Abstract
Stacking interactions between amino acids and bases are common in RNA–protein interactions. Many proteins that regulate mRNAs interact with single-stranded RNA elements in the 3′ UTR (3′-untranslated region) of their targets. PUF proteins are exemplary. Here we focus on complexes formed between a Caenorhabditis elegans PUF protein, FBF, and its cognate RNAs. Stacking interactions are particularly prominent and involve every RNA base in the recognition element. To assess the contribution of stacking interactions to formation of the RNA–protein complex, we combine in vivo selection experiments with site-directed mutagenesis, biochemistry, and structural analysis. Our results reveal that the identities of stacking amino acids in FBF affect both the affinity and specificity of the RNA–protein interaction. Substitutions in amino acid side chains can restrict or broaden RNA specificity. We conclude that the identities of stacking residues are important in achieving the natural specificities of PUF proteins. Similarly, in PUF proteins engineered to bind new RNA sequences, the identity of stacking residues may contribute to “target” versus “off-target” interactions, and thus be an important consideration in the design of proteins with new specificities.
Keywords: 3′ UTRs, RNA decay, RNA–protein interactions, translational regulation, stacking interactions
INTRODUCTION
RNA–protein interactions control the function and maturation of RNAs. As a result, they are critical in many biological contexts, from pattern formation to learning. Recognition of specific, unstructured RNA sequences requires discrimination of RNA bases rather than higher-order structural elements. Base recognition relies on hydrophobic interactions, hydrogen bonding, and van der Waals contacts. Stacking interactions, in which the aromatic RNA bases interact with one another or with amino acid side chains, are common. Here we examine the role of stacking interactions in the RNA-binding specificity of PUF proteins, an important class of mRNA regulators.
PUF family proteins bind to sequences in the 3′ UTRs of target RNAs and play important roles in stem cell maintenance and memory (Schweers et al. 2002; Wickens et al. 2002; Dubnau et al. 2003; Ye et al. 2004; Morrison and Kimble 2006; Kimble and Crittenden 2007; Chen et al. 2008). These proteins commonly reduce expression of the mRNA (for review, see Wickens et al. 2002; Goldstrohm et al. 2006; Chritton and Wickens 2010), although several instances of activation recently have been reported (Pique et al. 2008; Kaye et al. 2009; Suh et al. 2009). A single PUF protein recognizes a subset of mRNAs, from a few dozen to more than a thousand different messages (Gerber et al. 2004, 2006; Galgano et al. 2008; Morris et al. 2008; Kershner and Kimble 2010). In some instances, the collection of target mRNAs encodes proteins with related biological functions, enabling coordinate control (Gerber et al. 2004; Kershner and Kimble 2010). These PUF–RNA regulatory networks depend on the ability of the protein to discriminate only its own set of mRNAs, and not others.
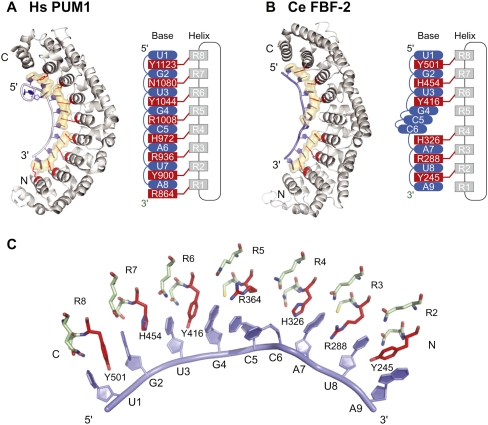
The PUF RNA-binding domain typically comprises eight imperfect α-helical structural repeats, which together form a crescent-shaped molecule (Fig. 1A; Zamore et al. 1997; Edwards et al. 2001; Wang et al. 2001). RNA binds on the concave face (Wang et al. 2002). The complex of Homo sapiens Pumilio1 (HsPUM1) with RNA is exemplary: the second of three α-helices in each PUF repeat binds one nucleotide (Fig. 1A; Wang et al. 2002). (α-Helices that contact RNA are referred to here as “RNA-recognition helices.”) In a typical recognition helix, three amino acid side chains interact with the base opposite: Two amino acid residues provide hydrogen-bonding and/or van der Waals contacts, while a third stacks between adjacent nucleotides (Wang et al. 2002). The three amino acid residues that contact RNA, in all eight repeats, are conserved among PUF proteins (Wang et al. 2002). The HsPUM1–RNA structure is prototypical and represents the simplest condition: Each RNA-recognition helix interacts directly with the base opposite.
FIGURE 1.
PUF protein–RNA interaction. (A) Hs PUM1: structure of human Pumilio1–RNA complex (PDB accession code 1M8Y) (Wang et al. 2002). (B) Ce FBF-2: structure of C. elegans FBF-2–RNA complex (PDB accession code 3K5Y) (Wang et al. 2009b). Eight RNA-recognition helices (gray in structures and diagrams) contribute side chains to contact RNA bases (blue). (Red) Stacking residues; interactions with base edges are not shown. (Beige shading) Amino acid side chains and RNA bases involved in stacking interactions. (C) Key interactions in the FBF-2–RNA complex. (Red) Stacking residues; (green) base edge interacting residues; (blue) RNA bases.
Crystal structures of several other PUF–RNA complexes reveal that specificity can be achieved by modifications of this scaffold. Different PUF proteins combine nearly identical sets of atomic contacts with structural distortions or additional protein-specific contacts to recognize distinct RNA targets (Wang et al. 2002; Opperman et al. 2005; Gupta et al. 2008; Miller et al. 2008; Wang et al. 2009b; Zhu et al. 2009). For example, the Caenorhabditis elegans PUF protein, FBF-2 (fem-3 binding factor-2), binds to nine nucleotides rather than eight. FBF-2 recognition sequences include an extra, central nucleotide relative to the 8-mer PUF consensus sequence recognized by HsPUM1 (Opperman et al. 2005; Gupta et al. 2008; Wang et al. 2009b). That nucleotide is flipped away from the protein, exposed to solvent (Fig. 1B; Wang et al. 2009b). Based on studies of chimeric proteins, the requirement for the extra base is thought to be due to locally flattened curvature of the FBF-2 RNA-binding surface opposite the flipped base. Flanking this distortion, the contacts between RNA and protein are nearly identical to those in HsPUM1. Similarly, a crystal structure of a yeast Puf4p–RNA complex contains a flipped base (Miller et al. 2008), as do complexes of HsPUM1 with non-cognate RNAs (Gupta et al. 2008). Alternatively, yeast Puf3p possesses a specific binding pocket for an additional cytosine residue, 5′ of the consensus PUF recognition sequence (Zhu et al. 2009). Contacts with the downstream consensus sequence again are similar to those in HsPUM1.
Stacking interactions pervade PUF–RNA complexes. For example, in FBF-2, amino acid side chains in multiple repeats interact with bases, while bases in positions 4, 5, and 6 stack with one another (Fig. 1B,C; Wang et al. 2009b). These interactions results in long columns of stacking bases and amino acid side chains along the entire length of the protein. The effects of these interactions on affinity and specificity for RNA have not been explored and are the subject of this study.
To understand the role of stacking interactions, we focused on complexes formed between FBF-2 and RNA. We combined in vivo selection experiments with site-directed mutagenesis, biochemistry, and structural analysis to determine how the identities of stacking amino acid residues influence binding.
RESULTS
Stacking interactions are required
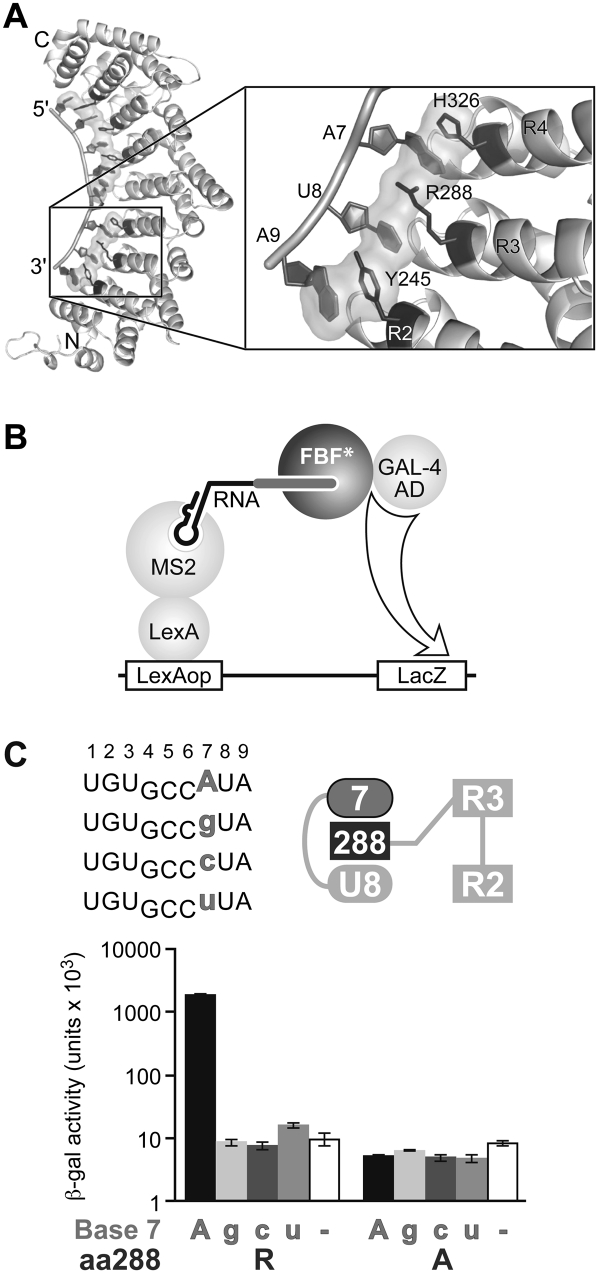
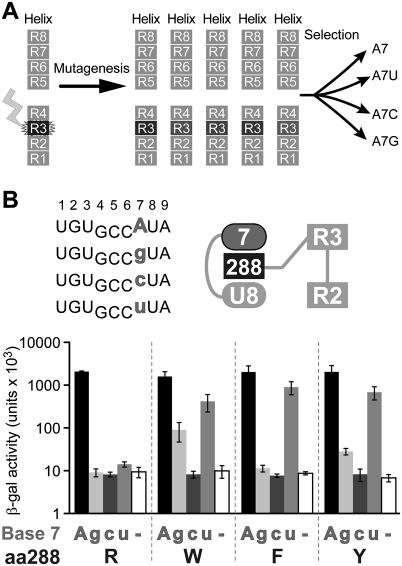
FBF-2 binds most tightly to an RNA site derived from the gld-1 3′ UTR, called the FBEa (FBF binding element), having the sequence UGUGCCAUA. (By convention, nucleotides in the RNA site are numbered +1 to +9, beginning with the UGU.) To probe the role of stacking interactions, we first focused on arginine 288 (R288) of FBF-2, which lies in the third PUF repeat, between an adenosine at position +7 and a uridine at +8. R288 forms a stacking, cation–π interaction with A7, and its aliphatic chain stacks with U8 (Fig. 2A). We analyzed an alanine substitution at this position 288, which should eliminate those interactions.
FIGURE 2.
Stacking interactions at Repeat 3 are required for binding in the yeast three-hybrid assay. (A) Structure of FBF-2, highlighting the region around R288 of Repeat 3 (Wang et al. 2009b). (B) Yeast three-hybrid assay used to screen binding of FBF-2 to various RNAs. An interaction between FBF-2 and the interrogated RNA will trigger expression of reporter gene LacZ. (C) Yeast three-hybrid assay with FBF-2 R288 (WT) or R288A. (Top left) Core elements of RNA sequences tested in the three-hybrid assay; (top right) diagram of RNA bases (ellipses) and stacking (dark gray rectangle) residue tested. RNAs with base substitutions (gray) at +7 are analyzed for binding with FBF-2 R288 (WT) and R288A in the three-hybrid assay. The WT RNA base is indicated in uppercase on the x-axis of the bar graph; mutant RNA bases in lowercase. β-Galactosidase activity reflects strength of the interactions (Hook et al. 2005).
We tested binding of the R288A protein to the gld-1 FBEa RNA sequence using the yeast three-hybrid assay (Fig. 2B). Wild-type FBF-2 (R288) and the R288A mutant protein were introduced into yeast containing plasmids encoding one of four RNAs. Each RNA possessed a different nucleotide at position +7: either A, G, C, or U (Fig. 2C). A positive RNA–protein interaction resulted in activation of the HIS3 and LacZ reporter genes (Hook et al. 2005; Stumpf et al. 2008); interactions were quantified by a β-galactosidase assay. Wild-type FBF-2 protein (R288) bound only to RNAs with an adenosine at position +7, as expected (Bernstein et al. 2005; Opperman et al. 2005). The R288A mutant protein bound poorly to each of the RNAs tested (Fig. 2C).
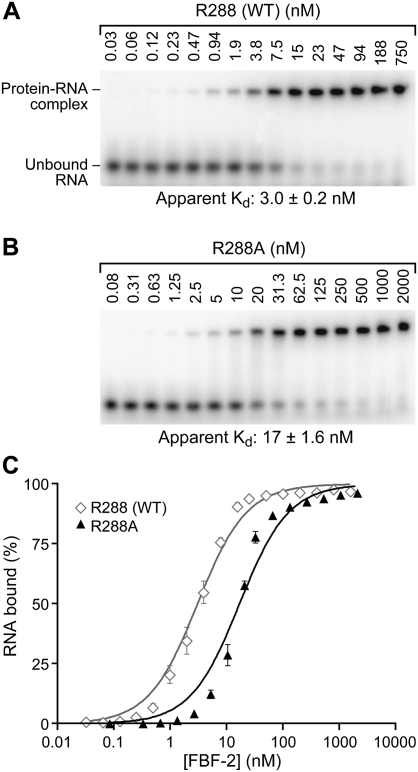
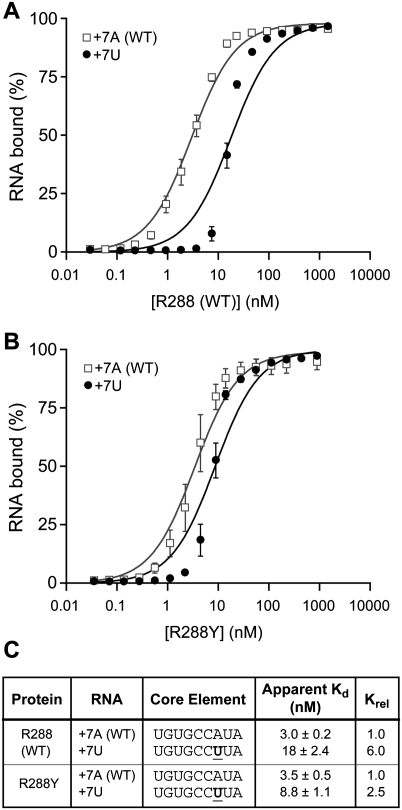
To quantify affinity of the R288A mutant protein for RNA, we compared R288 (WT) and R288A binding to the FBEa RNA element in vitro, using an electrophoretic mobility shift assay. The two proteins were fused to glutathione S-transferase (GST) and purified identically from Escherichia coli. In contrast to R288 (WT), where the protein is largely soluble in the lysate, the majority of the R288A mutant protein is insoluble; however, we obtained sufficient active protein to analyze binding in vitro (see Materials and Methods). FBF-2 R288 (WT) or FBF-2 R288A proteins were incubated with labeled, 13-nt RNAs (Fig. 3). FBF-2 R288 (WT) bound WT RNA with an apparent Kd of 3.0 ± 0.2 nM (Fig. 3A,C), while FBF-2 R288A bound WT RNA more weakly, with an apparent Kd of 17 ± 1.6 nM (Fig. 3B,C). We conclude that the arginine at position 288 is important for the affinity of the protein–RNA interaction. Structural analyses that are presented below (Fig. 7) reveal that changes in affinity with substitutions at position 288 are likely due to alterations in the stacking of R288 with RNA bases, rather than to altered charge–charge interactions.
FIGURE 3.
Stacking interactions at Repeat 3 are important for binding in vitro. Representative EMSAs testing binding affinity of purified FBF-2 R288 (WT) (A), and R288A (B) with gld-1 FBEa RNA. Protein concentrations are indicated above the gels. Apparent Kd values (indicated below the gel) were obtained by plotting fraction RNA bound versus protein concentration. The interpretation of the difference between the slopes of the R288 (WT) and R288A binding curves is unclear. (C) Quantitation of EMSA results in A and B.
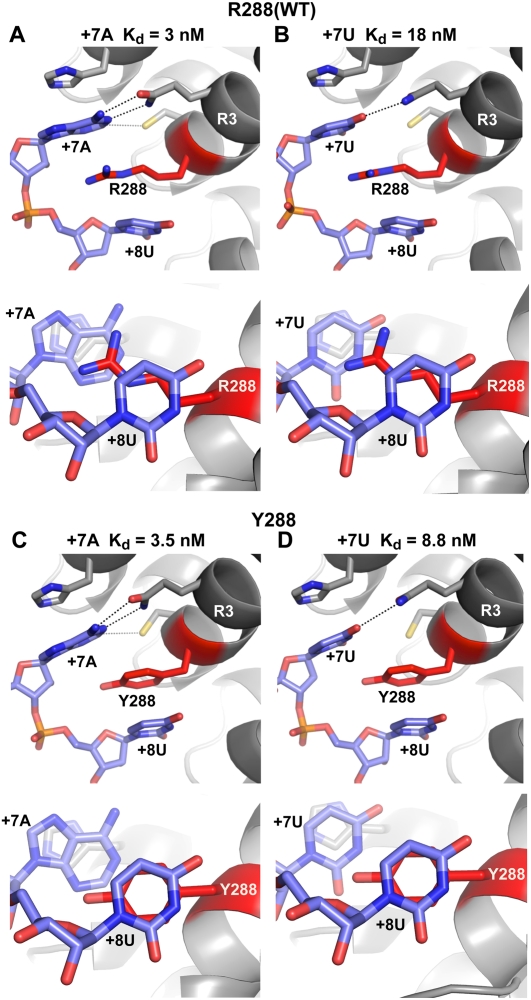
FIGURE 7.
Crystal structures of FBF-2–RNA complexes. Interaction of FBF-2 R288 (WT) with +7A (WT) RNA (A), and +7U RNA (B). Interaction of FBF-2 R288Y with +7A (WT) RNA (C), and +7U RNA (D). Structures are viewed laterally in the top panels in A–D; stacking interactions are viewed from the 3′ end of the RNA in the bottom panels. (Blue) RNA bases; (red) stacking residues at position 288. (Dashed lines) Interacting atoms. Apparent Kd values of each protein–RNA complex are indicated above each structure.
Selection of amino acid substitutions that alter RNA specificity
To test whether the identity of the amino acid residue at position 288 contributes to specificity for the RNA sequence, we performed a yeast three-hybrid selection experiment (Fig. 4A; Hook et al. 2005; Stumpf et al. 2008). Position 288 of FBF-2 was randomized by mutagenesis, and the library of resulting proteins was introduced into yeast containing plasmids encoding one of four RNAs. Each RNA possessed a different nucleotide at position +7, either A, G, C, or U. Proteins that interacted with the RNA activated the HIS3 reporter gene (Hook et al. 2005; Stumpf et al. 2008) and thus were identified by growth on selective media. Sequencing confirmed that the clones in the starting library were, indeed, randomized, and the number of transformants analyzed yielded a 99% confidence of complete coverage (data not shown).
FIGURE 4.
Identification of amino acid substitutions at position 288 of FBF-2 that alter RNA specificity. (A) Yeast three-hybrid selection used to identify stacking amino acid residues in FBF-2 Repeat 3 with altered RNA specificities. Position 288 of FBF-2 was randomized, and the library of mutants was screened with RNAs substituted at base +7. (B) Specificity testing of yeast 3-hybrid selected FBF-2 mutants with four different RNAs. (Top left) Core elements of RNA sequences tested in the three-hybrid assay; (top right) diagram of RNA bases (ellipses) and stacking residues (dark gray rectangle) tested. RNAs with base substitutions (gray) at +7 were analyzed for binding with WT FBF-2 (R288) and mutants (W, F, and Y) in the three-hybrid assay. β-Galactosidase activity reflects strength of the interactions (Hook et al. 2005).
The screen yielded three FBF-2 mutant proteins in which position 288 was altered to tyrosine (Y), tryptophan (W), or phenylalanine (F); the wild-type stacking residue, arginine (R), was isolated as well. No other mutations were isolated, suggesting that only the tyrosine, tryptophan, phenylalanine, and arginine substitutions provide sufficient affinity for selection in the three-hybrid assay (Fig. 2). However, it is formally possible that other substitutions cause misfolding.
The RNA-binding specificities of the R288 mutant proteins were analyzed using the yeast three-hybrid system (Fig. 4B). Wild-type FBF-2 protein (R288) bound only to RNAs with an adenosine at position +7. FBF-2 R288W exhibited broadened specificity, interacting with adenosine, guanosine, and uridine at position +7 (Fig. 4B). FBF R288F and R288Y accepted adenosine and uridine at position +7. We conclude that the identity of the stacking amino acid at position 288 influences specificity for the base opposite that RNA recognition helix.
Relative affinities in vitro
To quantify affinities of the mutant FBF proteins for RNA, we compared FBF-2 R288 (WT) and R288Y binding in vitro (Fig. 5). We analyzed RNAs with either a +7A (WT) or +7U because they bound differentially to R288 (WT) and R288Y proteins in the three-hybrid assay (Fig. 4B). The R288 (WT) protein bound the +7U RNA sixfold weaker than the +7A (WT) RNA, with apparent Kd's of 18 ± 2.4 nM and 3.0 ± 0.2 nM, respectively (Fig. 5A,C); in contrast, the R288Y protein bound the +7U only 2.5-fold weaker than the +7A (WT) RNA (Fig. 5B,C). The affinity of the R288Y protein for the +7A (WT) RNA remained the same as that of the R288 (WT) protein. We conclude that the arginine at 288 restricts specificity; the R288Y mutant protein is less selective, without perturbing affinity for the WT RNA sequence.
FIGURE 5.
Amino acid substitutions at position 288 of FBF-2 alter RNA specificity in vitro. (A) Base substitutions at position +7 of gld-1 RNA analyzed for binding with FBF-2 R288 (WT) in vitro using EMSAs. (B) Base substitutions at position +7 of gld-1 RNA analyzed for binding with FBF-2 R288Y in vitro using EMSAs. The interpretation of the difference between the slopes of the +7A and +7U binding curves is unclear. (C) Summary of binding affinities. Krel is the ratio of Kd relative to +7A (WT) RNA.
Specificity unaltered for neighboring bases
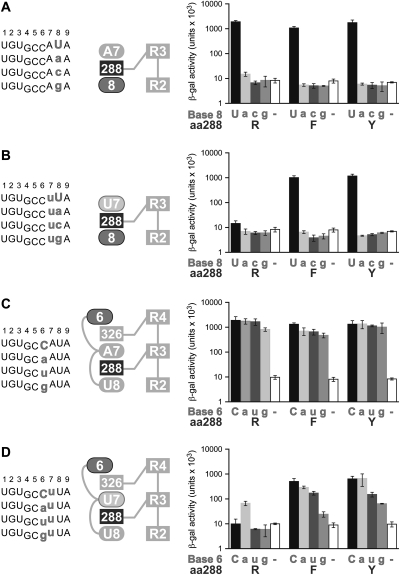
Since R288 lies between bases +7 and +8, we considered the possibility that an R288 substitution might affect specificity for position +8. We tested, in yeast three-hybrid assays, the binding of FBF-2 R288F and R288Y to RNAs possessing each of the four possible bases at +8 (Fig. 6A). The R288F and R288Y substitutions did not alter the nucleotide preference at the eighth position: Strict specificity for uridine at +8 was retained.
FIGURE 6.
RNA specificities for bases recognized by neighboring repeats are unaltered. (A–D) (left panel) RNA sequences tested; (middle panel) diagram of RNA bases (ellipses) and stacking residues (dark gray rectangles) tested. (Right panel) RNAs with base substitutions (gray) are analyzed for binding with FBF-2 R288 (WT) and mutants (F and Y) in the three-hybrid assay. β-Galactosidase activity reflects the strength of the interactions (Hook et al. 2005).
We also considered the possibility that the presence of a +7U rather than the wild-type +7A in an FBF-2 R288F or R288Y complex might alter specificity for the base at position +8. We therefore tested binding of FBF-2 R288F and R288Y to RNAs with a +7U and each of the four possible bases at +8 (Fig. 6B). As expected, FBF-2 R288 (WT) did not bind any of the RNAs with a +7U (Fig. 6B). The R288F and R288Y substitutions, which permit binding to RNAs with a +7U, did not alter the nucleotide preference at the eighth position; strict specificity for uridine at +8 was observed whether +7 was U (Fig. 6B) or A (Fig. 6A). We conclude that the preference at position +8 was unaffected by the identity of the stacking amino acid residue at position 288.
We also tested the effects of R288 substitutions on specificity at position +6. Wild-type FBF-2 (R288) does not contact position +6 in the RNA. Instead, the sixth base is flipped away from the protein, exposed to solvent (Fig. 6C). R288 (WT) and R288 mutant proteins (R288F and R288Y) bind similarly to RNAs with the different bases at +6 (Fig. 6C). A similar lack of specificity for +6 was seen with RNA carrying a +7U rather than +7A (Fig. 6D).
We conclude from this set of experiments (Fig. 6) that the R288Y protein's altered specificity is highly restricted. The tyrosine substitution affects discrimination only of the base at +7.
Structures of complexes with altered residues at amino acid 288
To analyze the structural basis of the specificities of the substitutions at amino acid residue 288, we determined the crystal structures of RNA complexes formed between FBF-2 R288Y with RNAs bearing either A or U at position +7 (Fig. 7). The other mutant proteins could not be obtained at sufficient purity and yield to permit structure determination.
As determined previously, the crystal structure of wild-type protein (R288) bound to wild-type RNA (+7A) exhibits parallel or stacked cation–π interactions between the side chain of the arginine and the adjacent adenosine at +7 and uridine at +8 (Fig. 7A; both side and axial views are shown for each structure). The interaction with R288 places +7A in a position to interact with the base-recognition side chains in Repeat 3 (Fig. 7A). With +7U, the stacked cation–π interaction of R288 with the base is minimal (Fig. 7B), and the uridine is placed opposite non-cognate amino acids, together resulting in the measured sixfold weaker interaction.
The structures of complexes with R288Y revealed differences in interactions with the +7 and +8 bases. With both wild-type (+7A) and mutant (+7U) RNAs, the aromatic ring of the tyrosine at 288 was stacked with the ring of the +8 base, a uridine, and made no contact with the base at +7 (Fig. 7C,D). With wild-type RNA, +7A retains interactions with the base-recognition side chains in Repeat 3 of R288Y and, together with the strong Y288/+8U interaction, yields a complex with similar Kd to wild type (Fig. 7A,C). With the mutant RNA (+7U), the Y288 ring also formed a strong stacking interaction with the uridine at +8, thus improving binding relative to R288 with the same RNA, although +7U was opposite non-cognate base recognition side chains in Repeat 3 (Fig. 7D).
We conclude that the stacking interactions observed in the wild-type protein balance tight binding and enhanced specificity for +7A. The R288Y protein retains affinity for the WT RNA sequence, but loses specificity.
Contributions of stacking interactions in Repeat 7
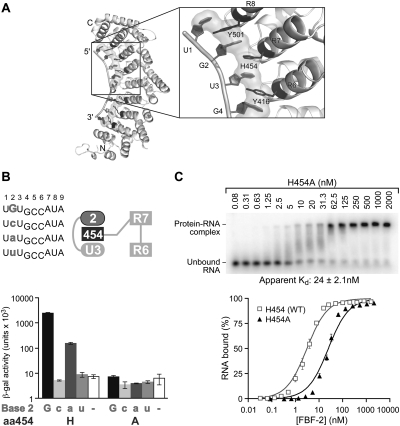
To determine whether an alanine substitution at another stacking amino acid position would affect RNA binding and specificity, we focused on Repeat 7, which contacts a guanosine at position +2 (Fig. 8A). A histidine residue, H454, lies between +2G and +3U in the structure of the wild-type complex (Fig. 8A,B; Wang et al. 2009b). A histidine residue at this position in Repeat 7 is unique to FBF-2 among PUF proteins with known crystal structures. We generated an alanine substitution at position 454 of FBF-2 and tested binding to RNAs with base substitutions at position +2 using the three-hybrid assay (Fig. 8B). FBF-2 H454 (WT) bound both purines; guanosine was preferred (Fig. 8B). In contrast, the H454A mutant protein bound none of the RNAs tested. We analyzed binding of WT RNA to both H454 (WT) and H454A proteins (Figs. 3A, 8C) using EMSAs. Similar to the R288A mutant, the H454A protein was considerably less soluble compared to WT FBF-2. Despite the low solubility of H454A, we obtained sufficient active protein for analysis in EMSAs. The H454 (WT) protein bound WT RNA with an apparent Kd of 3.0 ± 0.2 nM (Fig. 3A). In contrast, FBF-2 H454A bound WT RNA weaker, with an apparent Kd of 24 ± 2.1 nM (Fig. 8C). We infer that the H454 amino acid side chain contributes to affinity.
FIGURE 8.
Stacking interactions at Repeat 7 are important for binding. (A) FBF-2, highlighting the region around H454 of Repeat 7 (Wang et al. 2009b). (B) Specificity testing of FBF-2 H454 (WT) and H454A with four different RNAs. (Top left) Core elements of RNA sequences tested in the three-hybrid assay; (top right) a diagram of RNA bases (ellipses) and stacking residue (dark gray rectangle) tested. RNAs with base substitutions (gray) at +2 were analyzed for binding with FBF-2 H454 (WT) and H454A in the three-hybrid assay. The WT RNA base is indicated in uppercase on the x-axis of the bar graph and mutant RNA bases in lowercase. β-Galactosidase activity reflects strength of the interactions (Hook et al. 2005). (C) Representative EMSA of purified FBF-2 H454 (WT) and H454A with gld-1 FBEa RNA.
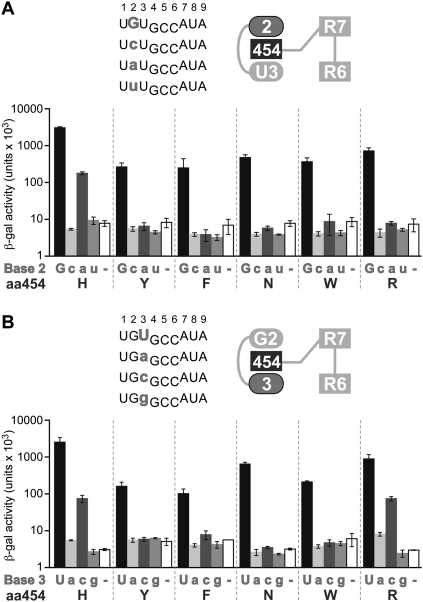
We next determined whether the identity of the amino acid residue at position 454 affected specificity of the interaction with the base +2 (Fig. 9). We substituted H454 with residues that were selected in screening at Repeat 3 or commonly found at this position in other PUF proteins (tyrosine [Y], phenylalanine [F], asparagine [N], tryptophan [W], and arginine [R]) and tested binding to RNAs with base substitutions at position +2 (Fig. 9A). Unlike FBF-2 H454 (WT), which bound both purines, all the H454 substitutions only bound RNAs with a guanosine at position +2. We conclude that the presence of histidine at position 454 broadens specificity for the base opposite the RNA-recognition helix.
FIGURE 9.
Stacking residue at Repeat 7 affects RNA specificity at RNA positions +2 and +3. RNAs with base substitutions (gray) at +2 (A) and at +3 (B) were analyzed for binding with FBF-2 H454 (WT) and mutants (Y, F, N, W, and R) in the three-hybrid assay. (Top left) Core elements of RNA sequences tested; (top right) diagram of RNA bases (ellipses) and stacking residues (dark gray rectangle) tested. The WT RNA base is indicated in uppercase on the x-axis of the bar graph; mutant RNA bases in lowercase. β-Galactosidase activity reflects the strength of the interactions (Hook et al. 2005).
Since H454 lies between bases +2 and +3, we considered the possibility that a H454 substitution might affect specificity for position +3. We tested, in yeast three-hybrid assays, the binding of FBF-2 H454 mutant proteins to RNAs possessing each of the four possible bases at +3 (Fig. 9B). FBF-2 H454 (WT) bound both pyrimidines, although uridine was preferred. H454R, which exhibited a narrowed base specificity at +2 (Fig. 9A), had a similar binding profile as FBF-2 H454 (WT). Unlike FBF-2 H454 (WT) and H454R, which bound both pyrimidines, the H454N mutant protein only bound RNAs with a uridine at position +3. Although all other H454 substitutions preferentially bind uridine at +3, we could not determine if there was truly a lack of binding to cytosine, or if the binding was below detectable limits in this assay. We conclude that the identity of amino acid residue 454 influences specificity at position +2 (by H454R), or at both +2 and +3 (by H454N).
DISCUSSION
Stacking interactions pervade PUF–RNA complexes, extending along the length of the protein–RNA interface, and influence both RNA-binding affinity and specificity. Alanine substitution experiments reveal that the stacking amino acids in FBF-2 Repeats 3 and 7 contribute to the protein's specificity for RNA sequence, as well as its affinity. Structural analysis reveals that different stacking arrangements can mediate the altered binding profiles of mutant RNAs and proteins.
In FBF-2, every base forms at least one stacking interaction with either another base or an amino acid side chain (Fig. 1; Wang et al. 2009b). Elimination of a single stacking interaction, as in R288A or H454A, decreases affinity sixfold to eightfold in vitro (Figs. 3, 8). Analogous data have been reported with certain other complexes: For example, alanine substitution of a stacking tyrosine in the MS2 coat protein–RNA complex reduces binding 160-fold, while substitution of a phenylalanine in the U1A–RNA complex is reported to reduce binding 10,000-fold (LeCuyer et al. 1996; Nolan et al. 1999). We suggest that the abundance of consecutive stacking interactions in the FBF-2–RNA complex mitigates the effects of single mutations, and local plasticity allows substitutions of stacking side chains with reduced effect on binding affinity.
In addition to their effects on affinity, stacking interactions in FBF-2 also contribute to RNA base specificity. Substitution of R288 with tyrosine, phenylalanine, or tryptophan allows recognition of different bases at the seventh position, broadening the protein's specificity. Structural analysis of the tyrosine-substituted complex suggests that the broader spectrum of base tolerance is due to stronger stacking interactions of the following base (eighth position) with the aromatic residues, which overcomes non-cognate base edge interactions of the seventh base.
The local structure of the stacking arginine in Repeat 3, R288, is conserved in PUF–RNA complexes. Comparison of complexes formed by FBF-2, yeast Puf3p, yeast Puf4p, and human Pumilio reveal that the contacts made by equivalent stacking arginines relative to the adjacent bases are very similar (RMSD = 0.2–1.0 Å over 27 atoms). Moreover, arginines from other RNA recognition helices, such as in Repeat 5 of human Pumilio, are similar. Thus our findings with R288 of FBF-2 apply broadly to base-stacking arginines among PUF proteins.
The stacking residues in each PUF repeat likely function in combination with neighboring amino acids to specify the base recognized by that RNA-recognition helix. Statistical analysis of other protein–RNA complexes suggests that the size and shape of binding pockets on the protein, coupled with a few direct contacts to the base, are critical (Auweter et al. 2006; Morozova et al. 2006). Base specificity of PUF proteins thus may also depend on the binding pocket created by the local environment of each RNA-recognition helix. Rationally designed changes in the identities of hydrogen-bonding amino acid residues can alter RNA-binding specificity in a predictable manner (Wang et al. 2002, 2009a; Opperman et al. 2005; Cheong and Hall 2006; Koh et al. 2009). However, the effects of those rationally designed substitutions act together with the identity of the stacking residue. Relative to the wild-type protein, substitutions of stacking residues can either broaden the range of acceptable bases, as seen with R288 mutations, or restrict it, as seen with H454.
The creation of PUF proteins with new specificities both by natural selection or rational design must balance specificity and binding affinity. In the evolution of PUF proteins with different specificities, the combinations of RNA-recognition side chains may be selected to increase or decrease specificity while maintaining the binding affinity needed for biological function. Optimizing target specificity and affinity involves the residues involved in stacking interactions. For rational design of PUF protein specificity, our results here suggest the selection of stacking residues is an important consideration. The identity of stacking residues in the protein can affect not only its affinity for the RNA, but also broaden the range of RNA sequences it will accept. These considerations bear directly on the design of proteins that bind the desired target RNAs without “off-target” effects.
MATERIALS AND METHODS
DNA constructs used in the yeast three-hybrid system
The RNA-binding region of FBF-2 (GenBank accession number NM_062819), comprising amino acids 121–632, was cloned into an activation domain vector, pACT2. The stacking residues analyzed, amino acids 288 and 454, were randomized by site-directed mutagenesis (QuikChange Method; Stratagene). The library, called FBF RCR 03/04, was transformed into DH5-α, and isolated plasmids were sequenced to determine the library's quality. The FBF RCR 03/04 library had ∼15%–18% contamination of wild-type FBF-2 protein. DNA oligonucleotides designed to express various RNA sequences were cloned into the XmaI and SphI sites of pIIIa MS2-2 as described previously (Hook et al. 2005).
Yeast three-hybrid assays
Three-hybrid assays were performed in the YBZ-1 yeast strain as described previously (Hook et al. 2005). Selection experiments were carried out using medium lacking histidine and in the presence of 1 mM 3-aminotriazole.
Protein construct and purification
The RNA-binding domain of FBF-2 (amino acids 164-575) was expressed as a glutathione S-transferase (GST) fusion protein in E. coli and purified using glutathione resin. The GST tag was removed by incubating with TEV protease, and the protein was further purified using a heparin column and a Superdex 200 column (Wang et al. 2009b). The monomer peak fractions were pooled and concentrated for electrophoretic mobility shift assays. FBF-2 mutants R288Y, R288A, and H454A were generated using the QuikChange site-directed mutagenesis kit (Stratagene). The mutant proteins were purified in the same way as the wild-type protein.
Crystallization, structure determination, and refinement
Crystals of the FBF-2/RNA complexes were grown by the hanging-drop vapor diffusion method at room temperature, against a crystallization solution of 100 mM Tris (pH 7.5), 8%–10% (w/v) polyethylene glycol 8000, 8% (v/v) ethylene glycol, similar to that for the wild-type protein reported previously (Wang et al. 2009b). Crystals were cryoprotected with 20% (v/v) glycerol. Diffraction data were collected from crystals at 100 K at the SER-CAT beamline 22-ID or 22-BM at the Advanced Photon Source, Argonne National Laboratory, or by using a home X-radiation source (Rigaku Micromax-007HF X-ray generator with Saturn 92 CCD detector, wavelength 1.5418 Å, NIH/NIEHS). The structures of the FBF-2/RNA complexes were determined by molecular replacement method using the FBF-2 coordinates from the FBF-2/FBE complex (PDB ID: 3K5Q) as the initial search model. Data collection and refinement statistics are shown in Table 1.
TABLE 1.
Crystallographic statistics of the FBF-2/RNA complexes
Electrophoretic mobility shift assay
One hundred picomolar 13-nt RNA oligonucleotides (Dharmacon), 32P 5′-end-labeled using T4 polynucleotide kinase, was combined with a range of protein concentrations in 10 mM HEPES (pH 7.4), 1 mM EDTA, 50 mM KCl, 2 mM DTT, 0.1 mg/mL BSA, 0.01% Tween 20 and 0.1 mg/mL yeast tRNA (Ambion) for 30 min at room temperature. Loading dye (5 μL of 6% glycerol and 0.06% Bromophenol Blue) was added to each 10-μL reaction before loading 5 μL on a pre-run non-denaturing 5% polyacrylamide gel (Bio-Rad). Gels were resolved in 1× TBE at 100 V for 30 min at 4°C. Gels were fixed (50% methanol and 10% acetic acid) and dried, followed by exposure to storage phosphor screens for 1–2 d. Storage phosphor screens were scanned using a Typhoon 9410 Workstation (GE Healthcare). The fraction of bound RNA relative to total RNA in the reaction was determined using ImageQuant (Amersham). The apparent Kd, concentration of protein at which half-maximal binding occurs, was determined using GraphPad Prism 4. The Kd values and standard errors reported here were from four independent experiments.
Dissociation constants were adjusted based on the percentage of active protein in each preparation (WT, 75%; R288Y, 45%). To determine the percentage of active protein, an RNA titration experiment was carried out. Increasing amounts of RNAs were incubated with a fixed amount of protein (WT, 50 fmol; R288Y, 80 fmol). The samples were incubated for 30 min at room temperature, and each reaction was spotted on a pre-wetted nitrocellulose filter (Millipore) that was part of a single filtration apparatus. A DEAE filter (Whatman) was placed under the nitrocellulose filter to bind free RNA. Vacuum was applied to the filtration apparatus, followed by a wash with 0.5 mL of binding buffer. Filters were dried at room temperature, and the amount of protein-bound RNA was determined by scintillation counting the nitrocellulose filters. Background counts were determined by setting up a parallel binding reaction without protein, and were subtracted from each filter. The total number of counts per reaction was determined by adding the counts from both filters. A curve of bound RNA (counts) versus total RNA (femtomoles) was plotted. The RNA is in excess where the curve plateaus, and the molar amount of bound RNA equals the molar amount of active protein.
ACKNOWLEDGMENTS
We thank members of the Wickens and Hall laboratories for discussions and suggestions. We appreciate the help of Laura Vanderploeg and Adam Steinberg in the preparation of figures. This work was supported in part by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences (to T.H.), and by extramural grants from the National Institutes of Health (to M.W.). Y.Y.K. was supported by the A*STAR National Science Scholarship from Singapore.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2540311.
REFERENCES
- Auweter SD, Oberstrass FC, Allain FH 2006. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res 34: 4943–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein D, Hook B, Hajarnavis A, Opperman L, Wickens M 2005. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA 11: 447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Li W, Zhang QS, Regulski M, Sinha N, Barditch J, Tully T, Krainer AR, Zhang MQ, Dubnau J 2008. Identification of synaptic targets of Drosophila Pumilio. PLoS Comput Biol 4: e1000026 doi: 10.1371/journal.pcbi.1000026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong CG, Hall TM 2006. Engineering RNA sequence specificity of Pumilio repeats. Proc Natl Acad Sci 103: 13635–13639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chritton JJ, Wickens M 2010. Translational repression by PUF proteins in vitro. RNA 16: 1217–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, et al. 2003. The Staufen/Pumilio pathway is involved in Drosophila long-term memory. Curr Biol 13: 286–296 [DOI] [PubMed] [Google Scholar]
- Edwards TA, Pyle SE, Wharton RP, Aggarwal AK 2001. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell 105: 281–289 [DOI] [PubMed] [Google Scholar]
- Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP 2008. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS ONE 3: e3164 doi: 10.1371/journal.pone.0003164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Herschlag D, Brown PO 2004. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol 2: E79 doi: 10.1371/journal.pbio.0020079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D 2006. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci 103: 4487–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M 2006. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol 13: 533–539 [DOI] [PubMed] [Google Scholar]
- Gupta YK, Nair DT, Wharton RP, Aggarwal AK 2008. Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity. Structure 16: 549–557 [DOI] [PubMed] [Google Scholar]
- Hook B, Bernstein D, Zhang B, Wickens M 2005. RNA–protein interactions in the yeast three-hybrid system: Affinity, sensitivity, and enhanced library screening. RNA 11: 227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye JA, Rose NC, Goldsworthy B, Goga A, L'Etoile ND 2009. A 3′UTR Pumilio-binding element directs translational activation in olfactory sensory neurons. Neuron 61: 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershner AM, Kimble J 2010. Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc Natl Acad Sci 107: 3936–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Crittenden SL 2007. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol 23: 405–433 [DOI] [PubMed] [Google Scholar]
- Koh YY, Opperman L, Stumpf C, Mandan A, Keles S, Wickens M 2009. A single C. elegans PUF protein binds RNA in multiple modes. RNA 15: 1090–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCuyer KA, Behlen LS, Uhlenbeck OC 1996. Mutagenesis of a stacking contact in the MS2 coat protein–RNA complex. EMBO J 15: 6847–6853 [PMC free article] [PubMed] [Google Scholar]
- Miller MT, Higgin JJ, Hall TM 2008. Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat Struct Mol Biol 15: 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova N, Allers J, Myers J, Shamoo Y 2006. Protein–RNA interactions: exploring binding patterns with a three-dimensional superposition analysis of high resolution structures. Bioinformatics 22: 2746–2752 [DOI] [PubMed] [Google Scholar]
- Morris AR, Mukherjee N, Keene JD 2008. Ribonomic analysis of human Pum1 reveals cis–trans conservation across species despite evolution of diverse mRNA target sets. Mol Cell Biol 28: 4093–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Kimble J 2006. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 441: 1068–1074 [DOI] [PubMed] [Google Scholar]
- Nolan SJ, Shiels JC, Tuite JB, Cecere KL, Baranger AM 1999. Recognition of an essential adenine at a protein–RNA interface: Comparison of the contributions of hydrogen bonds and a stacking interaction. J Am Chem Soc 121: 8951–8952 [Google Scholar]
- Opperman L, Hook B, DeFino M, Bernstein DS, Wickens M 2005. A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat Struct Mol Biol 12: 945–951 [DOI] [PubMed] [Google Scholar]
- Pique M, Lopez JM, Foissac S, Guigo R, Mendez R 2008. A combinatorial code for CPE-mediated translational control. Cell 132: 434–448 [DOI] [PubMed] [Google Scholar]
- Schweers BA, Walters KJ, Stern M 2002. The Drosophila melanogaster translational repressor Pumilio regulates neuronal excitability. Genetics 161: 1177–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf CR, Opperman L, Wickens M 2008. Chapter 14. Analysis of RNA–protein interactions using a yeast three-hybrid system. Methods Enzymol 449: 295–315 [DOI] [PubMed] [Google Scholar]
- Suh N, Crittenden SL, Goldstrohm A, Hook B, Thompson B, Wickens M, Kimble J 2009. FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics 181: 1249–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zamore PD, Hall TM 2001. Crystal structure of a Pumilio homology domain. Mol Cell 7: 855–865 [DOI] [PubMed] [Google Scholar]
- Wang X, McLachlan J, Zamore PD, Hall TM 2002. Modular recognition of RNA by a human Pumilio-homology domain. Cell 110: 501–512 [DOI] [PubMed] [Google Scholar]
- Wang Y, Cheong CG, Hall TM, Wang Z 2009a. Engineering splicing factors with designed specificities. Nat Methods 6: 825–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Opperman L, Wickens M, Hall TM 2009b. Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein. Proc Natl Acad Sci 106: 20186–20191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R 2002. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet 18: 150–157 [DOI] [PubMed] [Google Scholar]
- Ye B, Petritsch C, Clark IE, Gavis ER, Jan LY, Jan YN 2004. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr Biol 14: 314–321 [DOI] [PubMed] [Google Scholar]
- Zamore PD, Williamson JR, Lehmann R 1997. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA 3: 1421–1433 [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Stumpf CR, Krahn JM, Wickens M, Hall TM 2009. A 5′ cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc Natl Acad Sci 106: 20192–20197 [DOI] [PMC free article] [PubMed] [Google Scholar]