Abstract
Four decades after early in vitro assembly studies demonstrated that ribosome assembly is a controlled process, our understanding of ribosome assembly is still incomplete. Just as structure determination has been so important to understanding ribosome function, so too will it be critical to sorting out the assembly process. Here, we used a viable deletion in the yjeQ gene, a recognized ribosome assembly factor, to isolate and structurally characterize immature 30S subunits assembled in vivo. These small ribosome subunits contained unprocessed 17S rRNA and lacked some late ribosomal proteins. Cryo-electron microscopy reconstructions revealed that the presence of precursor sequences in the rRNA induces a severe distortion in the 3′ minor domain of the subunit involved in the decoding of mRNA and interaction with the large ribosome subunit. These findings suggest that rRNA processing events induce key local conformational changes directing the structure toward the mature assembly. We concluded that rRNA processing, folding, and the entry of tertiary r-proteins are interdependent events in the late stages of 30S subunit assembly. In addition, we demonstrate how studies of emerging assembly factors in ribosome biogenesis can help to elucidate the path of subunit assembly in vivo.
Keywords: ribosome assembly, 30S subunit, YjeQ protein, 17S rRNA, rRNA processing, GTPase
INTRODUCTION
The ribosome constitutes perhaps one of the most intricate macromolecular complexes in the cell and assembles with remarkable reliability. The high-resolution atomic models of the ribosome (Ban et al. 2000; Schluenzen et al. 2000; Wimberly et al. 2000; Harms et al. 2001; Yusupov et al. 2001; Schuwirth et al. 2005) provide the detailed structure of the end product of the assembly line. However, very little information about the assembly process itself can be interpreted from the inspection of these structures.
Ribosomal subunit assembly has been the subject of more than 40 yr of investigation that began with seminal studies by the Nomura laboratory in the 1960s to map the reconstitution of the 30S subunit in vitro (Hosokawa et al. 1966; Traub and Nomura 1968a,b, 1969). Interest in this field has resurfaced in the last several years, sparked by the high-resolution X-ray structures of the ribosome and by the development of new techniques for the study of the kinetics and mechanism of rRNA folding in subunit assembly in vitro (Talkington et al. 2005; Adilakshmi et al. 2008). Together these studies have considerably advanced our understanding of the fundamental driving forces of assembly, including RNA folding, protein binding order, and the possible existence of intermediates. However, just as structures of the intermediates in protein synthesis have transformed our understanding of ribosome function (Frank 2003; Mitra and Frank 2006), so too will structural studies constitute the next frontier in ribosome assembly.
The 70S ribosome in Escherichia coli is composed of a large 50S and a small 30S subunit. The 30S subunit consists of one 16S ribosomal RNA (rRNA) molecule and 21 ribosomal proteins (r-proteins), and the 50S subunit is made of two RNA molecules, the 23S and 5S rRNAs and 34 proteins. Bacterial ribosome assembly commences with the transcription of rRNA as a single precursor transcript containing the three rRNAs (Srivastava and Schlessinger 1990). Processing of the primary rRNA transcript occurs very rapidly and begins before transcription is completed. RNase III performs the primary processing that separates the three rRNAs. The resulting fragments are called precursor rRNAs and contain additional nucleotides at both their 5′ and 3′ ends (Srivastava and Schlessinger 1990). Subsequent secondary processing events cleave the extra nucleotides and produce the mature rRNA molecules. In particular, the precursor rRNA for the 30S ribosomal subunit (called 17S rRNA), contains an additional 115 and 33 nucleotides at the 5′ and 3′ ends, respectively. The coordinated action of RNase E and RNase G removes the precursor sequence at the 5′ terminus (Li et al. 1999). Processing of the 3′ end is less well characterized but may occur in a single cleavage step that removes the 33 extra nucleotides at this end of the molecule (Hayes and Vasseur 1976).
After transcription, the 17S rRNA is thought to form local secondary structures rapidly prior to trimming of the 5′ and 3′ precursor sequences. The initially folded precursor rRNA is quickly recognized and bound by the r-proteins. Vintage experiments by Nomura (Hosokawa et al. 1966; Traub and Nomura 1968a,b, 1969) and more recent experiments from the Williamson and Woodson laboratories (Talkington et al. 2005; Adilakshmi et al. 2008) have defined the hierarchy and kinetic pathway of binding of the 21 r-proteins of the 30S subunit to mature 16S rRNA in vitro. The r-proteins are designated as primary (bind directly to the rRNA), secondary (binding is dependent on primary binding r-proteins), or tertiary (binding is dependent on secondary binding r-proteins). Using pulse-chase labeling quantified by mass spectrometry, Williamson and colleagues (Talkington et al. 2005) have presented compelling evidence to suggest that protein binding to the rRNA drives conformational rearrangements that stabilize the native fold of the 30S subunit. This paradigm has been strengthened by Woodson and coworkers (Adilakshmi et al. 2008), who have used time-resolved X-ray hydroxyl radical foot-printing to show multiple early folding nucleation events and induced fit of protein–rRNA complexes. Binding of primary proteins appears to stabilize the local RNA structure and induces conformational changes that create new binding sites for secondary proteins. Thus in vitro studies using mature 16S rRNA suggest that assembly proceeds by an alternating series of RNA conformational changes and protein binding events until the mature structure is produced. A long-standing question, however, is the predictive strength of these in vitro experiments to the assembly processes occurring in vivo with freshly transcribed precursor rRNA and newly expressed ribosomal proteins.
As described in early studies by Nomura, the in vitro reconstitution of intact ribosomal subunits (and even functional ribosomes) is possible by providing the processed rRNA and r-proteins (Mizushima and Nomura 1970). However, in vitro ribosomal assembly requires conditions that are far from physiological and occurs much more slowly than in vivo. Indeed, trans-acting protein factors, including ribonucleases, helicases, rRNA (and r-protein) modification enzymes, and GTPases are increasingly recognized as having a role in subunit assembly in the cell (Wilson and Nierhaus 2007). While the details of their mechanism of action are largely unknown, the deletion of genes encoding many of these factors causes accumulation of precursor rRNAs (Wilson and Nierhaus 2007; Connolly and Culver 2009). Perhaps the most notable and enigmatic of these are the GTPases (Brown 2005) including the YjeQ protein from E. coli. YjeQ is a slow GTPase that is highly stimulated by purified 30S subunits (Daigle et al. 2002; Daigle and Brown 2004; Himeno et al. 2004). Disruption of the yjeQ gene in E. coli severely affects growth, likely due to the accumulation of ribosomal subunits (Daigle and Brown 2004; Himeno et al. 2004; Campbell et al. 2005; Campbell and Brown 2008). In addition, a large proportion of the 30S ribosomal subunits in a yjeQ deletion strain of E. coli (ΔyjeQ) are immature as they contain a precursor of the 16S rRNA molecule (Himeno et al. 2004). In vitro, recombinant YjeQ interacts strongly with the 30S ribosomal subunit (Daigle and Brown 2004; Himeno et al. 2004), and this interaction results in a 160-fold stimulation of the YjeQ GTPase activity (Daigle and Brown 2004). The binding location of YjeQ to the 30S subunit is still unclear, and existing studies do not agree. Two studies suggested that YjeQ binds in close contact with the A-site and its binding induces conformational changes around the decoding center (Himeno et al. 2004; Kimura et al. 2008). Conversely, a chemical genetic-based study (Campbell et al. 2005) concluded that YjeQ binds at a distal site on the ribosome, which is different from the A-site.
In the present work, we used the ΔyjeQ strain of E. coli to purify immature 30S subunits that were assembled in vivo. We showed by Northern blotting that the 16S rRNA of these small subunits was incompletely processed, and using isotope tagging proteomic methodology, we demonstrated that 30S isolated from the ΔyjeQ strain had an incomplete protein complement. The three-dimensional reconstruction obtained by cryo-electron microscopy (cryo-EM) of these 30S subunits revealed a profound distortion of helix 44 that is located in the proximity (∼25 Å) of the 3′ mature terminus of its 16S rRNA. Helix 44 is a structural element in the 30S subunit crucial to the decoding of mRNA and interaction with the large ribosome subunit. This finding indicates that processing of the 3′ precursor sequence of the 17S rRNA is critical to the development of a conformation compatible with 50S binding and protein synthesis. Our results highlight the importance of the rRNA processing events in the assembly process and confirm that the YjeQ protein is a bona fide ribosome assembly factor assisting in late stages of assembly and link its function to processing of the 17S rRNA.
RESULTS
Maturation of both the 5′ and 3′ termini of the 17S rRNA is delayed in an E. coli ΔyjeQ strain
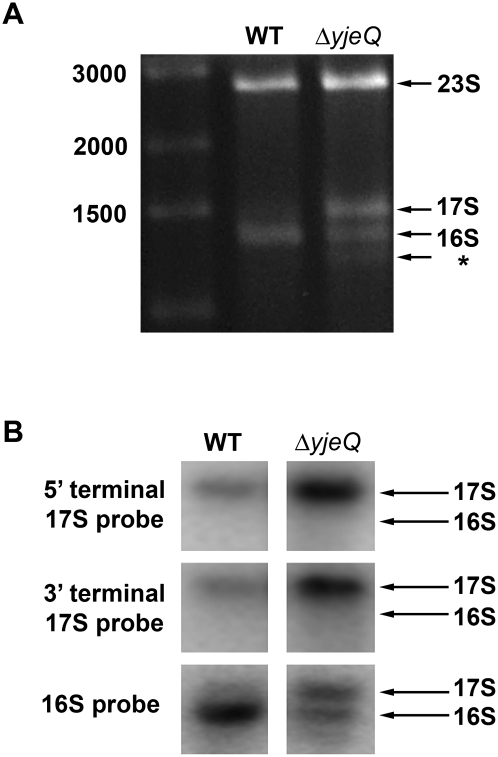
We isolated total cellular RNA from an E. coli deletion strain lacking the gene encoding the ribosome assembly factor YjeQ. The purified RNA was resolved and visualized in 0.7% modified agarose gels (Fig. 1A). In addition to the species corresponding to the mature 16S rRNA molecules seen in wild-type cells, we also observed a band of slower mobility corresponding to a precursor form of this molecule. A third band with faster mobility was also visible. The existence of this latter band is consistent with previous studies in ΔyjeQ cells (Himeno et al. 2004), and it may constitute either a 16S rRNA degradation product or an aberrantly processed rRNA produced in the absence of YjeQ.
FIGURE 1.
Characterization of the rRNA from E. coli ΔyjeQ strain. (A) Electrophoretic analysis of rRNA derived from the wild type and yjeQ-null strain cell extracts. The wild-type gel pattern exhibited only two bands with migration corresponding to mature 16S and 23S rRNA. Two additional bands were observed in the yjeQ deletion strain corresponding to a precursor form of 16S rRNA (17S label) and a previously identified species suspected to be a degradation product or an aberrantly processed rRNA (asterisk). The sizes of the fragments of the DNA ladder in the left lane of the gel are indicated. (B) The identity of the precursor 16S rRNA found in the yjeQ-null strain was confirmed by Northern blot analysis using radiolabeled DNA sequence-specific probes directed at the 5′- and 3′-terminal sequences of 17S rRNA. Hybridization of these probes with the rRNA precursor derived from the yjeQ deletion cell extract indicated that the species was indeed unprocessed 17S rRNA with intact 5′ and 3′ terminal sequences. Mobility of the 16S rRNA and 17S rRNA is indicated.
We used Northern hybridization to determine if the 5′ and 3′ precursor sequences were present in the immature 16S rRNA molecule accumulated in ΔyjeQ cells. Transferred membranes were complemented with end fragments of the 5′ and 3′ precursor sequences of 17S rRNA, and, indeed, both the 5′ and 3′ precursor sequences were found (Fig. 1B). These results indicate that in cells lacking the yjeQ gene, the secondary processing reactions leading to the production of mature 16S rRNA are delayed. Consequently, these bacteria accumulate unprocessed 17S rRNA molecules with precursor sequences at both, the 5′ and 3′, ends. Furthermore, our findings confirm that the YjeQ protein is a bona fide ribosome assembly factor with a role in processing the precursor 17S rRNA.
Protein complement of the immature 30S ribosomal subunits purified from E. coli ΔyjeQ cells
Next, we aimed to identify the protein complement present in the immature 17S rRNA–containing 30S subunits from ΔyjeQ cells and compared their relative protein levels with the mature, wild-type, 16S rRNA–containing 30S subunits using the iTRAQ (isobaric tag for relative and absolute quantification) method (Ross et al. 2004). Considering that ΔyjeQ cells have ∼30% of mature 16S rRNA–containing 30S subunits (Fig. 1A) and that they are mostly associated with large ribosomal subunits forming 70S ribosomes (Himeno et al. 2004), we purified the 30S fraction from the ΔyjeQ cells under nondissociating conditions in order to eliminate most of the mature 16S rRNA–containing 30S subunits and produce a preparation enriched in immature 17S rRNA–containing small subunits. Similarly, mature 16S rRNA–containing 30S subunits were prepared from wild-type E. coli for comparative purposes.
We performed a double duplex isobaric labeling experiment (Jiang et al. 2007) to find the proteins present in our two 30S subunit preparations. We identified 1207 peptides to give us 41 unique proteins identified by at least two different peptides at 95% confidence (Supplemental Table 1). The identification was comprehensive and included the 21 r-proteins of the 30S subunit (Fig. 2; Supplemental Table 1). We also identified 11 r-proteins from the 50S subunit and nine nonribosomal proteins in these purified preparations (Supplemental Fig. S1; Supplemental Table 1). Identification of these proteins suggests a minor contamination of our 30S subunit preparations with 50S subunits and other proteins, most of them unrelated to the ribosome.
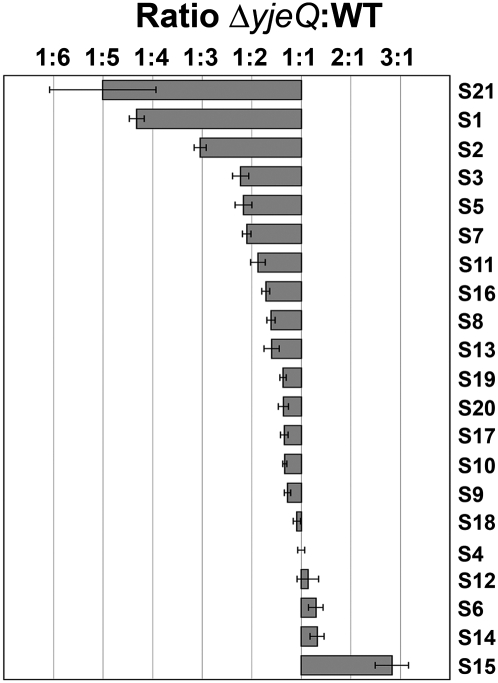
FIGURE 2.
iTRAQ analysis of the 30S subunits purified from ΔyjeQ cells. Small subunit r-proteins. The plot shows the relative levels of the small subunit r-proteins between the 30S subunit purified from ΔyjeQ and wild-type (WT) cells. Relative levels for each protein are expressed as the average ratio ΔyjeQ:WT obtained from two replicas of the experiment. Only peptides that identify proteins with ≥95% confidence were used for the calculation of these ratios. The error bars indicate the standard error of the mean for each ratio.
The iTRAQ double duplex isobaric labeling was also used to compare the relative amounts of the proteins in the 30S subunits purified from ΔyjeQ (largely immature 30S subunits) and wild-type cells (largely mature 30S subunits). Results from this analysis were expressed as the ΔyjeQ:WT and WT:ΔyjeQ ratios for the 30S subunit r-proteins and for the other identified proteins (Fig. 2; Supplemental Fig. S1; Supplemental Table 1). These figures and table provided two levels of analysis of the data. Only peptides that identified proteins with ≥95% confidence were used for the calculation of the ratios shown in Figure 2 and Supplemental Figure S1. Additional ratios are presented in Supplemental Table 1 that include some peptide identification data that lie outside of this strict confidence interval. Both analyses concluded that the 30S subunit r-protein complement was largely the same in the immature and mature 30S subunits purified from ΔyjeQ and wild-type cells, respectively. However, three r-proteins—S21, S1, and S2—were found to be significantly underrepresented in the immature 30S subunits (Fig. 2) with a high degree of confidence (P-values for these proteins in two biological replicas were ≤0.05; see Supplemental Table 1). In addition, r-proteins S3, S5, S7, S11, S16, S8, and S13 were found slightly underrepresented in the immature 30S subunits (Fig. 2). Interestingly, S15 was slightly underrepresented in the mature 30S subunit (Fig. 2). This was a curious result that may well be consistent with a primary role for S15 in ribosome assembly rather than ribosome function (Bubunenko et al. 2006; Sykes and Williamson 2009), and thus, binding to the immature structure could conceivably be tighter than to the mature structure. Accordingly, S15 may have dissociated during the purification process from some of the mature 30S subunits prepared from wild-type cells but not from the immature particles from ΔyjeQ cells, resulting in the underrepresentation of this protein in the former. Alternatively, it is possible that S15 was bound nonspecifically to the immature particles in superstoichiometric amounts and generated this result. Our data cannot rule out either of these two possibilities.
In conclusion, this analysis indicated that the protein complement of the immature 30S subunits purified from ΔyjeQ cells was similar to the mature small subunits except for the r-proteins S21, S1, and S2, which were significantly underrepresented. These findings suggest that in the absence of YjeQ, binding of these proteins is weaker and/or their entry in the assembling 30S subunit is delayed. Interestingly, these r-proteins are tertiary proteins that bind to the 30S subunit only at the latest stages of the assembly process, which indicates that while immature, these 30S particles are at a late stage in the subunit assembly process (Mizushima and Nomura 1970; Culver 2003).
The immature 30S subunit from ΔyjeQ cells features a distorted helix 44 and decoding center
The structure of the immature 30S subunit purified from ΔyjeQ cells was obtained (Fig. 3, upper right panel) using cryo-EM, image classification, and three-dimensional (3D) reconstruction techniques. To this end, 30S subunits projections were selected from the electron micrographs. The contamination of the immature 30S subunit preparation with 50S subunits revealed by the iTRAQ technique (Supplemental Fig. S1; Supplemental Table 1) was very minor, and the electron micrographs rarely showed any 50S subunit projections (Supplemental Fig. S2). These particles were easily excluded during the picking process. The supervised classification approach (Valle et al. 2002; Gao et al. 2004) described in Supplemental Figure S3 was first used to eliminate the projections representing mature 30S subunits (27.4%) from the data set. The structure of the immature 30S subunit (Fig. 3, upper right panel) was then obtained from the remaining projections in the data set, and the structure was refined to 11.6 Å resolution (Supplemental Fig. S4). In addition, a homogeneous preparation of mature 16S rRNA–containing 30S subunits purified from wild-type E. coli cells was used to produce a control cryo-EM reconstruction that we used to evaluate the structural defects in the immature structure (Fig. 3, upper left panel).
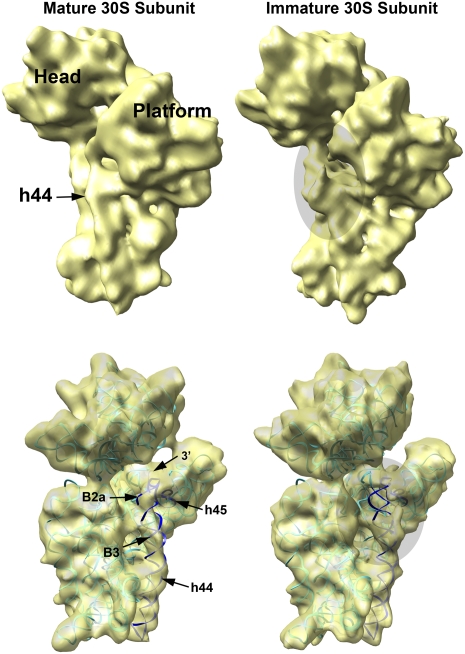
FIGURE 3.
3D reconstruction of the immature 30S ribosomal subunits purified from ΔyjeQ E. coli cells. Platform view of the cryo-EM maps of the immature (upper right) and mature (upper left) 30S subunits. The immature structure was produced from 42,873 projections of 30S subunits purified from ΔyjeQ cells after 16,180 projections (27.4% of the total) representing mature small subunits were removed from the data set using a supervised classification approach. The mature structure (left) was produced from 19,924 projection images obtained from a homogeneous sample of mature 16S rRNA-containing 30S subunits purified from wild-type cells. Main landmarks of the 30S subunit are indicated. In the panels below, the X-ray structure of the wild-type 30S subunit, shown as a ribbon representation, was fitted into the cryo-EM maps of both the mature (left) and immature (right) 30S subunits to illustrate their divergences. In the X-ray structure, the rRNA is shown in cyan except helices 44 and 45, which are dark blue. The r-proteins are not shown. Main landmarks of the 30S subunit indicated include helix 44 (h44), helix 45 (h45), the intersubunit bridges B2a (B2a) and B3 (B3) important for the interaction with the 50S subunit, and the 3′ terminus of the 16S rRNA molecule (3′). The shadowed ovals in the right panels show the area in the helix 44 and decoding center distorted in the EM map of the immature 30S subunit.
When compared to the control reconstruction, the structure of the immature 30S subunit showed the main landmarks existing in the mature subunit, suggesting that these particles represent a late stage of assembly in which the main structural domains of the 30S subunit have been assembled (Fig. 3, upper panels). However, the reconstruction of the immature subunit revealed that the conformation of the 3′ minor domain, including helix 44, was clearly distorted. In particular, the upper segment of helix 44, responsible for tRNA binding, was displaced outwardly, protruding from the interface of the 30S subunit that contacts the large 50S subunit (Fig. 3, lower right panel; Supplemental Movie S1, right panel). In the mature 30S subunit, this structural motif forms part of the decoding center and then further meanders toward helix 45 and the 3′ terminus of the 16S rRNA (Fig. 3, lower left panel; Supplemental Movie S1, left panel). In the immature structure, the distortion in helix 44 is likely also displacing helix 45 and the unprocessed 3′ terminus to an outward flexible position. These changes indicate that the decoding center and two inter-subunit bridges with the 50S subunit (B3 and B2a) are still not fully structured (Fig. 3, lower panels).
The very significant distortions observed in helix 44 and the decoding center of immature 30S subunits isolated from the yjeQ deletion strain suggest that the presence of the precursor sequences (in particular, the one at the 3′ end) imposes a nonfunctional conformation in the 30S subunit and that rRNA processing is required to gain a conformation compatible with 50S binding and protein synthesis.
Conformational differences in the immature 30S subunit relative to the mature structure
To obtain further details on the displacement of helix 44 as well as to analyze the conformational differences between the immature and mature 30S subunit structure, we calculated the “forward” difference map by subtracting the control map of the mature 30S subunit from wild-type cells (Fig. 3, upper left panel) from that of the immature 30S subunit from ΔyjeQ cells (Fig. 3, upper right panel). In addition, to differentiate those regions in the EM map of the immature 30S subunit in a different conformation from areas lacking or containing an additional density, we also calculated the “reverse” difference map (i.e., mature 30S subunit map minus immature 30S map). Overlaying these two maps allowed us to study the differences between the two structures (Fig. 4; Supplemental Movie S2).
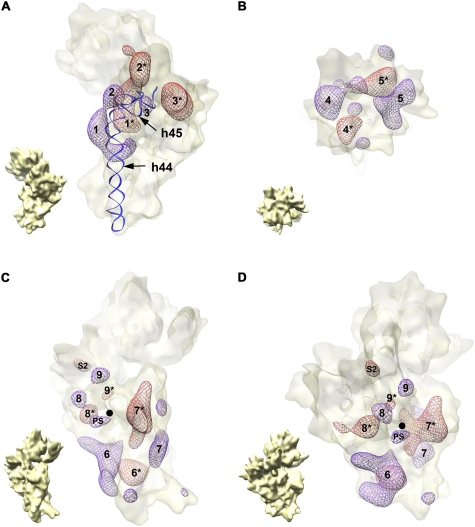
FIGURE 4.
Difference map analysis of the immature 30S subunit cryo-EM structure. Overlay of the forward and reverse difference maps. Densities from the forward (violet mesh) and reverse (red mesh) difference maps are shown contained in a semitransparent surface representation of the immature 30S subunit structure from ΔyjeQ cells. Matching densities are identified with the same number, and the density in the reverse map is identified by an asterisk (*). (A) The densities present in the 3′ minor (helices 44 and 45) and central domains of the two difference maps. Similarly, panel B displays only the densities present in the 3′ major domain of these difference maps, and panels C and D show two views of the densities existing on the 5′ domain. The small cryo-EM map in the bottom left corner of each panel is an aid for orientation and represents the immature 30S subunit in the same view as the difference maps in the corresponding panel. Helices 44 (h44) and 45 (h45) are also shown as a ribbon representation in A to illustrate the displacement of helix 44 in the immature 30S subunit structure. The 5′ end of the mature 16S rRNA is indicated with a black dot in C and D. The density labeled as “S2” in C and D indicates the amount of density corresponding to this protein missing in the immature structure. The density labeled as “PS” in the same panels represents the additional density existing in the immature structure due to the presence of the 5′ precursor sequence of the rRNA.
Most of the densities observed in the forward map had a nearby density in the reverse map (matching densities are identified with the same number in Fig. 4; the density in the reverse map is identified by an asterisk) highlighting movement and/or conformational variations in the immature subunit with respect to the mature structure. Consistent with the large distortion observed in the upper segment of helix 44 in the structure of the immature 30S subunit, the overlaid difference maps of the 3′ minor (helices 44 and 45) and central (platform) domains showed two matching densities (Fig. 4A, densities 1 and 1*) that describe the outward displacement of the helix in the immature subunit. Regarding the other three major domains of the small subunit (5′ [body], central [platform], 3′ major [head]), the surface rendering representation of the immature 30S subunit EM map showed that the structure of these domains in the immature subunit was very similar to the mature structure (Fig. 3, upper panels). However, we found nearby densities in the overlaid difference maps of all three domains (Fig. 4; Supplemental Movie S2), indicating that the relative orientation of the major structural domains in the immature subunit differs slightly from the mature subunit. This is consistent with a recently proposed model suggesting that folding of the different subunit domains occurs independently (Sykes and Williamson 2009) and that they are properly oriented at late stages of assembly by the formation of the central pseudoknot involving helix 2 and connecting the three major structural elements of the small subunit (Poot et al. 1998; Holmes and Culver 2004).
It is important to note that the nearby densities in the overlaid difference maps did not always match in size and shape, since parts of these densities were canceled out in the difference maps due to partial overlapping of the moved domains between the two structures. A proximity criterion was used to pair the densities between the forward and reverse difference maps (Fig. 4). The assignment of the pair 1 and 1* (Fig. 4, upper left panel) defining the movement of helix 44 between both structures was unambiguous, corroborating the distortion of this helix in the immature 30S subunit structure. Conversely, an alternative assignment of pairs was possible for some of the other densities. Nevertheless, a different assignment does not change our conclusion that the relative orientation of the major structural domains in the immature subunit differs slightly from the mature subunit.
The forward map failed to reveal any large densities that could be assigned to the 115 and 33 nucleotides of the 5′ and 3′ precursor sequences, respectively. Interestingly, we only observed a small density very close to the 5′ mature end of the rRNA that probably represents only the first few residues of the 5′ precursor sequence (Fig. 4C,D, density labeled as PS). Concerning the 3′ precursor sequence, the outward displacement of helix 44 probably displaced the helix 45 and the extra 33 nucleotides of the precursor sequence into the interface region, where it most likely adopted a flexible conformation. These results suggest that the precursor sequences are flexible and occupy a variable position in the structure, resulting in a smearing of their densities during the 3D reconstruction.
In addition, we expected to observe a density co-localizing with the S1, S21, and S2 r-proteins in the reverse map because they were found to be significantly underrepresented in the immature 30S subunits (Fig. 2). However, only S2 showed a corresponding density in the difference map (Fig. 4C,D, density labeled as S2). The size of this density was small compared to the size of the S2 protein, suggesting that there were still a significant number of immature subunits that contain this r-protein. A matching density corresponding to S21 and S1 was not observed in the reverse map. S21 is a r-protein located between the head and platform and probably not observed because the small size of this protein, S1, when present in the 30S subunit, fills the cleft region between the head and platform (Sengupta et al. 2001). However, a corresponding density was not observed in the EM maps of the immature and mature 30S subunits. This result was expected because the high salt conditions we used to purify the 30S subunits lead to a loss of the S1 protein. The small ΔyjeQ:WT ratio found for S1 in the iTRAQ experiment (Fig. 2; Supplemental Table 1) may reflect the fact that our purification conditions eliminated the S1 protein even more efficiently in the immature 30S subunits.
In summary, the difference map analysis presented here indicated that the relative orientation of the structural domains in the immature 30S subunit diverged somewhat from that of the mature subunit. Most importantly, it revealed that the immature 30S subunits did not constitute a structurally homogeneous population.
Helix 44 in the immature 30S subunit moves inward into its correct position during late stages of the maturation process
In order to structurally characterize the subpopulations of immature 30S subunits purified from ΔyjeQ cells, we further classified the projections that produced the 3D reconstruction of the immature 30S subunit (Fig. 3, upper right panel) using a maximum likelihood-based classification technique (Supplemental Fig. S5; Scheres et al. 2005a,b, 2007).
This procedure yielded three cryo-EM structures (Figs. 5, 6). The three EM maps were similar to the previously calculated 3D reconstruction of the immature 30S subunit (Fig. 3, upper right panel). Difference map analysis of each individual map with respect to the mature structure from wild-type cells showed that the relative orientation of the major structural domains in these three structures also diverged somewhat from that of the mature subunit (data not shown). However, they contained varying degrees of distortion in helix 44 (Fig. 5) and differed in the amount of density associated with the S2 r-protein in the neck of the subunit (Fig. 6). Conversely, a control classification of all the projections collected from the 30S subunit fraction purified from wild-type cells was performed using the same approach and rendered three very similar 3D reconstructions, all resembling the mature 30S subunit structure (Supplemental Fig. S6).
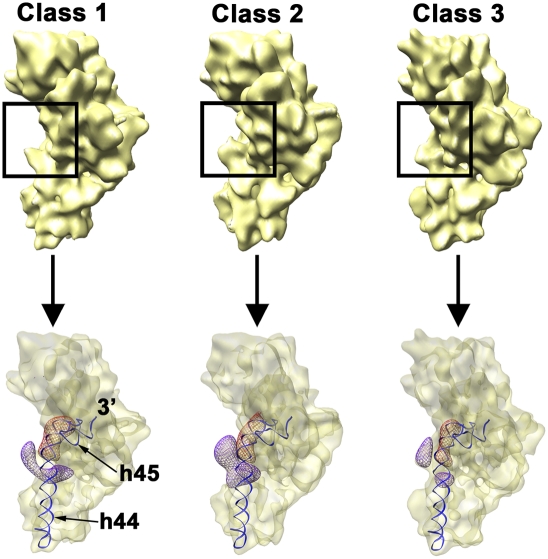
FIGURE 5.
Conformational variability of the immature 30S subunit from ΔyjeQ cells. Platform view of the three cryo-EM structures (labeled from class 1 to class 3) representing three subpopulations of immature 30S subunits present in ΔyjeQ cells (top). These 3D reconstructions were produced from 13,447 (class 1), 12,570 (class 2), and 11,931 (class 3) projections and refined to 14.1 Å (class 1), 15.4 Å (class 2), and 14.7 Å (class 3) resolution. The framed region in each map highlights the area of helix 44 that is distorted in the three subpopulations. The bottom panel illustrates the degree of distortion of helix 44 in each cryo-EM map. The density corresponding to the upper segment of helix 44 in each conformational state is shown as a violet mesh. The displacement of helix 44 in each conformation is shown by comparison to the position of the helix 44 in the mature 30S subunit shown as a red mesh. Helices 44 (h44) and 45 (h45) in the mature structure are also shown for reference.
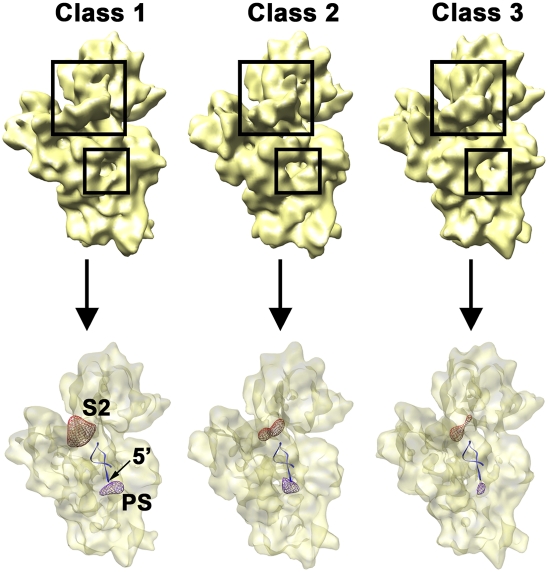
FIGURE 6.
Variability of the density representing the S2 r-protein and 5′ precursor sequences in the immature 30S subunit from ΔyjeQ cells. Back view of the three cryo-EM structures (labeled from class 1 to class 3) representing three subpopulations of immature 30S subunits present in ΔyjeQ cells (top). The framed regions highlight the location of the S2 r-protein and the 5′ mature end of the 16S rRNA. The dissimilar amount of density in these two locations in the cryo-EM maps is shown in the bottom panel. The density corresponding to the S2 r-protein (red mesh) from the reverse difference map (i.e., mature 30S subunit map minus immature 30S map) and the one corresponding to the 5′ precursor sequence (violet mesh) from the forward difference map (i.e., immature 30S subunit map minus mature 30S map) for each cryo-EM map are displayed within a semitransparent surface of the 30S subunit structure. The first one indicates the amount of density corresponding to the S2 r-protein missing in each immature structure. Conversely, the second one shows the additional density existing in each structure due to the presence of the 5′ precursor sequence. The first 20 nucleotides of the 5′ end of the mature 16S rRNA are shown as a ribbon representation.
Regarding the 3D reconstructions produced by the classification of the immature particles from ΔyjeQ cells, one of the maps presented the most severe distortion with respect to helix 44 (Fig. 5, upper panel, class 1 map, framed area) and showed this structural element protruding outward from the interfacial surface of the subunit. A second map presented a less severe distortion (Fig. 5, upper panel, class 3 map, framed area). The helix was still displaced outward but closer to the location in the mature structure. Finally, the third map showed an intermediate degree of distortion between the two previous structures (Fig. 5, upper panel, class 3 map, framed area). To observe the degree of displacement of helix 44 in these three structures, the forward and reverse difference maps from each structure were overlaid, and the corresponding densities for helix 44 were displayed (Fig. 5, bottom panel). This analysis revealed a progressive movement of helix 44 into correct position indicated by the decrease in size of the corresponding densities in the difference maps.
The density representing the S2 r-protein in the three maps of the immature 30S subunit was also variable (Fig. 6, large framed area). Interestingly, the map showing the largest distortion on helix 44 contained the least amount of density in this area (Fig. 6, class 1 map), indicating that the occupancy of S2 in this subpopulation was significantly decreased. This group of particles constitutes, in fact, the subpopulation causing the relative amount of S2 r-protein to be significantly underrepresented in the iTRAQ experiments (see above). Conversely, the EM map with helix 44 closer to the mature position showed comparable S2 density to the mature structure (Fig. 6, class 3 map). This result is consistent with our interpretation that this last structure represents an immature 30S subunit but it is closer to the mature state. In agreement with this statement is the finding that the amount of density that we have assigned to the 5′ precursor sequence in these structures (Fig. 5, small framed area) was largest in the EM map with the most severe distortion in helix 44 and lowest S2 density (class 1 map) and smaller in the structure with helix 44 and S2 densities similar to the mature particle (class 3 map).
In addition, one finding of the rRNA analysis performed on the ΔyjeQ cells was that apart from the 30S particles containing 17S rRNA, these cells also possessed degraded or aberrantly processed 16S rRNA, equivalent to ∼10% of the total rRNA (Fig. 1). We considered that these species may have been incorporated into 30S subunits. Interestingly, even when additional maximum likelihood–based classifications were performed assuming the existence of more than three classes in the data set, we consistently observed duplication in the obtained structures and it was possible to merge them into three, closely resembling the structures described above (data not shown). However, none of these classification experiments found a subpopulation that could be assigned to the particles containing the degraded rRNA. This result led us to conclude, first, that mainly three subpopulations of immature 30S subunit accumulate in ΔyjeQ cells and, second, that particles containing degraded rRNA may have diverged largely from the wild-type 30S subunit conformation. It is possible that these particles might have been excluded during the initial step of particle image selection, or, alternatively, they may not represent a structurally congruent class and thus, they were eliminated during the 3D reconstruction process.
In sum, through our particle classification approach, we have characterized three structures that highlight changing structural features spanning the immature and mature 30S subunits derived from a strain of E. coli that lacks the biogenesis factor YjeQ. Furthermore, the observed correlations between the distortion in helix 44 and the amount of density in the EM maps associated with S2 and the 5′ precursor sequence suggest that the processing of the rRNA precursor sequences, proper folding of the 3′ minor domain of the subunit (helices 44 and 45), and the entry of tertiary proteins are interdependent events in the late stages of 30S subunit assembly.
DISCUSSION
In the work reported here, we have made use of a viable deletion in the yjeQ gene to isolate and structurally characterize for the first time an in vivo assembled immature 30S subunit. Previous studies of 30S subunit assembly have largely been systematic in vitro investigations of rRNA folding and protein binding using processed 16S rRNA and r-proteins (Mizushima and Nomura 1970; Talkington et al. 2005; Adilakshmi et al. 2008). These works are responsible for the remarkable understanding that we have today of 30S assembly, not the least of which is the hierarchy of protein binding described by the Nomura assembly map. Perhaps the most important principle to emerge from recent studies is that r-proteins drive conformational rearrangements in the RNA and stabilize the native fold. The in vivo approach followed here has raised some additional guiding principles. These include the idea that a specific lesion in ribosome assembly such as that achieved with a deletion in yjeQ can lead to an immature 30S subunit of profoundly distorted 3D structure, incapable of participating in either protein synthesis or 50S binding. Furthermore, our results imply that rRNA processing events are key contributors to the final structure of the 30S subunit by defining a fold that is sterically capable of binding late subunit proteins.
We further conclude that ongoing structural studies will be critical to achieving precise understanding of the assembly process in three dimensions. To this end, as we demonstrate herein, emerging trans-acting factors in ribosomal subunit assembly represent an extraordinary opportunity to manipulate the assembly process in vivo in order to study subunit maturation in three dimensions using cryo-electron microscopy.
There have been a series of studies suggesting that the final rRNA processing events occur in polysomes (Mangiarotti et al. 1974; Hayes and Vasseur 1976). In contrast, the structure of the immature 30S subunit presented here suggests that processing of the 3′ precursor sequence of the 17S rRNA is a prerequisite to the formation of functional 70S ribosomes. Unfortunately, the precursor 16S rRNAs used in these early studies (Mangiarotti et al. 1974; Hayes and Vasseur 1976) were not fully characterized, and thus, it is unclear which of the precursor sequences were present in these precursor rRNA molecules. More recently, other studies have characterized precursor 16S rRNAs produced in E. coli strains containing mutations in the rne (encoding for RNase E) and rng genes (encoding for RNase G) and the incorporation of these precursor molecules into 70S ribosomes. One of the studies (Li et al. 1999) found that a strain lacking rng combined with a temperature-sensitive allele of rne produces a precursor 16S rRNA with 115 extra nucleotides in the 5′ end, but its 3′ end is fully processed. Interestingly, this precursor 16S rRNA is incorporated into 70S ribosomes. A second report (Roy-Chaudhuri et al. 2010) found that an rng deletion strain produces two precursor 16S rRNA forms. These two forms contain 115 (lp 16S rRNA) and 66 (sp 16S rRNA) extra nucleotides in the 5′ end, respectively, but only the sp 16S rRNA is incorporated into the 70S ribosomes. Unfortunately, in this later study (Roy-Chaudhuri et al. 2010), the presence of the precursor sequence in the 3′ end of lp and sp 16S rRNA was not investigated. Therefore, in light of the available evidence, our cryo-EM structure of the immature 30S subunit is not in disagreement with the idea that 5′ rRNA processing may occur in the context of the 70S ribosome or even later in the polysomes. However, we cannot rule out whether the 3′ rRNA processing occurs in the 70S ribosome. Nevertheless, recent work in our laboratory showed that while the sp 16S rRNA in the rng deletion strain is processed at the 3′ terminus, the lp 16S rRNA is not and still contains its precursor sequence in the 3′ end (Vivian Leong, unpubl.). Consequently, we favor the model in which processing of the 3′ precursor sequence occurs in the 30S subunit prior to association with the 50S subunit.
The data presented here also suggest that YjeQ has an indirect role in processing the precursor 16S rRNA, either by directly facilitating the interaction of the 30S subunit with these RNases or by stabilizing the 30S subunit in a specific conformation that ensures efficient processing. Most likely, YjeQ is mediating this process in conjunction with other trans-acting factors. A recent publication (Goto et al. 2011) has reported a functional interplay between YjeQ and RbfA during maturation of the 30S subunit. RbfA is a small protein that binds the 30S subunit at the junction of the head and body, and its binding alters the position of helices 44 and 45 (Datta et al. 2007). Interestingly, RbfA seems to promote the association of YjeQ to the 30S subunits, and binding of YjeQ stimulates the dissociation of RbfA from the small subunit (Goto et al. 2011).
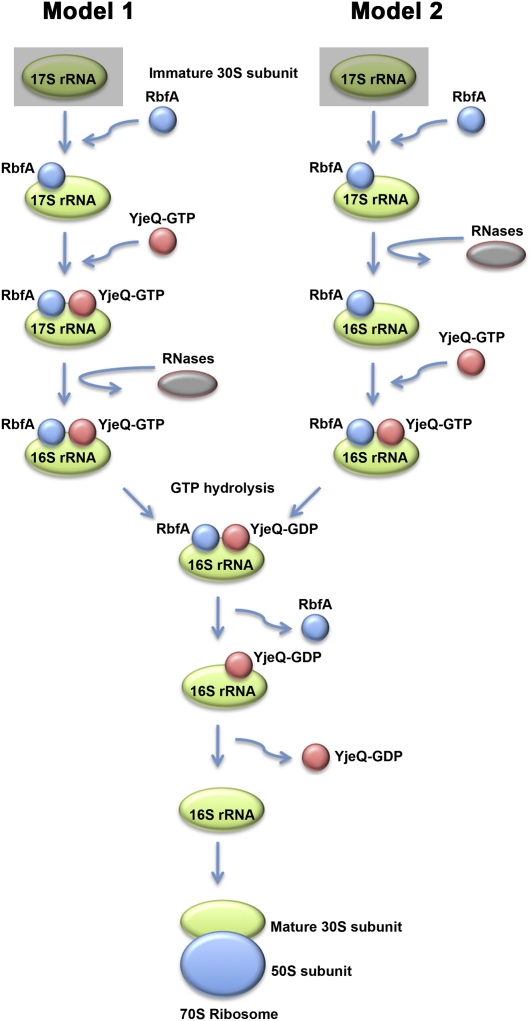
Taken together, this and previous studies of YjeQ appear consistent with the following events in the late stages of 30S assembly (Fig. 7, model 1): (1) RbfA binds first to an immature form of the 30S subunit; (2) this facilitates the entry of YjeQ-GTP into the complex either directly or after inducing a conformation that is then recognized by YjeQ; (3) this binding causes an additional conformational change in the immature particle; (4) the 30S subunit is recognized by RNases that process the 17S rRNA; (5) subsequent hydrolysis of GTP facilitates dissociation of RbfA; and (6) YjeQ-GDP dissociates from the mature 30S subunit, which is now in a conformation compatible for binding to the 50S subunit. Under this model, both RbfA and YjeQ would trigger specific conformations recognized by the RNases or directly recruit these enzymes to the maturing subunit. This order of events is somewhat different from that proposed recently by Goto et al. (2011), who suggested that processing of the 17S rRNA is triggered by RbfA before YjeQ binding. In our view, the latter model seems inconsistent with the accumulation of 17S rRNA containing 30S subunits in the ΔYjeQ strain, a finding that is reinforced by the data presented herein. Nevertheless, this accumulation could be explained as a consequence of impaired protein synthesis due to a shortage of mature ribosomes in these cells. Indeed, it has been reported that disturbances in protein synthesis arising from exposure to a 50S subunit-targeting antibiotic, such as puromycin, erythromycin, or chloramphenicol, also induce accumulation of 17S rRNA (Hosokawa and Nomura 1965; Himeno et al. 2004; Siibak et al. 2009). Thus it is conceivable that the structure of the immature 30S subunit purified from the ΔYjeQ strain may not represent the binding substrate for YjeQ but rather for RbfA. In any case, this cryo-EM reconstruction constitutes the first detailed structure of an immature 30S subunit prior to rRNA processing and containing 17S rRNA.
FIGURE 7.
Placement of the structurally characterized immature 30S subunits in the current biogenesis model of the 30S subunit. The diagram describes the likely order of events involving YjeQ in the late stages of assembly of the 30S subunit and processing of the 17S rRNA. A recent publication (Goto et al. 2011) has suggested that YjeQ mediates this process in conjunction with RbfA, and thus, this protein is included in the diagram. RbfA seems to bind first to an immature form of the 30S subunit facilitating the subsequent entry of YjeQ-GTP to the maturing subunit. At this moment, there is not enough experimental evidence yet to establish whether the processing of the 17S rRNA occurs before (model 2) or after (model 1) YjeQ binding to the immature 30S subunit. The cryo-EM reconstruction of the immature 30S subunit purified from the ΔYjeQ strain that is presented constitutes the structure of a 17S rRNA–containing 30S subunit prior to rRNA processing and could possibly represent the structure of the immature 30S subunit bound by RbfA (shadowed squares in models 1 and 2).
In addition to yjeQ and rbfA, there are several other genes encoding trans-acting factors that appear to have a role in assembly of the 30S subunit including era, rimM, and rimN (Wilson and Nierhaus 2007; Connolly and Culver 2009) with which YjeQ may also exert a functional interplay. era is of particular interest because of the genetic interaction of this gene with yjeQ: Overexpression of era suppresses defects in growth and ribosome maturation of a yjeQ-null mutant (Campbell and Brown 2008). Interestingly, a cryo-EM reconstruction of Thermus thermophilus Era protein bound to the mature 30S subunit reveals that this protein binds between the head and platform in a cavity formed by r-proteins S2, S7, S11, and S18 (Sharma et al. 2005). This binding site overlaps with the most severely distorted domain in the 30S subunit from cells lacking YjeQ. Perhaps Era performs its function in conjunction with YjeQ or YjeQ binds the 30S subunit in a location overlapping with the Era binding site. Additional structural, genetic, and biochemical studies of YjeQ and other assembly factors will ultimately describe the existing functional relationships between these factors. The characterization of immature subunits generated with genetic lesions in these factors will be an important approach in ongoing measures to understand bacterial ribosome biogenesis.
Finally, an important question in our present study is whether the obtained structures of immature 30S subunits constitute genuine intermediates of the ribosome assembly process or off-pathway assembly species generated in cells lacking the assembly factor YjeQ. In this context, it is worth considering a recent in vitro study (Talkington et al. 2005) suggesting that 30S subunit assembly does not proceed through a single pathway. Instead, there may be many parallel folding pathways that converge in the mature 30S subunit, and thus, it is also unclear whether the characterized structures may represent intermediates within the same or different assembly pathway. The yjeQ gene is dispensable to cells, and its loss leads not to cell death but to slow growth (Campbell et al. 2005). Perhaps the slow-growth phenotype originates from a slower uncatalyzed transformation of maturing species in the late stages of ribosome assembly. In this instance, we may well have characterized bona fide intermediates that accumulate in cells lacking YjeQ. Conversely, it is conceivable that YjeQ is a critical catalyst in a particular assembly pathway and that we have characterized off-pathway end products accumulated by the cell. Nevertheless, structural characterization of such species in ribosome assembly will surely also prove to be ultimately informative. Indeed, enzymologists have long endeavored to understand the structure of short-lived reaction intermediates by isolating and characterizing the by-products thereof. Thus while we cannot be definitive about the nature of the immature 30S subunits isolated and characterized in this study, these structures provide new and exciting insights into the process of ribosomal subunit assembly.
MATERIALS AND METHODS
Isolation and analysis of cellular rRNA
Overnight cultures of E. coli ΔyjeQ (EB1140) and the parental strain (EB334) were subcultured in LB media (1:100 dilution) and grown with shaking at 37°C to an optical OD600 of 0.2. Subsequently, 1 mL of culture was pelleted by centrifugation at 5000g for 10 min in an Eppendorf 5424 microcentrifuge. Cell pellets were resuspended in 100 μL of lysis buffer (25 mM Tris-HCl at pH 8.0, 10 mg/mL lysozyme, 1 mg/mL proteinase K, 0.15 mM calcium acetate). Lysis mixtures were incubated for 10 min at 22°C with shaking. Ribosomal RNA was then purified from cells using the RNeasy Mini Kit (QIAGEN) according to the manufacturer's protocols. Purified RNA was incubated with an equal volume of RNA loading buffer (50% [w/v] urea, 10% [w/v] sucrose, 1× TBE) for 10 min at 75°C. Samples were loaded onto a modified agarose gel comprised of 0.7% agarose and 0.9% Synergel (Diversified Biotech). RNA was separated by electrophoresis for 4 h at 4.5 V/cm. Gels were stained with ethidium bromide and visualized under UV light.
Northern hybridization analysis was performed for both the 5′ and 3′ precursor sequences of the 17S rRNA to determine its identity. Samples were transferred to Hybond-N+ charged nylon membranes (GE Healthcare) under alkaline conditions (0.1 M NaOH, 3 M NaCl) using the standard capillary method. Membranes were incubated in 50 mL of prehybridization buffer (0.5 M sodium phosphate at pH 7.2, 7% [w/v] SDS, 1 mM EDTA at pH 7.0) for 2 h at 32°C. Single-stranded DNA probes complementary to the 5′ or 3′ terminal precursor sequences (Sigma Canada) were labeled with 32P using T4 polynucleotide kinase (Fermentas). The sequence of the 5′ probe was 5′-TTAAGAATCCGTATCTTCGAGTGCCCACA-3′, and the sequence of the 3′ probe was 5′-TGTGTGAGCACTACAAAGTACGCTTCTTTAAGGTAAGG-3′.
Labeled probes (20 μg) were added to the prehybridization solutions and incubated with rotation for 20 h at 32°C. Subsequent washes with 1× SSC, 0.5× SSC, and 0.1× SSC in 1% SDS were performed for 15 min each at 50°C. Blots were visualized using a Typhoon Trio+ PhosphorImager (GE Healthcare). Northern blot analysis was also performed with a single-stranded DNA probe complementary to an internal fragment of 16S RNA as a control. The sequence of this probe was as follows: 5′- GGGCCATGATGACTTGACGTCATCCCCACC-3′. The method employed was identical to that used for the precursor probes.
Purification of 30S ribosomal subunits
Purified 30S subunits from E. coli ΔyjeQ (EB1140) and BW25113 parental (EB334) strains were prepared using centrifugations over sucrose cushions and gradients as previously described (Daigle and Brown 2004). For each strain, 1 L of Luria broth (LB) was inoculated with 10 mL of a saturated overnight culture. Cultures were grown at 37°C to an OD600 of 0.2. Cells were cooled down to 4°C, and all subsequent steps were conducted at this temperature.
Cultures were harvested by centrifugation at 8500g for 15 min, and the cell pellet was resuspended in buffer A (20 mM Tris-HCl at pH 7.5, 10.5 mM magnesium acetate, 100 mM NH4Cl, 0.5 mM EDTA, and 3 mM 2-mercaptoethanol). Cell lysis was performed by three consecutive passes of the cell suspension through a French pressure cell at 20,000 lb/in2. The cell lysate was spun at 30,000g for 45 min to clear cell debris. Recovered supernatant (S30 fraction) was overlaid on an equal volume of 1.1 M sucrose cushion made up in buffer B (20 mM Tris-HCl at pH 7.5, 10.5 mM magnesium acetate, 500 mM NH4Cl, 0.5 mM EDTA, and 3 mM 2-mercaptoethanol) and centrifuged at 100,000g for 16 h. The ribosomal pellet was gently washed, and sucrose was removed by resuspension in buffer C (10 mM Tris-HCl at pH 7.5, 10.5 mM magnesium acetate, 100 mM NH4Cl, 0.5 mM EDTA, and 7 mM 2-mercaptoethanol). The crude ribosomes were pelleted at 100,000g for 16 h.
To obtain the 30S fraction from the E. coli ΔyjeQ strain, the crude ribosome pellet was resuspended in buffer E (10 mM Tris-HCl at pH 7.5, 10 mM magnesium acetate, 60 mM NH4Cl, 3 mM 2-mercaptoethanol) (nondissociating conditions). In the case of the 30S fraction from the parental strain, the crude ribosome pellet was resuspended in buffer F (10 mM Tris-HCl at pH 7.5, 1.1 mM magnesium acetate, 60 mM NH4Cl, 0.5 mM EDTA, and 2 mM 2-mercaptoethanol) (dissociating conditions). In both cases, a portion of the subunit suspension (50–60 A260 units) was layered onto a 32-mL 10%–30% (wt/vol) sucrose gradient made up in buffer E (for nondissociating conditions) or F (for dissociating conditions) and centrifuged at 43,000g for 16 h using a Beckman SW32 Ti rotor. Gradients were fractionated using an AKTAprime purification system (GE Healthcare), and the elution peaks corresponding to 30S, 50S, and 70S (in the case of nondissociating conditions) particle peaks were monitored by absorbance at A260. The 30S ribosomal subunits were then recovered by centrifugation at 100,000g for 16 h, and the pellet was resuspended in buffer E (10 mM Tris-HCl at pH 7.5, 10 mM magnesium acetate, 60 mM NH4Cl, 3 mM 2-mercaptoethanol) and stored at −80°C until further use. Quantification of the 30S subunits was accomplished by absorbance at 260 nm (1 A260 unit is equivalent to 69 pmol of 30S).
iTRAQ analysis
Biological replicates of 30S particles from E. coli ΔyjeQ (EB1140) and BW25113 parental (EB334) strains were purified separately using nondissociating conditions and analyzed independently. Total protein was measured using a 2D Quant kit (GE Healthcare). A total of 25 μg from each 30S subunit preparation was precipitated with 80% ice-cold acetone and washed twice with 100% acetone. The pellet was resuspended in 21 μL of 0.5 M triethylammonium bicarbonate (TEAB), 0.01% SDS, and proteins were reduced with tris(2-carboxyethyl)phosphine (TCEP); cysteines were blocked with methylmethanethiosulfate (MMTS) and digested with trypsin as described previously (Jiang et al. 2006). Labeling with the iTRAQ reagents was according to the manufacturer's recommendations (Applied Biosystems). For each biological replicate, the peptides derived from the parental and ΔyjeQ 30S subunits were labeled with two different iTRAQ reagents. The iTRAQ reagents 115 and 117 were used to label tryptic peptides from independently isolated ΔyjeQ 30S subunit preparations, and 114 and 116 were used to label tryptic peptides from independently isolated parental 30S subunits. The iTRAQ reagent was reconstituted in ethanol and added directly to the protein digest (70% ethanol, final concentration). The mixture was incubated for 1 h at room temperature, and the reaction was stopped by adding 250 μL of 0.1% trifluoroacetic acid (TFA; Fisher Science). Reaction mixtures from both labeling experiments were combined, dried by lyophilization, and stored at −20°C.
iTRAQ-labeled peptides were resuspended in buffer A (5 mM KH2PO4 at pH 2.7, 30% acetonitrile) and applied to a strong cation exchange chromatography (SCX) using a Paradigm MG4 HPLC system (Michrom BioResources, Inc.) on a packed polysulfoethyl aspartamide column, 200 × 2.1 mm, 5 μm, 200 Å (Poly LC, Inc.). Peptides were eluted using a linear gradient with B (buffer A with 500 mM KCl) using a flow rate of 200 μL/min over 50 min. Two-minute fractions were collected throughout and pooled into 12 fractions. For RP-HPLC-MS analysis, fractions were dried and redissolved in 45 μL of 0.1% TFA, desalted, and concentrated on a reversed-phase cartridge (Zorbax C18) calibrated with 0.1% TFA as described previously (Chen et al. 2006). Peptides were eluted with a linear gradient using 0.1% TFA with 90% acetonitrile using a flow rate of 4 μL/min for 90 min and spotted at 20-sec intervals onto a stainless steel MALDI target plate (192 wells; Applied Biosystems).
The MS and MS/MS spectra were acquired on an Applied Biosystems 4800 Proteomics Analyzer (TOF/TOF; Applied Biosystems/MDX Sciex) in positive ion reflection mode (selected mass range of 800–3000 m/z) with a 200-Hz Nd:YAG laser operating at 355 nm. Accelerating voltage was 20 kV with 450-nsec delay. For MS/MS spectra, the collision energy was 6 keV, and the collision gas was air. The instrument was calibrated using a default mixture of seven peptides. Typical MS spectra were obtained with the minimum possible laser energy to maintain the best resolution. MS/MS spectra were collected for up to 12 of most intense MS peaks (minimum signal-to-noise ratio of 60) from each well.
The peak lists were generated from the acquired MS/MS spectra with the Protein Pilot software (Applied Biosystem, version 3.0) using default parameters. Acquired MS/MS spectra were searched against a concatenated (forward and reverse) E. coli K12 protein sequence database downloaded as a FASTA file from UniProtKB/SWISS-PROT (http://www.uniprot.org/taxonomy/complete-proteomes) on April 10, 2008. The Protein Pilot software with default settings and the Paragon search engine (Applied Biosystems, version 3.0) was the software used to perform all the database searches. Proteins were identified with 1% local false discovery rate calculated by a module built-in in the software. Peptides for which one or more of the iTRAQ values were absent and proteins identified by only a single distinct peptide were discarded. Peak areas for all of the reporter ions (m/z 114.1, 115.1, 116.1, and 117.1) were extracted and automatically corrected using Protein Pilot (Applied Biosystems, version 3.0). The MS/MS data obtained were deposited into TRANCHE (https://proteomecommons.org/tranche/) with the Tranche Hash: dDZes+9slItFdCPmF8YepUC7GZEGcVekFr284mjTK4lsoEtAC1HZThAr5WcXUfce+JeZ1BYon2HV3tLR42W0aBiwCFsAAAAAAGmUKQ==.
The ΔyjeQ:WT (wild type) and WT:ΔyjeQ protein ratios as well as the P-values in Supplemental Table 1 were generated with the Protein Pilot software. The ratios in Figure 2 and Supplemental Figure S1 were generated manually, as we have done previously (Jiang et al. 2006, 2007). In this case, only peptides that identify proteins with ≥95% confidence were used for the calculation. Peptide ratios for each replicate (115/114 and 117/116) were combined and then averaged. To obtain protein ratios, the average from all peptide ratios was obtained after removal of outliers using a Grubbs test (http://www.graphpad.com/quickcalcs/Grubbs1.cfm). The entire data set was normalized at the level of the proteins by adjusting the median of the protein ratio to 1:1.
Cryo-electron microscopy, image classification, and 3D reconstruction
For cryo-EM, holey carbon grids (400 mesh copper) containing an additional continuous thin (5–10 nm) layer of carbon were previously washed with acetone vapor for 15 min and glow discharge in air for 30 sec (Aebi and Pollard 1987). Then, 3.5-μL aliquots containing 30S subunits purified from ΔyjeQ or wild-type cells were applied to the grid for 1 min. The 30S subunits were in buffer E and at a concentration of ∼60 nM. Grids were blotted for 7 sec and vitrified by rapidly plunging into liquid ethane at −180°C with a Vitrobot (FEI). Data acquisition was performed under low dose conditions (∼10 e−/Å2) on a JEOL 2010F FEG microscope operated at 200 kV with a Gatan 914 side-entry cryo-holder and at a nominal magnification of 50,000×. A total number of 72 and 29 electron micrographs were collected from the samples containing 30S subunits purified from ΔyjeQ and wild-type cells, respectively. The defocus range of the images was from −0.65 to −3.9 μm. The micrographs were digitized with a step size of 12.7 μm in a Nikon Supercool Scan 9000 producing images with a sampling value of 2.54 Å/pixel.
Projections representing 30S subunits purified from ΔyjeQ and wild-type cells were picked using Boxer (Ludtke et al. 1999). The contrast transfer function of the micrographs was estimated using CTFFIND software (Mindell and Grigorieff 2003) and corrected using Xmipp software package (Scheres et al. 2008). Image classification was performed using supervised (Valle et al. 2002; Gao et al. 2004) and 3D maximum likelihood–based (Scheres et al. 2005b, 2007) methods. Detailed procedures regarding these methods are included in Supplemental Figures S3 and S5.
The 3D reconstructions were calculated using 3D projection alignment procedures as implemented in the Xmipp software package (Scheres et al. 2008). The reference map used to refine the 3D reconstruction of both the mature and immature 30S subunits was the X-ray structure of the E. coli 30S subunit (PDB ID: 2Z4K) (Borovinskaya et al. 2007) low-pass-filtered to 25 Å resolution. In each refinement, sets of projections were calculated from the reference map using an angular spacing that ranged from 15° to 2° during the multiple cycles of refinement. The correct handedness of the structures was imposed by the X-ray crystal structure of the 30S subunit from E. coli (PDB ID: 2Z4K). Resolution of the cryo-EM maps was estimated by calculating two maps following the last cycle of refinement from the even- and odd-numbered particles. Then Fourier shell correlation was calculated between both maps, and the resolution was estimated using an FSC value of 0.5. These values were used to low-pass-filter the refined cryo-EM maps.
Difference maps between the immature and mature 30S subunit structures were calculated using Xmipp software package (Scheres et al. 2008). To this end, maps were subtracted after alignment of the two structures and normalization to a comparable grayscale.
Docking of X-ray crystallographic structures and structure visualization
Fitting of the X-ray crystallographic structure of the E. coli 30S subunit (PDB ID: 2Z4K) (Borovinskaya et al. 2007) was performed using rigid-body fitting of the entire X-ray structure of the 30S subunit onto the EM maps as implemented in the Situs software (Wriggers et al. 1999). Visualization of the fitted atomic structure and the cryo-EM density maps and difference maps was done with UCSF Chimera software (Pettersen et al. 2004).
Accession numbers
The EM maps of the immature 30S subunit purified from ΔyjeQ E. coli cells and the mature 30S subunit purified from wild-type E. coli cells have been deposited in the Electron Microscopy Data Bank (EMDB IDs: 1774 and 1775).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We are grateful to the staff at the Canadian Centre for Electron Microscopy. We also acknowledge Vivian Leong for technical support, Sjors Scheres for assistance with the maximum likelihood–based classification programs implemented in Xmipp, and the Supercomputing Center of Galicia (CESGA) for the use of their high-performance computers. J.O. is a Canadian Institutes of Health Research (CIHR) New Investigator and also acknowledges support from an Early Researcher Award from the Ministry of Research and Innovation. This work was supported by a grant from the National Institutes of Health (Grant # GM077628) to J.R.M. and a grant from M. Ciencia e Innovacion (BFU2010-21886) to J.M.B. E.D.B. is a Canada Research Chair and supported for this work by an operating grant from the Canadian Institutes of Health Research (MOP-6492). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2509811.
REFERENCES
- Adilakshmi T, Bellur DL, Woodson SA 2008. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature 455: 1268–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi U, Pollard TD 1987. A glow discharge unit to render electron microscope grids and other surfaces hydrophilic. J Electron Microsc Tech 7: 29–33 [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920 [DOI] [PubMed] [Google Scholar]
- Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JH 2007. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat Struct Mol Biol 14: 727–732 [DOI] [PubMed] [Google Scholar]
- Brown ED 2005. Conserved P-loop GTPases of unknown function in bacteria: an emerging and vital ensemble in bacterial physiology. Biochem Cell Biol 83: 738–746 [DOI] [PubMed] [Google Scholar]
- Bubunenko M, Korepanov A, Court DL, Jagannathan I, Dickinson D, Chaudhuri BR, Garber MB, Culver GM 2006. 30S ribosomal subunits can be assembled in vivo without primary binding ribosomal protein S15. RNA 12: 1229–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell TL, Brown ED 2008. Genetic interaction screens with ordered overexpression and deletion clone sets implicate the Escherichia coli GTPase YjeQ in late ribosome biogenesis. J Bacteriol 190: 2537–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell TL, Daigle DM, Brown ED 2005. Characterization of the Bacillus subtilis GTPase YloQ and its role in ribosome function. Biochem J 389: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Walker AK, Strahler JR, Simon ES, Tomanicek-Volk SL, Nelson BB, Hurley MC, Ernst SA, Williams JA, Andrews PC 2006. Organellar proteomics: Analysis of pancreatic zymogen granule membranes. Mol Cell Proteomics 5: 306–312 [DOI] [PubMed] [Google Scholar]
- Connolly K, Culver G 2009. Deconstructing ribosome construction. Trends Biochem Sci 34: 256–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver GM 2003. Assembly of the 30S ribosomal subunit. Biopolymers 68: 234–249 [DOI] [PubMed] [Google Scholar]
- Daigle DM, Brown ED 2004. Studies of the interaction of Escherichia coli YjeQ with the ribosome in vitro. J Bacteriol 186: 1381–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle DM, Rossi L, Berghuis AM, Aravind L, Koonin EV, Brown ED 2002. YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with circularly permuted G-motifs and marked burst kinetics. Biochemistry 41: 11109–11117 [DOI] [PubMed] [Google Scholar]
- Datta PP, Wilson DN, Kawazoe M, Swami NK, Kaminishi T, Sharma MR, Booth TM, Takemoto C, Fucini P, Yokoyama S, et al. 2007. Structural aspects of RbfA action during small ribosomal subunit assembly. Mol Cell 28: 434–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J 2003. Toward an understanding of the structural basis of translation. Genome Biol 4: 237 doi: 10.1186/gb-2003-4-12-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Valle M, Ehrenberg M, Frank J 2004. Dynamics of EF-G interaction with the ribosome explored by classification of a heterogeneous cryo-EM dataset. J Struct Biol 147: 283–290 [DOI] [PubMed] [Google Scholar]
- Goto S, Kato S, Kimura T, Muto A, Himeno H 2011. RsgA releases RbfA from 30S ribosome during a late stage of ribosome biosynthesis. EMBO J 30: 104–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107: 679–688 [DOI] [PubMed] [Google Scholar]
- Hayes F, Vasseur M 1976. Processing of the 17-S Escherichia coli precursor RNA in the 27-S pre-ribosomal particle. Eur J Biochem 61: 433–442 [DOI] [PubMed] [Google Scholar]
- Himeno H, Hanawa-Suetsugu K, Kimura T, Takagi K, Sugiyama W, Shirata S, Mikami T, Odagiri F, Osanai Y, Watanabe D, et al. 2004. A novel GTPase activated by the small subunit of ribosome. Nucleic Acids Res 32: 5303–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KL, Culver GM 2004. Mapping structural differences between 30S ribosomal subunit assembly intermediates. Nat Struct Mol Biol 11: 179–186 [DOI] [PubMed] [Google Scholar]
- Hosokawa K, Nomura M 1965. Incomplete ribosomes produced in chloramphenicol- and puromycin-inhibited Escherichia coli. J Mol Biol 12: 225–241 [DOI] [PubMed] [Google Scholar]
- Hosokawa K, Fujimura RK, Nomura M 1966. Reconstitution of functionally active ribosomes from inactive subparticles and proteins. Proc Natl Acad Sci 55: 198–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Datta K, Walker A, Strahler J, Bagamasbad P, Andrews PC, Maddock JR 2006. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J Bacteriol 188: 6757–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Sullivan SM, Walker AK, Strahler JR, Andrews PC, Maddock JR 2007. Identification of novel Escherichia coli ribosome-associated proteins using isobaric tags and multidimensional protein identification techniques. J Bacteriol 189: 3434–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Takagi K, Hirata Y, Hase Y, Muto A, Himeno H 2008. Ribosome-small-subunit-dependent GTPase interacts with tRNA-binding sites on the ribosome. J Mol Biol 381: 467–477 [DOI] [PubMed] [Google Scholar]
- Li Z, Pandit S, Deutscher MP 1999. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J 18: 2878–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W 1999. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128: 82–97 [DOI] [PubMed] [Google Scholar]
- Mangiarotti G, Turco E, Ponzetto A, Altruda F 1974. Precursor 16S RNA in active 30S ribosomes. Nature 247: 147–148 [DOI] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N 2003. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142: 334–347 [DOI] [PubMed] [Google Scholar]
- Mitra K, Frank J 2006. RIBOSOME DYNAMICS: Insights from atomic structure modeling into cryo-electron microscopy maps. Annu Rev Biophys Biomol Struct 35: 299–317 [DOI] [PubMed] [Google Scholar]
- Mizushima S, Nomura M 1970. Assembly mapping of 30S ribosomal proteins from E. coli. Nature 226: 1214. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Poot RA, van den Worm SH, Pleij CW, van Duin J 1998. Base complementarity in helix 2 of the central pseudoknot in 16S rRNA is essential for ribosome functioning. Nucleic Acids Res 26: 549–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. 2004. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3: 1154–1169 [DOI] [PubMed] [Google Scholar]
- Roy-Chaudhuri B, Kirthi N, Culver GM 2010. Appropriate maturation and folding of 16S rRNA during 30S subunit biogenesis are critical for translational fidelity. Proc Natl Acad Sci 107: 4567–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Valle M, Carazo JM 2005a. Fast maximum-likelihood refinement of electron microscopy images. Bioinformatics 21: 243–244 [DOI] [PubMed] [Google Scholar]
- Scheres SH, Valle M, Nunez R, Sorzano CO, Marabini R, Herman GT, Carazo JM 2005b. Maximum-likelihood multi-reference refinement for electron microscopy images. J Mol Biol 348: 139–149 [DOI] [PubMed] [Google Scholar]
- Scheres SH, Gao H, Valle M, Herman GT, Eggermont PP, Frank J, Carazo JM 2007. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat Methods 4: 27–29 [DOI] [PubMed] [Google Scholar]
- Scheres SH, Nunez-Ramirez R, Sorzano CO, Carazo JM, Marabini R 2008. Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc 3: 977–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, et al. 2000. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102: 615–623 [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310: 827–834 [DOI] [PubMed] [Google Scholar]
- Sengupta J, Agrawal RK, Frank J 2001. Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc Natl Acad Sci 98: 11991–11996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma MR, Barat C, Wilson DN, Booth TM, Kawazoe M, Hori-Takemoto C, Shirouzu M, Yokoyama S, Fucini P, Agrawal RK 2005. Interaction of Era with the 30S ribosomal subunit implications for 30S subunit assembly. Mol Cell 18: 319–329 [DOI] [PubMed] [Google Scholar]
- Siibak T, Peil L, Xiong L, Mankin A, Remme J, Tenson T 2009. Erythromycin- and chloramphenicol-induced ribosomal assembly defects are secondary effects of protein synthesis inhibition. Antimicrob Agents Chemother 53: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AK, Schlessinger D 1990. Mechanism and regulation of bacterial ribosomal RNA processing. Annu Rev Microbiol 44: 105–129 [DOI] [PubMed] [Google Scholar]
- Sykes MT, Williamson JR 2009. A complex assembly landscape for the 30S ribosomal subunit. Annu Rev Biophys 38: 197–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkington MW, Siuzdak G, Williamson JR 2005. An assembly landscape for the 30S ribosomal subunit. Nature 438: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub P, Nomura M 1968a. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc Natl Acad Sci 59: 777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub P, Nomura M 1968b. Structure and function of Escherichia coli ribosomes. I. Partial fractionation of the functionally active ribosomal proteins and reconstitution of artificial subribosomal particles. J Mol Biol 34: 575–593 [DOI] [PubMed] [Google Scholar]
- Traub P, Nomura M 1969. Studies on the assembly of ribosomes in vitro. Cold Spring Harb Symp Quant Biol 34: 63–67 [DOI] [PubMed] [Google Scholar]
- Valle M, Sengupta J, Swami NK, Grassucci RA, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J 2002. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J 21: 3557–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN, Nierhaus KH 2007. The weird and wonderful world of bacterial ribosome regulation. Crit Rev Biochem Mol Biol 42: 187–219 [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339 [DOI] [PubMed] [Google Scholar]
- Wriggers W, Milligan RA, McCammon JA 1999. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J Struct Biol 125: 185–195 [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896 [DOI] [PubMed] [Google Scholar]