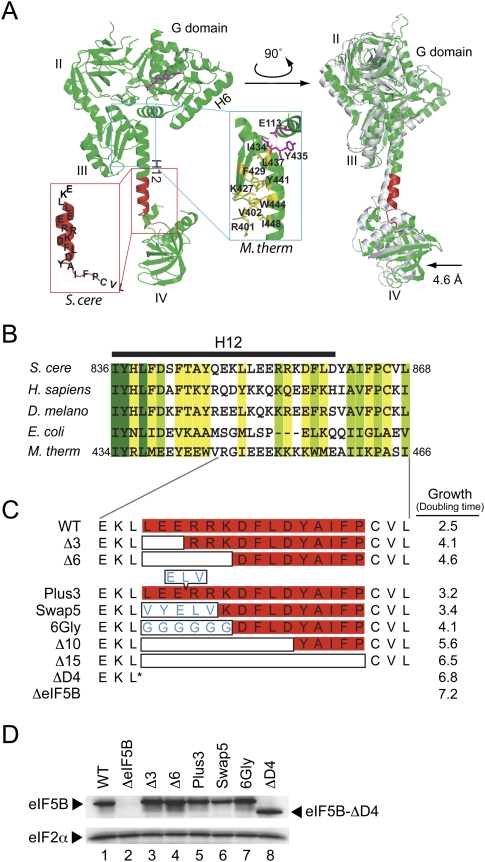
FIGURE 1.
Mutational analysis of helix H12 in yeast eIF5B. (A) Ribbon representation of X-ray crystal structure of aIF5B (M. thermoautotrophicum) in complex with GTP (Protein Data Bank [PDB] code 1G7T) (Roll-Mecak et al. 2000). The four domains of the protein (G, II, III, and IV) and helices (H6 and H12) are labeled. The structure on the right is rotated about the vertical axis to better visualize the lever-type domain IV movement in the presence of GTP (colored) versus GDP (gray; PDB code 1G7S). The inset in the blue box highlights the interactions between top of H12 and the G domain and domain III in aIF5B (labels refer to M. thermoautotrophicum residues). The inset in the red box depicts the corresponding H12 region in yeast (S. cerevisiae) eIF5B that was subjected to mutational analysis. (B) Sequence alignments of helix H12 region of eukaryotic, bacterial, and archaeal eIF5B/IF2. Identical residues within the aligned sequences are shown in dark green, conserved residues are shown in yellow green, and weakly conserved residues (conserved in three out of five residues sequences) are in yellow. Residue numbers are shown for S. cerevisiae and M. thermoautotrophicum, and the position of helix H12 is denoted by the black bar above the sequences. (C) Schematic representation of helix H12 mutants. Open boxes indicate deleted residues, whereas amino acid residues within the boxes indicate insertions (Plus3) or substitutions (Swap5 and 6Gly). Doubling times in SD medium of yeast strain J111 expressing the indicated H12 mutant are shown on the right. (D) Western blot analysis of eIF5B expression. Whole cell extracts prepared from yeast transformants in panel C were subjected to immunoblot analysis using anti-eIF5B or anti-eIF2α antiserum as previously described (Choi et al. 2000).
In yeast, eIF5B is encoded by the nonessential FUN12 gene (Choi et al. 1998; Pestova et al. 2000). Deletion of FUN12 causes a severe slow-growth phenotype, impairs the derepression of GCN4 expression in response to amino acid starvation conditions, and enhances leaky scanning wherein ribosomes scan past an AUG codon without initiating translation (Choi et al. 1998; Shin et al. 2002). These phenotypes of cells lacking eIF5B are also observed in cells expressing GTPase-deficient mutant forms of eIF5B (Choi et al. 1998; Lee et al. 2002; Shin et al. 2002, 2007, 2009). Previous studies showed that GTP hydrolysis by eIF5B is not required for subunit joining but is required to release the factor from 80S ribosomes following subunit joining (Shin et al. 2002). Consistently, purified eIF5B binds to 80S ribosomes in a GTP-dependent manner and ribosomes stimulate the GTPase activity of eIF5B. The C-terminal amino acids of eIF1A bind in a groove in domain IV of eIF5B (Marintchev et al. 2003), and this interaction is thought to help recruit and position eIF5B on the 40S ribosome (Choi et al. 2000; Acker et al. 2006, 2009; Fringer et al. 2007). Likewise, GTP hydrolysis by eIF5B promotes the rapid release of eIF1A (Fringer et al. 2007; Acker et al. 2009). Whereas domain IV of eIF5B is important for subunit joining, it is not clear whether the length or rigid helical structure of helix H12 is important for subunit joining, ribosome binding, or eIF5B ribosome-dependent GTP hydrolysis. To answer these questions, we generated a series of eIF5B mutants with alterations in helix H12 and examined the properties of the mutants using both in vivo and in vitro assays of eIF5B function. Our results indicate that helix H12 is not critical for eIF5B binding to the ribosome or for GTP hydrolysis with purified ribosomes; however, the length and rigidity of helix H12 are important for formation of stable 80S ribosomal complexes.
RESULTS
Mutations in eIF5B helix H12 impair yeast cell growth
To examine the functional significance of α-helix H12, mutations targeting H12 were introduced into a FUN12 plasmid designed to express eIF5B396-1002 from the native FUN12 promoter. Previous studies demonstrated that removal of the N-terminal region of yeast eIF5B does not impair its function in vivo (Choi et al. 2000; Shin et al. 2002). Moreover, as it has not been possible to produce recombinant versions of full-length eIF5B, the use of the N-terminally truncated mutants allows direct comparison of the results from in vivo and in vitro experiments. As shown in Figure 1A, Glu113 in the aIF5B G-domain hydrogen bonds with the residues at the top of helix H12, and the N-terminal half of the helix packs against domain III, forming a series of hydrophobic interactions (Fig. 1A, blue inset). Whereas the first two residues at the N terminus of helix H12 are universally conserved during evolution, the remainder of the H12 sequence is less conserved. However, the hydrophobic nature of the residues interacting with domain III has been retained in aIF5B, eIF5B, and IF2 (Fig. 1B). To specifically address the importance of the length and rigidity of helix H12, and to avoid complications associated with disruption of contacts with domain III and the G domain, the mutational analysis was restricted to the C-terminal half of the helix (Fig. 1A, red inset). Note that in the crystal structure of aIF5B helix H12 is connected to the N-terminal β-strand S24 in domain IV by a five-residue unstructured segment corresponding to the yeast eIF5B sequence Y-A-I-F-P (Fig. 1A, inset in red box, residues 861–865).
Mutations were designed to alter both the length and rigidity of helix H12 (Fig. 1C). To assess the importance of the length of helix H12, the mutations Δ3 (residues 851–853 in yeast eIF5B), Δ6 (residues 851–856), or Δ10 (residues 851–860) were designed to delete 3, 6, or 10 residues, respectively, from the helix (Fig. 1C). In addition, the mutation Δ15 was designed to remove the entire C-terminal portion of helix H12 plus the five-residue coil linking H12 to domain IV (residues 851–865; Fig. 1C). To extend the length of helix H12, three residues (E-L-V, residues 167–169) comprising one turn of helix H6 in M. thermoautotrophicum aIF5B were inserted in helix H12 after Glu853, generating the mutant Plus3. To disrupt the helical structure of H12, the six residues from Leu851 to Lys856 were substituted with glycine, creating the mutant 6Gly. As a control for the deletion and 6Gly mutants, and to test whether the amino acid sequence of helix H12 is important, the residues Leu851 to Arg855 were substituted with the helix forming residues Val165 to Val169 from helix H6 of M. thermoautotrophicum aIF5B, generating the mutant Swap5 (Fig. 1C). Finally, eIF5B was truncated after residue Leu850 to remove the C-terminal half of helix H12 and all of domain IV in the mutant ΔD4 (Fig. 1C).
When expressed in the eIF5BΔ (fun12Δ) strain J111, the Plus3 and Swap5 mutants grew slightly slower than a strain expressing WT eIF5B (see doubling times in Fig. 1C and growth assays in Fig. 2A). The progressive shortening of helix H12 by 3, 6, 10, and 15 residues led to decreasing yeast cell growth with the Δ15 mutant (doubling time 6.5 h) growing nearly as poorly as the eIF5B deletion strain (doubling time 7.2 h). The growth defects were not due to impaired eIF5B expression as the mutants were expressed to the same level as WT eIF5B (Fig. 1D; note that the Swap5 mutant protein was less abundant than WT eIF5B; however, this mutant grew nearly as well as the WT strain). Not surprisingly, deletion of the entire C-terminal portion of eIF5B in the ΔD4 mutant resulted in a growth phenotype similar to the eIF5BΔ strain, as was previously observed with deletion of the C-terminal helix H14 (Fringer et al. 2007). Like the Δ3 mutant, disrupting the helical nature of H12 by the 6Gly mutant partially impaired yeast cell growth. As the 6Gly and Swap5 mutants both impair yeast cell growth, albeit to different extents, it could be argued that the amino acid sequence of H12 is important. However, biochemical analyses of these mutants, described below, indicate that the 6Gly mutation has a significantly more severe impact on eIF5B function. Thus the defects associated with the 6Gly mutation likely reflect more than a change in the amino acid sequence of this region of eIF5B. Taken together, the results of the growth tests indicate that both the length and helical nature of H12 are important; however, the relatively good growth of the Swap5 mutant indicates that the amino acid sequence of this portion of helix H12 may not be critical.
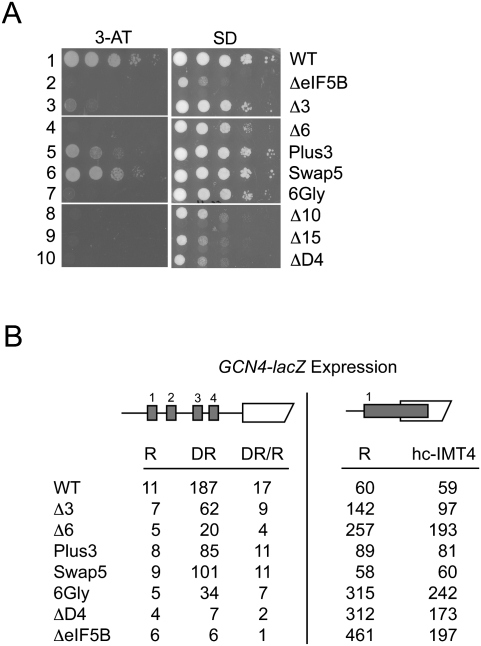
FIGURE 2.
Translational control of GCN4 expression in eIF5B mutant strains. (A) Growth phenotypes. Derivatives of the ΔeIF5B strain J111 transformed with plasmids expressing wild-type eIF5B (WT), empty vector (ΔeIF5B) or various H12 mutants, as indicated, were grown to saturation, and 5 μl of serial dilutions (of OD600 = 1.0, 0.1, 0.01, 0.001, and 0.0001) were spotted on minimal medium supplemented with essential nutrients (SD) or medium containing 10 mM 3-aminotriazole (3-AT). Plates were incubated 3 d at 30°C. (B) Analysis of GCN4-lacZ expression. The wild-type GCN4-lacZ plasmid p180 (Hinnebusch 1985) or a derivative in which an extended version of uORF1 overlaps the GCN4 AUG start codon (pM226) (Grant et al. 1994) were introduced into derivatives of strain J111 described in panel A. Cells were grown and β-galactosidase activities were determined as described previously (Hinnebusch 1985). R, cells were grown under nonstarvation conditions in SD medium where GCN4 expression is repressed; DR, cells were grown under amino acid starvation conditions (SD + 10 mM 3-aminotriazole) where GCN4 expression is derepressed. The β-galactosidase activities are the averages of three independent transformants and have standard errors of 20% or less.
Helix H12 mutants impair GCN4 translational control
Translation of the yeast GCN4 mRNA, encoding a transcriptional activator of genes encoding amino acid biosynthetic enzymes, is very sensitive to perturbation of general translation initiation (Hinnebusch 2005). Previously, we showed that yeast cells lacking eIF5B or expressing GTPase-defective eIF5B mutants failed to grow under amino acid starvation conditions imposed by the histidine biosynthesis inhibitor 3-aminotriazole (3-AT, see Fig. 2A, row 1 versus 2). This growth defect is due to impaired derepression of GCN4 expression under starvation conditions as detected using a GCN4-lacZ reporter assay (see Fig. 2B, left panel, cf. WT and ΔeIF5B; Choi et al. 1998; Shin et al. 2002). As shown in Figure 2A and consistent with their modest affects on yeast cell growth, the Plus3 and Swap5 mutants weakly impaired growth on 3-AT medium (rows 5 and 6). The Δ3 mutant significantly impaired growth on 3-AT medium (Fig. 2A, row 3), whereas the larger truncation mutants as well as the deletion of domain IV and the 6Gly mutant conferred a 3-AT-sensitive phenotype (Fig. 2A, rows 4, 7–10).
Analysis of GCN4-lacZ expression in the helix H12 mutant strains supported the results of the 3-AT growth tests. In WT cells, GCN4-lacZ expression was derepressed 17-fold under amino acid starvation (3-AT) conditions (Fig. 2B, left panel, see DR/R ratio). The Swap5, Plus3, and Δ3 mutants, which showed weak to moderate growth on 3-AT medium, derepressed GCN4-lacZ expression at least ninefold; whereas the 3-AT-sensitive Δ6, 6Gly, and ΔD4 mutants showed less than sevenfold derepression (Fig. 2B, left panel). Taken together, these results indicate that in these strains at least a ninefold induction of GCN4-lacZ expression under amino acid starvation conditions is required to show detectable growth on 3-AT medium. It is noteworthy that the Δ6 and 6Gly mutants have modest impacts on yeast cell growth (see Fig. 2A, right panel, and Fig. 1B, doubling times), yet severely impair GCN4-lacZ expression and confer a 3-AT-sensitive phenotype. These results indicate that the proper length and integrity of the eIF5B H12 structure are required for proper translational regulation of GCN4 expression.
The GCN4 mRNA contains four short upstream open reading frames (uORFs) in its 5′ leader sequence that control access of ribosomes to the AUG start codon of GCN4. Translational control of GCN4 is dependent on ribosomes translating the first uORF followed by regulated reinitiation at the subsequent uORFs (Hinnebusch 2005). Failure to translate uORF1 impairs GCN4 expression. In order to assess the efficiency of translation initiation at the uORF1 AUG codon, we used a GCN4-lacZ construct in which the DNA sequence between uORF1 and uORF4 was removed and the uORF1 stop codon was mutated such that uORF1 overlaps the GCN4 ORF (Fig. 2B, right panel). On this mutant GCN4-lacZ mRNA, ribosomes will scan the leader, translate uORF1, and then terminate 3′ of the GCN4 AUG start codon. As ribosomes would have to scan 130 nts in a 3′ to 5′ direction and bypass four AUG codons to access the GCN4 start codon following translation of the extended uORF1 (Abastado et al. 1991; Grant et al. 1994), GCN4-lacZ expression was low in the strain expressing wild-type eIF5B (Fig. 2B, row 1). In contrast, in cells lacking eIF5B or expressing the Δ3, Δ6, 6Gly, or ΔD4 mutants, GCN4-lacZ expression from this uORF1-extended construct was increased ∼2.4–7.5-fold (Fig. 2B). This increased leaky scanning phenotype in these mutants is likely contributing to the defective GCN4-lacZ expression from the construct with the WT leader (Fig. 2B, left panel) and indicates that alterations to the helix H12 length or flexibility impairs the ability of the 43S complex to recognize an AUG start codon. Interestingly, overexpression of IMT4 encoding tRNAiMet lowered GCN4-lacZ expression from the uORF1-extended construct in strains expressing the eIF5B mutants, but not WT eIF5B (Fig. 2B, right panel). These results suggest that the helix H12 mutations might affect the stability of Met-tRNAiMet binding to the ribosome.
Helix H12 mutations do not affect the GTP switch properties of eIF5B
Previously, we showed that a GTP/GDP switch regulates the ribosome-binding activity of eIF5B: Ribosome-binding affinity is high in the presence of GTP and low in reactions containing GDP (Shin et al. 2002). As the lever-type domain movements of helix H12 during GTP binding and hydrolysis might be predicted to modulate the ribosomal binding affinity of eIF5B, we examined the effect of the helix H12 mutations on the GTP-switch properties of eIF5B. First, the relative GTP-binding affinity of each mutant protein was measured by monitoring the ability of GTP to compete with fluorescently labeled Mant-GDP for binding to eIF5B. As shown in Figure 3A, the H12 mutations did not significantly alter the binding affinity for GTP, consistent with the notion that helix H12 mutations did not affect the structural integrity of the GTP-binding domain.
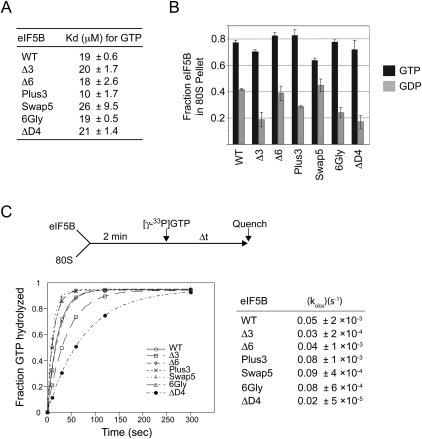
FIGURE 3.
Analysis of GTP and ribosome binding and GTPase activities of eIF5B mutant proteins. (A) Kd values and standard deviations for GTP binding to eIF5B mutants from three independent measurements. (B) Ribosome binding assay. Purified WT or helix H12 mutants of eIF5B were mixed in the presence of GTP or GDP, as indicated, with purified yeast 80S ribosomes, and then loaded onto a 10% sucrose cushion. Following centrifugation, the amounts of eIF5B recovered in the pellet and supernatant fractions were determined by SDS-PAGE and quantitative densitometry. The data presented represent the fraction of eIF5B present in the ribosomal pellet and are representative of results from at least three independent experiments ± standard error. (C) Ribosome-dependent GTPase assay. As shown in the reaction scheme (top), 1 μM eIF5B was incubated with 0.4 μM 80S ribosomes for 2 min. Following addition of 50 nM [γ-33P]GTP, aliquots from the reactions were analyzed at the indicated times by thin-layer chromatography. The data presented are representative of results from at least three independent experiments. Data were fit to the single exponential expression A[1 − exp(−kt)], in which A is the amplitude, k is the rate constant, and t is time. Fits were performed using KaleidaGraph (Synergy Software), and values in parentheses are fit errors.
Next, the ribosomal binding affinities of the helix H12 mutants were tested using a sucrose cushion assay. Briefly, a WT or mutant form of eIF5B was mixed with purified yeast 80S ribosomes in the presence of GDPNP (slowly hydrolyzable GTP analog) or GDP, and then the mixture was layered on a 10% sucrose cushion and subjected to centrifugation to pellet the ribosomes and associated proteins. As quantified in Figure 3B, a larger fraction of WT eIF5B pelleted with ribosomes in reactions containing GDPNP (GTP) than in reactions containing GDP. Interestingly, similar switchlike properties were observed for all of the eIF5B helix H12 mutant proteins (Fig. 3B). Surprisingly, even the mutant ΔD4, which lacks the entire C-terminal domain including half of helix H12 and all of domain IV, displayed GTP/GDP switch properties similar to WT eIF5B (Fig. 3B). Thus, conformational changes occurring in the first three domains (G-domain, domain II, and domain III, including the N-terminal portion of H12 that packs against domain III) of eIF5B are sufficient to confer the GTP switch property governing ribosome association of eIF5B.
In addition to GTP-dependent ribosome-binding activity, eIF5B possesses ribosome-dependent GTPase activity. To assess eIF5B GTPase activity uncoupled from translation initiation, WT eIF5B and the various mutant proteins were mixed with purified yeast 80S ribosomes and [γ-33P]GTP, and then reaction products were analyzed by thin-layer chromatography and quantified (Fig. 3C, upper panel). The WT eIF5B hydrolyzed GTP with an apparent rate constant of 0.05 s−1, and similar rates of hydrolysis were observed for the Δ3 and Δ6 mutants. The Plus3, Swap5, and 6Gly mutants displayed elevated GTPase rates and even the mutant ΔD4 retained significant GTPase activity (Fig. 3C). These results are consistent with the idea that the critical binding and GTPase-activating contacts between eIF5B and the ribosome are mediated through the G-domain, domain II, and/or domain III of the factor. While the yeast growth assays indicate that the length and flexibility of helix H12 and the presence of domain IV are critical for eIF5B function in vivo, the translation defect associated with these mutations is apparently not associated with eIF5B ribosome-binding or GTPase activities.
eIF5B Δ6 and 6Gly mutations affect 80S complex stability
To directly assess the impact of the helix H12 mutations on eIF5B function in translation initiation, we used a light scattering assay to monitor eIF5B-stimulated subunit joining activity (Acker et al. 2009). Following the scheme depicted in Figure 4A, 48S complexes were formed by mixing eIF2–GTP–Met-tRNAiMet ternary complex (TC) with 40S subunits, eIF1, eIF1A, and an unstructured model mRNA. Subunit joining was then initiated by adding 60S subunits, eIF5, WT or mutant forms of eIF5B, and GTP. Joining of the 60S subunit was monitored by following the increased intensity of light scattered by the newly formed 80S ribosomes. As this experiment requires the use of substantial amounts of purified initiation factors, we limited the analysis to two of the most severe mutants, Δ6 and 6Gly, and we used WT eIF5B and the Swap5 mutant as controls. With WT eIF5B, subunit joining occurred with a rate constant of 0.083 ± 0.008 s−1 and relative amplitude of 0.76 ± 0.02 (Fig. 4B). The extent of 80S formation, as monitored by the amplitude of light scattering during the subunit joining reaction, was not significantly altered for the Swap5, 6Gly, or Δ6 mutants. This indicates that the 80S complexes formed by these helix H12 mutants are relatively stable. Interestingly, the rates (kobs) of subunit joining were significantly decreased by each of the helix H12 mutations (Fig. 4B). Even the Swap5 mutant, that modestly impairs yeast cell growth (Fig. 2A), produces a fourfold decrease in the rate of subunit joining. The 6Gly and Δ6 mutations were more severe, decreasing the rate of subunit joining by six- and 13-fold, respectively (Fig. 4B). Taken together, these data suggest that a certain threshold (perhaps greater than fourfold) defect in the rate of subunit joining is necessary to cause a significant growth defect. Alternatively, as described below, the growth defects of the 6Gly and Δ6 mutants might reflect additional functional impairments of these mutants. The significant defects of the 6Gly and Δ6 mutants indicate that length and rigidity of helix H12 are important for the rate of subunit joining.
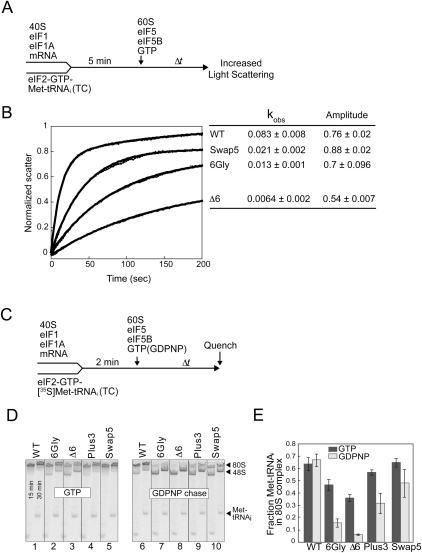
FIGURE 4.
Helix H12 mutants of eIF5B impair 80S complex stability. (A) Experimental scheme for subunit joining monitored by light scattering. (B) Subunit joining assay for WT eIF5B or the Swap5, 6Gly, or D6 mutant in the presence of GTP. Observed rate constant (kobs) and amplitude for each reaction are shown at the right end of the corresponding curve. Data are means ± standard error from at least two independent experiments. (C) Experimental scheme for 80S complex formation assay. (D,E) Results from 80S complex formation assay. 48S complexes were formed using limiting amounts of eIF2 ternary complexes containing [35S]Met-tRNAiMet and then subunit joining was monitored in the presence of either GTP (left panel) or GDPNP (right panel). Reactions were stopped at 15 and 30 min and loaded onto a running native gel. Following phosphorimage analysis of the gel (D), the amounts of [35S]Met-tRNAiMet that were free or bound to 43S or 80S complexes at 30 min were quantified, and the fraction of [35S]Met-tRNAiMet in 80S complexes was calculated (E). Data represent the mean of three independent experiments ± standard error.
To further monitor the subunit-joining activity of the eIF5B helix H12 mutants and to assess the stability of the 80S products of subunit joining, a native gel assay was used to monitor 80S complex formation under steady-state conditions. As depicted in the scheme in Figure 4C, 48S complexes were formed using limiting amounts of TC containing [35S]Met-tRNAiMet, and then subunit joining was initiated by adding 60S subunits, eIF5, WT or mutant forms of eIF5B, and either GTP (Fig. 4D, left panel) or GDPNP (Fig. 4D, right panel). In reactions containing GTP, the WT eIF5B and the Plus3 and Swap5 mutants stimulated nearly complete conversion of the 48S complexes to 80S complexes (Fig. 4D, lanes 1,4,5). In contrast, the 6Gly and Δ6 mutants were less effective in promoting 80S complex formation as noted by the presence of unreacted 48S complexes in these reactions (Fig. 4D, lanes 2–3). The fraction of total [35S]Met in each reaction present in 80S complexes versus 48S complexes or free Met-tRNAiMet was calculated and presented in Figure 4E (dark bars). The quantification reveals that the eIF5B 6Gly and Δ6 mutations impair 80S complex formation by 30% and 45%, respectively, compared to WT eIF5B. Although the light-scattering assay (Fig. 4B) indicated that the 6Gly and Δ6 mutants have a defect in the rate of subunit joining, the 30-min incubation time used for the 80S complex formation assay should be sufficient to allow completion of the subunit joining reaction. Considering the similar levels (amplitude) of subunit joining by WT eIF5B and the 6Gly and Δ6 mutants in the light-scattering assay (Fig. 4B), but the reduced association of [35S]Met-tRNAiMet with 80S complexes formed by the 6Gly and Δ6 mutants in the native gel assays, it seems that the [35S]Met-tRNAiMet is not stably bound to the 80S complexes formed by these eIF5B mutants. Accordingly, the [35S]Met-tRNAiMet dissociates from the 80S complexes formed by the 6Gly and Δ6 mutants of eIF5B, rebinds to eIF2 to form TC, which then binds to 40S subunits accounting for the increased 48S complexes observed in assays with these mutants (Fig. 4D, lanes 2–3).
To further examine the stability of Met-tRNAiMet binding to the 80S complexes formed following subunit joining, the assay was modified by the addition of an excess amount of GDPNP during subunit joining (Fig. 4D, GDPNP chase). In this revised protocol TC is formed with GTP, but subunit joining is performed in the presence of GDPNP. As GTP hydrolysis is required for eIF5B release following subunit joining, the use of GDPNP is expected to lock eIF5B on the 80S complexes. In reactions containing WT eIF5B, equal amount of [35S]Met-tRNAiMet were bound to 80S complexes in reactions containing GTP or GDPNP, consistent with the notion that WT eIF5B promotes stable binding of Met-tRNAiMet to 80S ribosomes (Fig. 4E). In contrast, the GDPNP chase resulted in a dramatic decrease in [35S]Met-tRNAiMet binding to 80S complexes formed by the 6Gly and Δ6 mutants of eIF5B (Fig. 4D, right panel; Fig. 4E). The corresponding increase in [35S]Met-tRNAiMet binding to 48S complexes in reactions containing the eIF5B 6Gly and Δ6 mutants (Fig. 4D, lanes 7–8) is consistent with the idea that [35S]Met-tRNAiMet dissociated from the 80S complexes and was recycled to form dead-end 48S complexes containing eIF2-GDPNP-[35S]Met-tRNAiMet TC. We conclude that altering the length (Δ6 mutant) or flexibility (6Gly mutant) of eIF5B helix H12 impairs the stability of Met-tRNAiMet binding to the 80S product of subunit joining. Consistent with this idea, and as described above, overexpression of tRNAiMet resulted in a significant decrease in GCN4-lacZ expression from the uORF1-extended construct in strains expressing the deleterious eIF5B mutants (Δ3, Δ6, 6Gly, ΔD4), but not in strains expressing WT eIF5B or the mild Plus3 or Swap5 mutants (Fig. 2B, right panel). These results further support the notion that altering the length or flexibility of eIF5B helix H12 lowers the stability of Met-tRNAiMet binding to the 80S product of subunit joining.
DISCUSSION
The chalice-shaped structure of aIF5B raised questions regarding the functional role of the stem helix H12. Studies on eIF5B have demonstrated that the cup of the chalice, consisting of the G domain, β-barrel domain II and domain III, performs important roles in regulating eIF5B function in response to GTP/GDP binding and in binding the ribosome (Shin et al. 2002, 2007, 2009; Unbehaun et al. 2007). Likewise, domain IV, the base of the chalice, interacts with eIF1A and is important for binding eIF5B to the ribosome (Marintchev et al. 2003; Acker et al. 2006, 2009; Fringer et al. 2007; Unbehaun et al. 2007). However, the function of helix H12 connecting these two regions of the protein has not been determined. The helix could simply act as a tether maintaining a physical connection between the cup and base of the chalice. Alternatively, helix H12 might function as a ruler to maintain proper spacing between the cup and base of the chalice. Finally, based on the motion of domain IV in response to GTP versus GDP binding to aIF5B, it was proposed that helix H12 functioned as a rigid lever arm (Roll-Mecak et al. 2000). We performed in vivo and in vitro structure-function studies using helix H12 mutants to help clarify the function of this helix. Our results indicate that helix H12 is more than simply a tether, and that both the length and rigidity of H12 are important for eIF5B function.
Shortening the helix by two turns (Δ6) or increasing its flexibility by replacing six consecutive residues by Gly had little impact on the GTP- or ribosome-binding function or the uncoupled GTPase activity of eIF5B (Fig. 3). However, these two mutations impaired yeast cell growth (Figs. 1, 2), and the subunit joining and 80S complex formation activities of eIF5B (Fig. 4). The fact that the Swap5 mutant partially impaired yeast cell growth and the rate of subunit joining could argue that the defects associated with the 6Gly mutation were due to the importance of the H12 amino acid sequence. At odds with this interpretation, the 6Gly mutant was substantially more defective than the Swap5 mutant in the 80S complex formation assay (Fig. 4D,E). Taken together, we conclude that unlike the Swap5 mutation, the 6Gly mutation alters the structure and sequence of helix H12. While the subunit joining and 80S complex formation assays are ostensibly measuring the same activity of eIF5B, it is noteworthy that whereas the subunit joining assay simply measures 80S complex levels, the 80S complex formation assay monitors the binding of Met-tRNAiMet to 80S complexes. Notably both the 6Gly and Δ6 mutations showed substantial defects in the rate of subunit joining (Fig. 4B) and in the 80S complex formation assay (especially in the presence of GDPNP) (Fig. 4D). We conclude that both the rigidity and the length of helix H12 are critical for the initial joining of the 60S subunit to a 48S complex. Because the Δ6 mutation impaired the rate of subunit joining, we conclude that helix H12 functions, in part, as a ruler to promote 60S subunit joining. While it is possible that the 80S complex adopts different conformations with the eIF5B mutants in the presence of GTP versus GDPNP, the defective association of Met-tRNAiMet with the 80S complexes formed with the 6Gly and Δ6 mutants indicates that both the length and rigidity of helix H12 are important to stably lock the Met-tRNAiMet in the P site of the 80S complex.
At this time we cannot determine whether this defect in stable Met-tRNAiMet binding is due to an altered structure of the 80S complex or due to lack of a specific contact between eIF5B and the Met-tRNAiMet. Domain IV of IF2, the bacterial ortholog of eIF5B, binds directly to the fMet portion of fMet-tRNAfMet (Guenneugues et al. 2000; Krafft et al. 2000; Spurio et al. 2000; Szkaradkiewicz et al. 2000). Whereas eIF5B has not been shown to directly and stably bind Met-tRNAiMet, eIF5B has been found to substitute for eIF2 and promote Met-tRNAiMet binding during translation of certain viral mRNAs (Pestova et al. 2008; Terenin et al. 2008). Moreover, aIF5B has been reported to protect Met-tRNAiMet from deacylation (Guillon et al. 2005). By analogy with bacterial IF2, perhaps domain IV of eIF5B contacts the Met-tRNAiMet on the 80S ribosome and stabilizes its binding. Accordingly, shortening or increasing the flexibility of helix H12 impairs this ability of eIF5B to stabilize Met-tRNAiMet binding. Alternatively, the length and rigidity of helix H12 might be important for eIF5B to trigger structural changes in the ribosome that stabilize Met-tRNAiMet binding in the P site.
While no structure of intact bacterial IF2 has been reported, structures of individual domains corresponding to aIF5B domain III (Wienk et al. 2005) and domain IV (Meunier et al. 2000) have been resolved. The region of IF2 corresponding to helix H12 in aIF5B was not resolved in either of these structures and was proposed to adopt a different structure based on homology modeling (Marzi et al. 2003). Consistent with this notion, a cryo-EM structure of IF2 bound to the 30S ribosomal subunit suggested that the residues corresponding to helix H12 in aIF5B form an unstructured loop connecting the regions corresponding to domains III and IV, which following a shift of domain III now lie adjacent to one another (Simonetti et al. 2008). Our data suggest that this proposed structural reconfiguration of IF2 is not conserved in eIF5B. The 6Gly mutation, which would be compatible with the proposed random coil structure of H12 in the cryo-EM structure of IF2, severely impaired eIF5B function. Moreover, the Swap5 mutation, designed to maintain the helical nature of H12, retained near WT eIF5B activity both in vivo and in the 80S complex formation assay in vitro. It will be interesting to test a similar Gly substitution mutant in IF2 to determine whether IF2 and eIF5B have different requirements for the connection between the cup and base of the chalice-shaped factors.
The crystal structure of aIF5B revealed structural transitions in the factor upon GTP versus GDP binding (Roll-Mecak et al. 2000). The transitions are initiated in the GTP-binding site and include reorientation of the Switch 2 element to contact the nucleotide and bound Mg2+ ion. The structural transitions in the G domain trigger movements in the other domains of aIF5B. Specifically, domain II rotates 8° toward the G domain triggering a 7° rotation of domain III. These domain movements in the cup of the chalice are then transmitted to the N terminus of helix H12. Acting like a pendulum, a 1 Å movement at the top of the 40 Å long helix H12 results in an ∼4–5 Å swing of domain IV. Similar domain reorientations were also observed in the cryo-EM images of 30S initiation complexes containing IF2•GDP versus IF2•GTP (Myasnikov et al. 2005; Simonetti et al. 2008). Mutational and biochemical studies on yeast eIF5B revealed that GTP hydrolysis was required for eIF5B and eIF1A dissociation from the ribosome following subunit joining (Shin et al. 2002, 2007, 2009; Acker et al. 2009). Thus, GTP hydrolysis functions as a regulatory switch governing the ribosome association of eIF5B (Shin et al. 2002). Taken together, it seemed reasonable to propose that the domain reorientations in eIF5B including the swing of domain IV were critical to govern the ribosome binding of the factor. However, here we show that the helix H12 mutants retain the GTP switch governing ribosome binding and the uncoupled ribosome-dependent GTPase activity. Moreover, ribosome binding by the ΔD4 mutant, completely lacking the C-terminal part of helix H12 and the entire domain IV, is still controlled by the GTP/GDP switch. Thus, the GTP-regulated switch for ribosome binding resides in the G domain and/or domains II and III of eIF5B. Consistent with this notion, we previously characterized an interaction between domain II of eIF5B and the 18S rRNA helix h5. Mutations in eIF5B or the ribosome that disrupt this interaction suppress eIF5B mutants that lack GTPase activity and thus have constitutively high ribosome binding affinity (Shin et al. 2009). Perhaps the rotation of domain II in WT eIF5B upon GTP hydrolysis disrupts this and similar interactions with the body of the 40S subunit to weaken ribosome binding and enable dissociation of the factor following subunit joining. Future experiments will investigate both the eIF5B–ribosome contacts that are disrupted upon GTP hydrolysis to enable eIF5B dissociation from the ribosome following subunit joining, and how the length and rigidity of helix H12 contribute to stable Met-tRNAiMet association with the 80S product of subunit joining.
MATERIALS AND METHODS
Plasmids and strains
The plasmid pC1285 (Shin et al. 2007) carrying an N-terminally deleted version of the yeast FUN12 gene encoding eIF5B with a deletion of residues 28 to 396 was used for all in vivo studies. Saccharomyces cerevisiae strain J111 (MATα ura3-52 leu2-3 leu2-112 fun12Δ) (Choi et al. 2000) was used for the examination of genetic phenotypes of eIF5B mutants. For the purification of wild-type or mutant forms of GST-eIF5B396-1002 from yeast, the eIF5B gene encoding residues 396–1002 was cloned to the yeast expression vector pEG-KT (Mitchell et al. 1993) and the resulting plasmids were introduced into the wild-type yeast strain H1511 (MATα ura3-52 leu2-3 leu2-112 trp1-63).
Biochemical analyses
For Western analysis of eIF5B, whole cell extracts were prepared using trichloroacetic acid extraction as previously described (Reid and Schatz 1982). The purification of GST-eIF5B fusion proteins was performed using Glutathione Sepharose 4B affinity chromatography as described previously (Shin and Dever 2007). Measurements of Kd values for GTP binding to eIF5B were performed using the fluorescent guanine nucleotide analog N-methylanthraniloyl (Mant)-labeled GDP (Molecular Probes and Invitrogen). Competition experiments with varying concentrations of GTP for the measurement of Kd values were performed as previously described (Shin et al. 2007). Ribosome binding assays for the measurement of eIF5B binding to 80S ribosomes, and ribosome-dependent uncoupled GTPase assays were performed as previously described (Shin and Dever 2007).
Ribosomal subunit joining assay
The kinetic parameters of ribosomal subunit joining were measured using a light scattering assay on an SX.180MV-R stopped-flow fluorometer (Applied Photophysics), as previously described (Acker et al. 2009). The final concentrations of components were as follows: 100 nM each of eIF1, eIF2, and Met-tRNAiMet; 50 nM eIF1A and 40S subunits; 1 μM mRNA; 250 nM eIF5; 500 nM eIF5B; 100–1200 nM 60S subunits; and 1 mM GTP•Mg2+. Briefly, 43S–mRNA complexes were formed by mixing preformed eIF2–GTP–Met-tRNAiMet ternary complexes with eIF1, eIF1A, mRNA, and 40S subunits. The 43S–mRNA complexes were then incubated with an initiation complex mix containing eIF5, eIF5B, and 60S subunits. The progress of subunit joining was monitored using a stopped-flow fluorometer by measuring the intensity of 435 nm light scattered at a right angle to the incident beam as a function of time. Native gel assays monitoring 80S complex formation were performed as described previously (Acker et al. 2009). The concentrations of the components in the 80S complex formation assay were as follows: 800 nM eIF2; 200 μM GTP•Mg2+ (for TC formation); 0.5 nM [35S]Met-tRNAiMet; 800 nM eIF1; 400 nM eIF1A; 800 nM mRNA; 400 nM of 40S and 60S subunits; 800 nM eIF5; 500 nM eIF5B; and 2 mM GTP•Mg2+ (for chase). The limiting Met-tRNAiMetconcentrations were used to achieve pseudo-first order conditions and to ensure single-round turnover in the reactions.
ACKNOWLEDGMENTS
We thank Alan Hinnebusch, Antonina Roll-Mecak, and our colleagues in the Dever, Lorsch, and Hinnebusch laboratories for advice and helpful discussions. This work was supported in part by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health (T.E.D.), and by American Cancer Society grant RSG-03-156-01-GMC (J.R.L.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2412511.
REFERENCES
- Abastado JP, Miller PF, Jackson BM, Hinnebusch AG 1991. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol Cell Biol 11: 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acker MG, Shin BS, Dever TE, Lorsch JR 2006. Interaction between eukaryotic initiation factors 1A and 5B is required for efficient ribosomal subunit joining. J Biol Chem 281: 8469–8475 [DOI] [PubMed] [Google Scholar]
- Acker MG, Shin BS, Nanda JS, Saini AK, Dever TE, Lorsch JR 2009. Kinetic analysis of late steps of eukaryotic translation initiation. J Mol Biol 385: 491–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J 2005. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 121: 703–712 [DOI] [PubMed] [Google Scholar]
- Choi SK, Lee JH, Zoll WL, Merrick WC, Dever TE 1998. Promotion of Met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science 280: 1757–1760 [DOI] [PubMed] [Google Scholar]
- Choi SK, Olsen DS, Roll-Mecak A, Martung A, Remo KL, Burley SK, Hinnebusch AG, Dever TE 2000. Physical and functional interaction between the eukaryotic orthologs of prokaryotic translation initiation factors IF1 and IF2. Mol Cell Biol 20: 7183–7191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fringer JM, Acker MG, Fekete CA, Lorsch JR, Dever TE 2007. Coupled release of eukaryotic translation initiation factors 5B and 1A from 80S ribosomes following subunit joining. Mol Cell Biol 27: 2384–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CM, Miller PF, Hinnebusch AG 1994. Requirements for intercistronic distance and level of eIF-2 activity in reinitiation on GCN4 mRNA varies with the downstream cistron. Mol Cell Biol 14: 2616–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenneugues M, Caserta E, Brandi L, Spurio R, Meunier S, Pon CL, Boelens R, Gualerzi CO 2000. Mapping the fMet-tRNAfMet binding site of initiation factor IF2. EMBO J 19: 5233–5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon L, Schmitt E, Blanquet S, Mechulam Y 2005. Initiator tRNA binding by e/aIF5B, the eukaryotic/archaeal homologue of bacterial initiation factor IF2. Biochemistry 44: 15594–15601 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG 1985. A hierarchy of trans-acting factors modulate translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol 5: 2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450 [DOI] [PubMed] [Google Scholar]
- Krafft C, Diehl A, Laettig S, Behlke J, Heinemann U, Pon CL, Gualerzi CO, Welfle H 2000. Interaction of fMet-tRNA(fMet) with the C-terminal domain of translational initiation factor IF2 from Bacillus stearothermophilus. FEBS Lett 471: 128–132 [DOI] [PubMed] [Google Scholar]
- Lee JH, Choi SK, Roll-Mecak A, Burley SK, Dever TE 1999. Universal conservation in translation initiation revealed by human and archaeal homologs of bacterial translation initiation factor IF2. Proc Natl Acad Sci 96: 4342–4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Pestova TV, Shin BS, Cao C, Choi SK, Dever TE 2002. Initiation factor eIF5B catalyzes second GTP-dependent step in eukaryotic translation initiation. Proc Natl Acad Sci 99: 16689–16694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marintchev A, Kolupaeva VG, Pestova TV, Wagner G 2003. Mapping the binding interface between human eukaryotic initiation factors 1A and 5B: A new interaction between old partners. Proc Natl Acad Sci 100: 1535–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi S, Knight W, Brandi L, Caserta E, Soboleva N, Hill WE, Gualerzi CO, Lodmell JS 2003. Ribosomal localization of translation initiation factor IF2. RNA 9: 958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier S, Spurio R, Czisch M, Wechselberger R, Guenneugues M, Gualerzi CO, Boelens R 2000. Structure of the fMet-tRNAfMet-binding domain of B. stearothermophilus initiation factor IF2. EMBO J 19: 1918–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Marshall TK, Deschenes RJ 1993. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast 9: 715–722 [DOI] [PubMed] [Google Scholar]
- Myasnikov AG, Marzi S, Simonetti A, Giuliodori AM, Gualerzi CO, Yusupova G, Yusupov M, Klaholz BP 2005. Conformational transition of initiation factor 2 from the GTP- to GDP-bound state visualized on the ribosome. Nat Struct Mol Biol 12: 1145–1149 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CUT 2000. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403: 332–335 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Lorsch JR, Hellen CUT 2007. The Mechanism of Translation Initiation in Eukaryotes. In Translational control in biology and medicine (ed. Mathews M, et al. ), pp. 87–128 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU 2008. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J 27: 1060–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid GA, Schatz G 1982. Import of proteins into mitochondria. Extramitochondrial pools and post-translational import of mitochondrial protein precursors in vivo. J Biol Chem 257: 13062–13067 [PubMed] [Google Scholar]
- Roll-Mecak A, Cao C, Dever TE, Burley SK 2000. X-ray structures of the universal translation initiation factor IF2/eIF5B. Conformational changes on GDP and GTP binding. Cell 103: 781–792 [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A, Shin BS, Dever TE, Burley SK 2001. Engaging the ribosome: Universal IFs of translation. Trends Biochem Sci 26: 705–709 [DOI] [PubMed] [Google Scholar]
- Shin BS, Dever TE 2007. Molecular genetic structure-function analysis of translation initiation factor eIF5B. Methods Enzymol 429: 185–201 [DOI] [PubMed] [Google Scholar]
- Shin BS, Maag D, Roll-Mecak A, Arefin MS, Burley SK, Lorsch JR, Dever TE 2002. Uncoupling of initiation factor eIF5B/IF2 GTPase and translational activities by mutations that lower ribosome affinity. Cell 111: 1015–1025 [DOI] [PubMed] [Google Scholar]
- Shin BS, Acker MG, Maag D, Kim JR, Lorsch JR, Dever TE 2007. Intragenic suppressor mutations restore GTPase and translation functions of a eukaryotic initiation factor 5B switch II mutant. Mol Cell Biol 27: 1677–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin BS, Kim JR, Acker MG, Maher KN, Lorsch JR, Dever TE 2009. rRNA suppressor of a eukaryotic translation initiation factor 5B/initiation factor 2 mutant reveals a binding site for translational GTPases on the small ribosomal subunit. Mol Cell Biol 29: 808–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti A, Marzi S, Myasnikov AG, Fabbretti A, Yusupov M, Gualerzi CO, Klaholz BP 2008. Structure of the 30S translation initiation complex. Nature 455: 416–420 [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurio R, Brandi L, Caserta E, Pon CL, Gualerzi CO, Misselwitz R, Krafft C, Welfle K, Welfle H 2000. The C-terminal subdomain (IF2 C-2) contains the entire fMet-tRNA binding site of initiation factor IF2. J Biol Chem 275: 2447–2454 [DOI] [PubMed] [Google Scholar]
- Szkaradkiewicz K, Zuleeg T, Limmer S, Sprinzl M 2000. Interaction of fMet-tRNAfMet and fMet-AMP with the C-terminal domain of Thermus thermophilus translation initiation factor 2. Eur J Biochem 267: 4290–4299 [DOI] [PubMed] [Google Scholar]
- Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN 2008. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat Struct Mol Biol 15: 836–841 [DOI] [PubMed] [Google Scholar]
- Unbehaun A, Marintchev A, Lomakin IB, Didenko T, Wagner G, Hellen CU, Pestova TV 2007. Position of eukaryotic initiation factor eIF5B on the 80S ribosome mapped by directed hydroxyl radical probing. EMBO J 26: 3109–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienk H, Tomaselli S, Bernard C, Spurio R, Picone D, Gualerzi CO, Boelens R 2005. Solution structure of the C1-subdomain of Bacillus stearothermophilus translation initiation factor IF2. Protein Sci 14: 2461–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]