Abstract
Self-cleaving glmS ribozymes selectively bind glucosamine-6-phosphate (GlcN6P) and use this metabolite as a cofactor to promote self-cleavage by internal phosphoester transfer. Representatives of the glmS ribozyme class are found in Gram-positive bacteria where they reside in the 5′ untranslated regions (UTRs) of glmS messenger RNAs that code for the essential enzyme L-glutamine:D-fructose-6-phosphate aminotransferase. By using comparative sequence analyses, we have expanded the number of glmS ribozyme representatives from 160 to 463. All but two glmS ribozymes are present in glmS mRNAs and most exhibit striking uniformity in sequence and structure, which are features that make representatives attractive targets for antibacterial drug development. However, our discovery of rare variants broadens the consensus sequence and structure model. For example, in the Deinococcus-Thermus phylum, several structural variants exist that carry additional stems within the catalytic core and changes to the architecture of core-supporting substructures. These findings reveal that glmS ribozymes have a broader phylogenetic distribution than previously known and suggest that additional rare structural variants may remain to be discovered.
Keywords: glucosamine-6-phosphate (GlcN6P), glutamine synthetase, Infernal, phosphoglucosamine mutase, riboswitch, self-cleaving ribozyme
INTRODUCTION
Riboswitches are regulatory elements that are typically found in the 5′ UTRs of mRNAs and that bind small molecules to control gene expression (Roth and Breaker 2009; Serganov 2009; Ames et al. 2010, 2011). Most metabolite-responsive riboswitches control the expression of genes whose protein products function in pathways that affect the concentration of the ligand sensed by the riboswitch. Each distinct riboswitch class is defined by its highly conserved ligand-binding aptamer that controls gene expression using an expression platform that commonly regulates transcription, translation, or alternative splicing of mRNAs (Barrick and Breaker 2007).
An unusual riboswitch class that senses glucosamine-6-phosphate (GlcN6P) uses its ligand as a cofactor to promote RNA self-cleavage (Winkler et al. 2004; Klein and Ferré-D'Amaré 2006; Cochrane et al. 2007). Examples of glmS ribozymes have been shown to be highly selective for GlcN6P and exhibit rate enhancements of greater than six orders of magnitude for RNA cleavage via internal phosphoester transfer (McCarthy et al. 2005; Wilkinson and Been 2005). Although GlcN6P-induced ribozyme action cleaves the mRNA, the scission is made upstream of the adjoining glmS open reading frame (ORF) and does not interfere with the coding region. However, in Bacillus subtilis, ribozyme self-cleavage triggers the action of the nuclease RNase J1 that rapidly degrades the glmS mRNA and thereby down-regulates GlmS protein production (Collins et al. 2007).
GlmS enzymes initiate the UDP-N-acetylglucosamine (UDP-GlcNAc) biosynthetic pathway by converting fructose-6-phosphate and glutamine into GlcN6P and glutamate. In both Gram-positive and Gram-negative bacteria, the glmS gene is essential for the production of peptidoglycan and extracellular lipopolysaccharides (Collins et al. 2007; Görke and Vogel 2008), though the mechanisms of glmS gene regulation differ. In certain Gram-positive bacteria, glmS ribozymes repress the production of GlmS protein by monitoring the concentration of its key enzymatic product, GlcN6P. Previously known examples of glmS ribozymes were identified by using comparative sequence analysis algorithms and were exclusively found in Gram-positive bacteria (Barrick et al. 2004; Winkler et al. 2004; Roth et al. 2006; Gardner et al. 2009). Each bacterium that contains a glmS ribozyme carries a single representative exclusively located in the 5′ UTR of its glmS gene. In contrast, the Gram-negative bacterium Escherichia coli has been shown to regulate the glmS gene by using the GlmY and GlmZ small RNAs (sRNAs) (Görke and Vogel 2008; Fröhlich and Vogel 2009).
RNA regulatory systems such as glmS ribozymes are candidates for targeting by antibacterial compounds that misregulate riboswitch-mediated gene expression (Blount and Breaker 2006). A number of studies have been conducted on glmS ribozymes to reveal key structural and functional details. For example, sequence and structural comparisons, mutational analyses, and crystallography experiments have shown that the secondary and tertiary structures for glmS ribozymes are complex and highly conserved (Wilkinson and Been 2005; Klein and Ferré-D'Amaré 2006; Roth et al. 2006; Soukup 2006; Cochrane et al. 2007). The ribozyme has also been subjected to directed evolution in an attempt to change substrate specificity or conserved nucleotides within the ribozyme, but all active ribozyme variants retained the catalytic core consensus sequence (Link et al. 2006). These results suggest that the core will remain highly conserved even in distantly related bacteria, and therefore analogs that trigger ribozyme action in one representative are likely to promote catalysis in distant homologs.
Although the precise mechanism of RNA cleavage by glmS ribozymes remains unresolved (Klein and Ferré-D'Amaré 2006; Brooks and Hampel 2009; Cochrane et al. 2009; Banáš et al. 2010), key aspects of its function have been established. The RNA promotes an internal phosphoester transfer reaction (Winkler et al. 2004) to yield RNA termini identical to those of other small self-cleaving ribozymes (Ferré-D'Amaré and Scott 2010). Magnesium ions are important for glmS ribozyme structure formation, but do not participate directly in catalysis (Roth et al. 2006; Klawuhn et al. 2010). A key requirement is the presence of a primary amine on the GlcN6P cofactor (Winkler et al. 2004; McCarthy et al. 2005; Lim et al. 2006), which is in position to protonate the 5′ oxyanion leaving group during the phosphoester transfer reaction (Klein and Ferré-D'Amaré 2006; Cochrane et al. 2007; Xin and Hamelberg 2010).
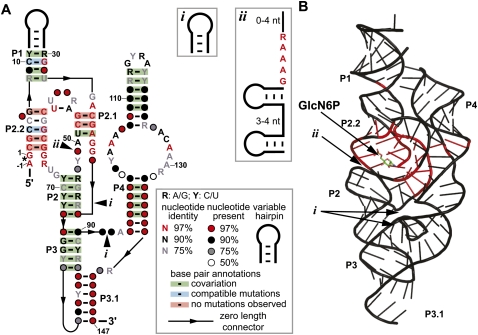
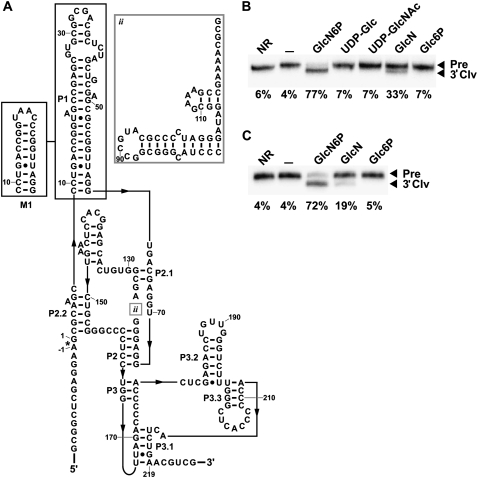
The extraordinary demands on the catalytic core to selectively bind GlcN6P, properly position this cofactor at the active site, and promote RNA cleavage may greatly restrict the sequence and structural diversity of glmS ribozymes. In this work, we sought to identify all glmS ribozyme representatives encoded in existing bacterial DNA sequence databases, including any distant variants that would reveal natural changes to the catalytic core or to structural support architectures. We utilized a bioinformatics search strategy that has previously yielded different types of riboswitch or ribozyme structural variants (Barrick et al. 2005; Kim et al. 2007; Weinberg et al. 2008; Weinberg and Breaker 2011; Perreault et al. 2011). We also used a different search strategy that has previously uncovered several new noncoding RNAs (ncRNAs) as well as a new class of S-adenosylmethionine-binding riboswitches (Meyer et al. 2009; Poiata et al. 2009). Our searches revealed additional consensus glmS ribozymes that were previously unknown, glmS ribozymes with novel genetic associations, and new types of structural variants. We also discovered a glmS ribozyme that lacks a support structure for the catalytic core that is found in all other known glmS ribozymes. Despite the variations in genetic contexts and structures of these glmS ribozymes, binding and functional specificity for GlcN6P is retained. These newfound RNAs were used to generate a revised consensus sequence and secondary structure model (Fig. 1A).
FIGURE 1.
Revised consensus model for glmS ribozymes and its relationship to an atomic-resolution structure model. (A) Consensus sequence and secondary structure model for glmS ribozymes. The image was drawn using the R2R program (Weinberg and Breaker 2011) and the output was modified to conform to a previously published glmS ribozyme structural model (Barrick and Breaker 2007). The asterisk denotes the site of ribozyme self-cleavage (Winkler et al. 2004). Optional hairpin (i) or optional hairpins and conserved nucleotides (ii) are present in some representatives. When provided, numbered nucleotides match a previously published structure (Klein and Ferré-D'Amaré 2006). (B) An atomic-resolution structure model (Protein Databank reference 3B4C) (Klein et al. 2007) of a glmS ribozyme with nucleotide identities that are at least 97% conserved shaded red. Arrows designate the locations of newly identified optional structures i and ii. The cofactor GlcN6P is depicted in ball-and-stick form.
RESULTS AND DISCUSSION
An expanded collection of consensus glmS ribozymes
Additional representatives of a known class of structured RNAs can be identified by their similarity to a consensus sequence and structural model. To search for more glmS ribozymes, we first created an updated consensus model by using all representatives from the Rfam database (Gardner et al. 2009). This model was then used to define the search parameters for the discovery of additional representatives by analyzing genomic DNA from the RefSeq38 and metagenomic databases with the Infernal 1.0 program (Nawrocki et al. 2009). This algorithm is used to search for nucleotide sequences that conform to a consensus model, wherein close matches are assigned low “E-values” and increasingly poor matches are assigned increasingly higher “E-values” (Infernal 1.0 user manual).
As additional glmS ribozyme representatives were identified, the consensus model was manually revised and new searches were conducted using revised search parameters until no more examples were uncovered. We also conducted searches based on a simplified consensus model wherein P3, P3.1, and P4 substructures (Fig. 1) were removed. These substructures are not essential for catalysis, but have been shown to promote higher rate constants for RNA cleavage (Winkler et al. 2004). This consensus model simplification permits any RNAs that carry the ribozyme core but that lack these supporting substructures to be scored higher by the Infernal 1.0 algorithm. This approach uncovered additional glmS ribozyme sequences in DNA from partially assembled genomes and metagenomic sequence data sets in which the catalytic core of the ribozyme was within a contig, but the sequenced fragment lacked regions P3 through P4.
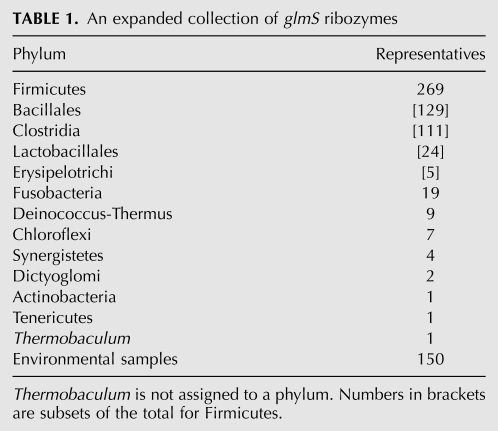
These combined searches increased the number of glmS ribozyme representatives from 160 to 463. Numerous additional examples were identified in expected phyla as well as some phyla not previously known to carry glmS ribozymes (Table 1). Slightly more than two-thirds of these representatives were found via Infernal 1.0. These are believed to be true hits because they carry all the sequence and structural hallmarks of known glmS ribozymes. Furthermore, when candidates are ranked by “E-value,” we found a stark boundary between all candidates affiliated with glmS or GlcN6P-related ORFs and those that are not. The sequences of a few glmS ribozymes from each phylum were used in BLAST searches of genomic DNA that was not within the sequence databases originally examined, which yielded slightly less than one-third of the glmS ribozymes identified.
TABLE 1.
An expanded collection of glmS ribozymes
Firmicutes and other Gram-positive bacteria still represent the largest collection of organisms that have the glmS ribozyme, in agreement with previous observations (Collins et al. 2007; Görke and Vogel 2008). We also found additional instances of glmS ribozymes in several other phyla that were previously not known to carry this ribozyme class (Supplemental Table 1). Though the number of Gram-negative species that have glmS ribozymes is small in comparison to the Gram-positive species with this RNA, it does indicate that the glmS ribozyme has a broader distribution than previously thought. All currently sequenced members of the phyla Fusobacteria and Deinococcus-Thermus appear to have the glmS ribozyme, but this may change due to the current paucity of sequenced organisms within each phylum. We identified the first instance of a glmS ribozyme occurring in a member of the Mollicutes class of the Tenericutes phylum. These organisms are notable for their lack of cell walls (Brown et al. 2007) and for their lack of lipid A in the extracellular lipopolysaccharide layer (Madigan et al. 2008), both of which require GlcN6P for their synthesis. Although the presence of a glmS ribozyme and gene in an organism that lacks major products that require GlcN6P is rare, organisms from other phyla possess the glmS gene, though without the glmS ribozyme, and share the same cell wall deficits as Tenericutes (Jenkins et al. 2002). We also found glmS ribozymes in a wide assortment of metagenomic sequences, including those from the human gut microbiome, various Antarctic sources, soil samples, hot springs, whale fall, freshwater samples, hypersaline mats, and terephthalate-degrading communities. These findings suggest that the glmS ribozyme class is among the most common of riboswitch classes in bacteria (Breaker 2010).
Distinct structural features outside the binding pocket of some glmS ribozymes
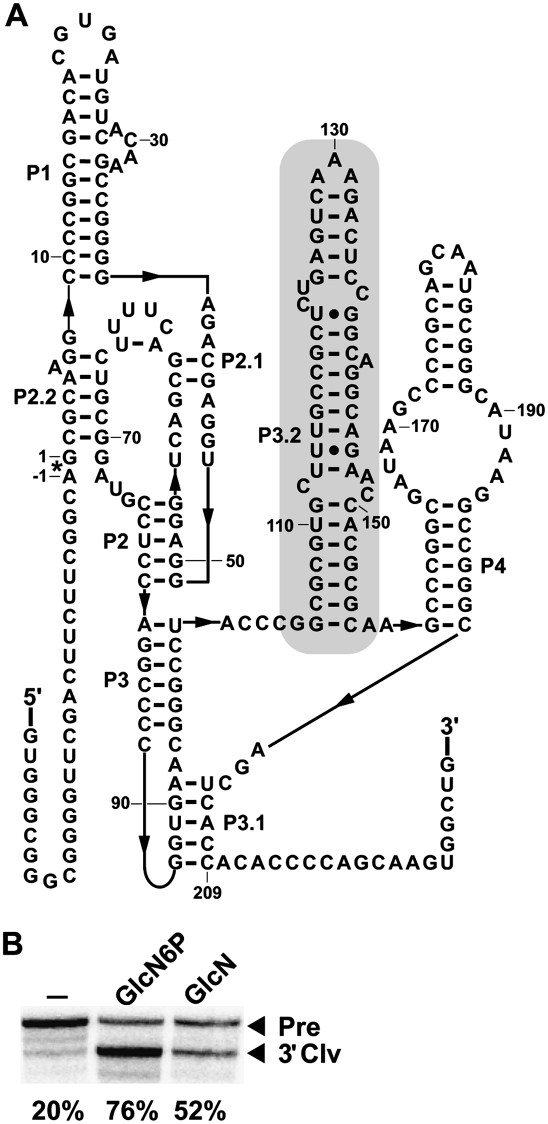
Most of the glmS ribozyme representatives identified carry the conserved sequences and substructures that were known to be characteristic of this ribozyme class. A few exceptions include a very small number of Clostridia examples that possess a hairpin that is found between P2.1 and P2 (Fig. 1). Also, it was previously noted (Roth et al. 2006) that a single glmS ribozyme representative was present in the Deinococcus-Thermus phylum that lacks a consensus P4 stem. This latter find prompted us to reexamine possible distinct sequence and structural features near the 3′ terminus of each example. Our search revealed five additional glmS ribozymes from various species within the Deinococcus-Thermus phylum, including Deinococcus radiodurans, Deinococcus geothermalis, Deinococcus desertii, Meiothermus ruber, and Meiothermus silvanus.
Within these examples, there is a hairpin of variable length in the region between P3 and P4 (i box in Fig. 1; shaded region in Fig. 2A). This hairpin had been previously mentioned as a possible novel P4 (Roth et al. 2006), but our new bioinformatics searches were able to identify a consensus P4 and the functionally important P3.1 pseudoknot (Wilkinson and Been 2005). An example of this structural arrangement, as adopted by the glmS ribozyme from D. radiodurans (Fig. 2A), is functional when exposed to GlcN6P at a concentration of 200 μM. Also, lower activity is observed with glucosamine (GlcN) (Fig. 2B), which was previously known to less-efficiently trigger glmS ribozyme self-cleavage (Winkler et al. 2004; Lim et al. 2006; Soukup 2006).
FIGURE 2.
Structure and activity of the glmS ribozyme from D. radiodurans. (A) The sequence and proposed secondary structure of the glmS ribozyme from D. radiodurans. The shaded region (P3.2) corresponds to a variable hairpin (i). (B) A D. radiodurans glmS ribozyme construct was analyzed using an in vitro transcription ribozyme cleavage assay. The reactions were incubated without an additive (–), or in the presence of 200 μM GlcN6P or glucosamine (GlcN), and the resulting precursor (Pre) RNAs were separated from 3′ cleavage products (3′ Clv) using denaturing PAGE. The percentages of the glmS ribozymes undergoing self-cleavage are presented below each lane and were derived from the single experiments depicted.
Distinct structural features within the binding pocket of some glmS ribozymes
Our bioinformatics searches revealed two additional structural features within the binding pocket of some glmS ribozymes. First, all glmS ribozymes that are from organisms in the order Deinococcales contain a guanosine residue within the 3′ portion of P2.1, instead of the uridine that pairs to the adenosine residue at the 5′ portion of P2.1 (uridine 51 in Fig. 1A; guanosine 57 in Fig. 2B). The guanosine residue still contains the keto group that is important for ribozyme function (Cochrane et al. 2009) and does not appear to interfere with ribozyme function in D. radiodurans (Fig. 2B). Outside of the Deinococcus-Thermus phylum, this variation occurs four times and is distributed amongst several phyla.
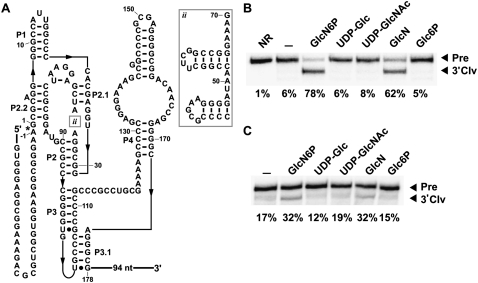
Second, our bioinformatics searches within the Meiothermus genus also revealed an additional module that is positioned just before the catalytic core (ii box in Fig. 1) exclusively in M. ruber, M. silvanus, Thermus thermophilus, Thermus aquaticus, and Truepera radiovictrix. This newly discovered feature consists of two variable hairpins, with a short linker between the two hairpins, followed by the conserved sequence GAAAR. The loop in the 3′ hairpin is almost always a tetraloop, some of which are of the GNRA or UNCG types. Due to close proximity of this new insertion to the binding pocket for GlcN6P, we felt it was necessary to determine whether the insertion is an adaptation for a different cofactor. This was assessed by examining the activity of a glmS ribozyme from T. thermophilus HB8 (Fig. 3A) with GlcN6P, GlcN, and several other related compounds. Thermales inhabit high-temperature environments, and therefore our assays were conducted at 65°C (Fig. 3B) as well as 37°C (Fig. 3C). At 65°C, the ribozyme cleaves to ∼78% completion within 5 min of exposure to 200 μM GlcN6P compared to 62% completion with 200 μM GlcN. No cleavage was detected with other ligands tested under similar conditions (Fig. 3B). Poorer yields were generated at 37°C with GlcN6P and GlcN, while still showing no cleavage with the other ligands that were tested (Fig. 3C). Although this variant rejects other ligands, the core insertion may have reduced its ability to discriminate strongly against GlcN as do other glmS ribozymes.
FIGURE 3.
Structure and activity of the glmS ribozyme from T. thermophilus. (A) The sequence and proposed secondary structure of the glmS ribozyme from T. thermophilus HB8. (B) Self-cleaving ribozyme assays using purified internally 32P-labeled RNAs with various ligands. Reactions were conducted at 65°C in absence of ligand (–) or with 200 μM of the compounds noted. NR indicates no reaction, and other notations are as described in Figure 2B. (C) Transcription/cleavage assays were conducted with the T. thermophilus glmS ribozyme as described in Figure 2B.
The glmS ribozyme found within T. radiovictrix, a thermophilic Deinococcales bacterium, is unusual for several reasons. This RNA possesses an insertion in the catalytic core that is similar to the one seen in the T. thermophilus glmS ribozyme, except that four additional nucleotides are present after the GAAAR sequence (Fig. 4A). The RNA carries a guanosine residue within the catalytic core that replaces an otherwise highly-conserved uridine, which is typical of Deinococcales species as stated above. Also notable is the region between P2.1 and P2.2, which is longer than in any other glmS ribozyme. Several differences are evident outside the catalytic core region. A sequence corresponding to the consensus P4 stem, otherwise present in every glmS ribozyme, is not observed in the entire intergenic region upstream of the glmS gene. However, there are two possible smaller stems located between P3 and P3.1. Furthermore, there is a GNRA tetraloop within the P1 stem, which is also about twice the normal length of P1 stems of glmS ribozymes. Despite the unique features of the T. radiovictrix representative, this RNA retains ribozyme activity in the presence of GlcN6P at 50°C (Fig. 4B) that is similar to glmS ribozymes with more conventional architectures. The glmS ribozyme cleaves to a slightly reduced extent at 37°C, but still maintains the same selectivity for GlcN6P (data not shown).
FIGURE 4.
Structure and activity of the wild-type and a mutant glmS ribozyme from T. radiovictrix. (A) The sequence and proposed secondary structure of the glmS ribozyme from T. radiovictrix. A portion of the P1 stem was deleted to yield construct M1, which removes nucleotides 20–52 and changes the loop sequence from GCGA to UAAC. The nucleotide positions within this RNA correspond to the numbering scheme in the wild-type construct. (B) Wild-type ribozyme self-cleavage assays conducted at 50°C as described for Figure 3B. Other annotations are as described for Figure 2B. (C) M1 ribozyme self-cleavage assays conducted as described in B.
The extended P1 stem and associated GNRA tetraloop led us to hypothesize that this glmS ribozyme may utilize this structure to support catalytic core folding and therefore replace the function of the missing P4 stem. To test this hypothesis, we removed half of the P1 stem and replaced the GNRA tetraloop with UAAC (Fig. 4A), which mimics the architecture of a GNRA tetraloop (Hendrix et al. 2005). Despite these drastic changes, the mutated glmS ribozyme from T. radiovictrix maintains the same relative activity at 50°C as the wild-type construct (Fig. 4C). This finding implies that the P1 helix does not serve as a support structure for the catalytic core.
Although some other glmS ribozymes are inactive at 60°C (Tinsley et al. 2007; Furchak et al. 2008), the glmS ribozyme from T. thermophilus requires elevated temperatures for proper function. It is possible that high G-C content (∼75%), coupled with the newly discovered insertion from Thermales and T. radiovictrix, are two adaptations that the glmS ribozyme utilizes in all but one thermophilic organism within the Deinococcus-Thermus phylum to maintain function at high temperatures. This is in contrast to the glmS ribozyme from Thermoanaerobacter tengcongensis, which has a lower G-C content (∼51%) and does not have any additional insertions (Barrick et al. 2004; Klein and Ferré-D'Amaré 2006). Due to the divergence between T. radiovictrix and members of the Thermales order (Albuquerque et al. 2005), it is likely that the ancestral form of the glmS ribozyme within the Deinococcus-Thermus phylum possessed the newly found insertion seen in Thermales and T. radiovictrix.
Distinct genomic locations of two glmS ribozymes
The glmS ribozyme was named due to its invariant presence in the 5′ UTRs of glmS genes (Winkler et al. 2004). However, two of the numerous RNA representatives found in the current study that correspond to the glmS ribozyme consensus are associated with other genes. One example is present upstream of the glmM ORF of Dethiobacter alkaliphilus, a Firmicute found in soda lakes (Sorokin et al. 2008), and the other example is located upstream of the glnA gene of Sebaldella termitidis, a Fusobacterium in the termite gut (Harmon-Smith et al. 2010). Both organisms also have glmS ribozymes in the 5′ UTRs of their glmS mRNAs, making these the first known instances of multiple glmS ribozymes within the same organism.
The D. alkaliphilus GlmM protein, phosphoglucomutase, converts GlcN6P into glucosamine-1-phosphate (GlcN1P), and therefore it functions one step after GlcN6P synthesis in this biochemical pathway. The S. termitidis GlnA protein, glutamine synthetase, converts L-glutamate into L-glutamine, which is an immediate precursor to GlcN6P. Although the functions of these gene products are only one step removed from the function of GlmS, and the RNA motifs associated with these genes conform to the consensus glmS ribozyme sequence and secondary structure, their unusual genomic locations raised the possibility that they may be triggered by a cofactor other than GlcN6P. Therefore, we examined activity of the D. alkaliphilus glmS ribozyme associated with the glmM ORF using GlcN1P or GlcN6P, and we examined the activity of the S. termitidis glmS ribozyme associated with the glnA ORF using L-glutamate, L-glutamine, or GlcN6P. Both RNAs only exhibit ribozyme activity in the presence of GlcN6P (data not shown), demonstrating that these RNAs retain their original cofactor specificity despite their unusual genomic locations.
Many other riboswitch classes that control various components of the synthesis or transport of specific metabolites are far more commonly associated with closely related steps in biochemical pathways compared to glmS ribozymes (Wang and Breaker 2008; Roth and Breaker 2009). Thus, the unusual placement of glmS ribozymes in the 5′ UTRs of the glmM and glnA mRNAs was considered more carefully. Location of a glmS ribozyme upstream of the glmM ORF is particularly noteworthy, since glmS ribozymes are normally expected to deactivate gene expression (Winkler et al. 2004; Collins et al. 2007). Production of GlmM is expected to be activated when GlcN6P is plentiful, and therefore it is possible that glmS ribozyme action will activate expression of the glmM ORF in D. alkaliphilus, thus allowing GlcN6P to be utilized in subsequent anabolic reactions.
Since GlcN6P synthesis consumes large amounts of nitrogen in the form of glutamine in bacteria (Milewski 2002), GlcN6P may be a useful ligand for S. termitidis cells to detect nitrogen availability. The regulation of the glnA mRNA by the glmS ribozyme suggests that this extra level of genetic control may be involved in nitrogen metabolism and the nitrogen starvation response in S. termitidis as glutamine levels are used by the bacterial cell as an indicator of nitrogen starvation (Hu et al. 1999). Regulation of the glnA mRNA via a glmS ribozyme could provide a signal that nitrogen in general and glutamine in particular is plentiful because GlcN6P is abundant.
Implications of the consensus and distribution of glmS ribozymes
It is striking that the main features of the catalytic core of glmS ribozymes do not mutate throughout the many examples that we have uncovered. The nucleotides of glmS ribozymes identified to date that are in direct contact with the GlcN6P retain their identities as had been revealed previously via directed evolution experiments on a population of glmS ribozyme variants (Link et al. 2006). The P2.2 and P2.1 stems, as well as other nucleotides that contact GlcN6P, remain highly conserved among every glmS ribozyme example that has been uncovered so far (Fig. 1A). It seems reasonable to speculate that variants close in sequence-space to glmS ribozymes but that have altered cofactor specificity (e.g., another primary amine-containing metabolite) would be exploited by some organisms if such RNAs were accessible and useful. However, our findings support the hypothesis that the catalytic core of this ribozyme class cannot tolerate mutations without loss of function.
In contrast, there is some sequence and structural variability permitted outside of the catalytic core and near the binding pocket of glmS ribozymes (Fig. 1). The most unusual of these structures is carried by the T. radiovictrix glmS ribozyme, which has eliminated the otherwise highly conserved P4 helix. Despite the differing architectures within the T. radiovictrix glmS ribozyme, the catalytic core of the ribozyme still functions normally. Thus, it is not clear why this alternative structure is so rare.
The expanded distribution of glmS ribozymes identified in this study suggests that it may be possible to develop a class of antibiotics that exclusively targets glmS ribozymes within a broad range of Gram-positive pathogenic bacteria. Representatives are present in many important human pathogens such as Bacillus anthracis, Clostridium botulinum, Clostridium difficile, Listeria monocytogenes, Staphylococcus aureus, and others (Supplemental Table 1). The key regulatory function and highly conserved catalytic core of glmS ribozymes are features that create an attractive target for the development of antibacterial compounds. Inhibition of glmS gene expression has been shown to result in marked differences in bacterial cellular morphology and cell viability (Milewski 2002; Collins et al. 2007). Therefore, an analog of GlcN6P that triggers ribozyme action and suppresses glmS gene expression should function as an antibacterial agent against many Gram-positive bacteria. Analogs based on GlcN6P have been shown to trigger ribozyme function in vitro (Lim et al. 2006), and the remarkable constancy within the catalytic core of glmS ribozymes may allow the synthesis of additional analogs that broadly trigger glmS ribozyme self-cleavage in cells.
MATERIALS AND METHODS
Bioinformatics searches
The microbial sets from RefSeq38 and environmental data sets (Perreault et al. 2011) were used in our searches. Searches were conducted by raising the maximally accepted E-value to 100, 500, 1000, and 5000, and were conducted in both global and local modes (Nawrocki et al. 2009). As some environmental sample hits do not yield complete ribozyme structures, we counted partial hits in our data set provided that the hits had either P2.2 through the end of P2 without a genetic context or had every component of the ribozyme from the start of P2.1 to the end of P3.1 and were positioned in 5′ UTRs of glmS genes. When additional glmS structural variations were verified or detected (see Results), we changed the consensus secondary structure lines within the glmS ribozyme Stockholm alignment to mirror the new structures. A new Stockholm alignment was made for each new class of variation using only structures that contained the variance in question and rerun in Infernal, as mentioned above. To validate output from our Infernal searches, the glmS crystal structures and sequences (Klein and Ferré-D'Amaré 2006; Barrick and Breaker 2007; Cochrane et al. 2007) were compared with our results to verify new hits. Proximity of a hit to the glmS gene was also used as a validation criterion, though this was not a strict requirement. For a full list of our findings, see Supplemental Table 1.
Chemicals, DNA oligonucleotides, ribozyme constructs
The compounds glucosamine-6-phosphate, glucosamine-1-phosphate, glucosamine, glucose-6-phosphate, L-glutamate, L-glutamine, UDP-glucose, and UDP-N-acetylglucosamine were obtained from Sigma-Aldrich.
DNA templates for in vitro transcription of glmS ribozymes were created by PCR amplification from genomic DNA or from synthetic DNAs. The RNAs produced do not necessarily carry 5′ termini that are biologically relevant, but have been chosen to assure the difference in size between the precursor RNAs and 3′ cleavage fragments were sufficient to permit easy separation by PAGE. The glmS ribozyme from D. alkaliphilus was constructed de novo using the following DNAs:
Primer 1, 5′-TAATACGACTCACTATAGGGATGATTATCGACAGCTGCATAGCGAAGCGC;
Primer 2, 5′-CCTCCCCCTCGTCAACACGCAAAACACGTGTTCTGGCGCTTCGCTATGCA;
Primer 3, 5′-TTGACGAGGGGGAGGTTTATCGAATTTTTCGGCGGATGCCTCCCGGCCTG;
Primer 4, 5′-CACTCATCGGTTTTGTAGGTTCCTTACGGCCGTGGCAGGCCGGGAGGCAT;
Primer 5, 5′-CAAAACCGATGAGTGATCAGCGGGACAAAGGGGTTTGCAGTGGCTTATTT; and
Primer 6, 5′-AACACAAATAAGCCACTGCA.
The glmS ribozyme from S. termitidis was constructed de novo using the following DNAs:
Primer 1, 5′-TAATACGACTCACTATAGGGTAGGGATTAAAAAAAATGAAAGCGCTAGGG;
Primer 2, 5′-TCTCTCCTCGTCAACTAAAATATTTGGTATTTTAGCCCTAGCGCTTTCAT;
Primer 3, 5′-GTTGACGAGGAGAGAGAATTATCGAGACTATCAGCGGGTATTCTCTGGGT;
Primer 4, 5′-TTATAAGGTTTGTTATTATCTTTTAGGTTGTTACAACCCAGAGAATACCC;
Primer 5, 5′-TAACAAACCTTATAAGTAATTATATGAACAAAAATATCTTAGTTACTAAT; and
Primer 6, 5′-ATTAGTAACTAAGATATTTTTGTTC.
The glmS ribozyme from T. radiovictrix was constructed de novo using the following primers:
Primer 1, 5′-TAATACGACTCACTATAGGGGCGGCTCGAGGAAGCGCAAGCCCTGACCGG;
Primer 2, 5′-CGCTCGCAGCTAGAGCGTCGCCGCACGCTCGGCTACCGGTCAGGGCTTGC;
Primer 3, 5′-CTCTAGCTGCGAGCGCCGGTTAGGTGACGAGGTGGAGGGCCCTACGGGCG;
Primer 4, 5′-TTTTCGGCTTTCGCCTATCCCTAGGGGCGTACGGCCGCCCGTAGGGCCCT;
Primer 5, 5′-GGCGAAAGCCGAAAACGCGAGCGGTGTCACGAGGCACCTCAAGTCTGCGG;
Primer 6, 5′-AACAGGTCTCGAGTGGGGGTCTAACCAGGAGGGGCCCGCAGACTTGAGGT;
Primer 7, 5′-CACTCGAGACCTGTTGGGTCTTTGGGCTCACCCCCCAACTCTGAACGTCG; and
Primer 8, 5′-CGACGTTCAGAGTTGGGGGGTGAGCCCAAA.
The P1 stem of the T. radiovictrix glmS ribozyme was mutated by replacing Primer 2 with the primer 5′-ACCTCGTCACCTAACCGGGTTACCGGTCAGGGCTTGC. The T. radiovictrix glmS ribozyme was also amplified by PCR using Primers 1 and 8 using genomic DNA obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ). The glmS ribozyme from D. radiodurans was amplified from genomic DNA obtained from the American Type Culture Collection (ATCC) with the following DNAs:
Primer 1, 5′-TAATACGACTCACTATAGGGTGGGCGGGGTTCGACTTCTTCGGGC; and
Primer 2, 5′-CAGCCACTTGCTGGGGTGTGGTGAGCT.
The glmS ribozyme from T. thermophilus HB8 was amplified from genomic DNA isolated from T. thermophilus using the following DNAs:
Primer 1, 5′-TAATACGACTCACTATAGGGTGGGAGGCGGAAAGACGCGT; and
Primer 2, 5′-CCTACGTAGCCTACGATCCCGCACAT.
All synthetic DNAs were obtained from Sigma-Aldrich.
In vitro transcription and RNA labeling
Up to 400 ng of template dsDNAs were transcribed in a 30 μL in vitro transcription reaction containing 80 mM HEPES-KOH (pH 7.5 at 23°C), 40 mM DTT, 24 mM MgCl2, 2 mM spermidine, 2.5 mM each of the four ribonucleoside 5′ triphosphates (NTPs), and 10 units/μL bacteriophage T7 RNA polymerase. 10 μCi [α-32P] GTP was added to the reaction to yield internally radiolabeled RNA transcripts. Reactions were incubated for 2 h at 37°C and RNA products were purified using denaturing (8 M urea) 6% polyacrylamide gel electrophoresis (PAGE). A band containing the appropriate RNA was excised from the gel and RNA was recovered by soaking in a solution containing 10 mM HEPES-KOH (pH 7.5 at 23°C), 200 mM NaCl, and 1 mM EDTA (pH 8.0 at 23°C). HEPES buffer, and not Tris-HCl, was used due to the ability of Tris to weakly trigger glmS ribozyme self-cleavage (Roth et al. 2006; Soukup 2006). The RNAs were precipitated by adding 0.1 volume of 3 M sodium acetate (pH 5.2 at 23°C) and 2.5 volumes of cold (−20°C) ethanol, pelleted by centrifugation, and the resulting pellet was washed with 70% cold ethanol, resuspended in water, and stored at −20°C until used.
In vitro transcription ribozyme cleavage assays
Ribozyme cleavage activity during in vitro transcription used the transcription protocols as above except for the addition of 200 μM of the ligand indicated. The gels were dried and images were visualized using a Storm 820 PhosphorImager (GE Healthcare) on PhosphorImager cassettes (Molecular Dynamics).
Ribozyme cleavage assays
A maximum of 10 nM internally 32P-labeled ribozyme RNA was added to a reaction containing 50 mM HEPES-KOH (pH 7.5 at 23°C), 50 mM MgCl2, and 200 mM KCl. This mixture was allowed to incubate for 5 min at a temperature noted for each experiment to allow for the ribozyme to appropriately fold. 200 μM of ligand or water were then added and allowed to incubate at the desired temperature for 5 min. The products were then separated by denaturing 6% PAGE. The gels were then dried and imaged, as mentioned above.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Dr. Jonathan Perreault, Dr. Zasha Weinberg, and the rest of the members of the Breaker lab for helpful comments and advice. We thank Dave Bulkley and Thomas Steitz (Yale University) for the gift of T. thermophilus HB8. We thank Nick Carriero and Rob Bjornson for assisting our use of the Yale Life Sciences High Performance Center (NIH Grant RR19895-02). P.J.M. is supported by the NIH Training Grant T32GM007499. This work was supported by the NIH Grant PO1 GM022778-34 and by the Howard Hughes Medical Institute. R.R.B. is a Howard Hughes Medical Institute Investigator.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2590811.
REFERENCES
- Albuquerque L, Simões C, Nobre MF, Pino NM, Battista JR, Silva MT, Rainey FA, da Costa MS 2005. Truepera radiovictrix gen. nov., sp. nov., a new radiation resistant species and the proposal of Trueperaceae fam. nov. FEMS Microbiol Lett 247: 161–169 [DOI] [PubMed] [Google Scholar]
- Ames TD, Breaker RR 2011. Eubacterial riboswitches responsive to glutamine. RNA Biol (in press) [Google Scholar]
- Ames TD, Rodionov DA, Weinberg Z, Breaker RR 2010. A eubacterial riboswitch class that senses the coenzyme tetrahydrofolate. Chem Biol 17: 681–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banáš P, Walter NG, Šponer J, Otyepka M 2010. Protonation states of the key active site residues and structural dynamics of glmS riboswitch as revealed by molecular dynamics. J Phys Chem 114: 8701–8712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Breaker RR 2007. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol 8: R239 doi: 10.1186/gb-2007-8-11-r239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, et al. 2004. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci 101: 6421–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Sudarsan N, Weinberg Z, Ruzzo WL, Breaker RR 2005. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA 11: 774–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount KF, Breaker RR 2006. Riboswitches as antibacterial drug targets. Nat Biotechnol 24: 1558–1564 [DOI] [PubMed] [Google Scholar]
- Breaker RR 2010. Riboswitches and the RNA world. In RNA Worlds (ed. Atkins JF et al. ), pp. 63–77 Cold Spring Harbor Laboratory Press, NY [Google Scholar]
- Brooks KM, Hampel KJ 2009. A rate-limiting conformational step in the catalytic pathway of the glmS ribozyme. Biochemistry 48: 5669–5678 [DOI] [PubMed] [Google Scholar]
- Brown DR, Whitcomb RF, Bradbury JM 2007. Revised minimal standards for description of new species of the class Mollicutes (division Tenericutes). Int J Syst Evol Microbiol 57: 2703–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane JC, Lipchock SV, Strobel SA 2007. Structural investigation of the glmS ribozyme bound to its catalytic cofactor. Chem Biol 14: 97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane JC, Lipchock SV, Smith KD, Strobel SA 2009. Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme. Biochemistry 48: 3239–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JA, Irnov I, Baker S, Winkler WC 2007. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev 21: 3356–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR, Scott WG 2010. Small self-cleaving ribozymes. In RNA Worlds (ed. Atkins JF et al. ). Cold Spring Harbor Laboratory Press, NY: doi: 10.1101/cshperspect.a003574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich KS, Vogel J 2009. Activation of gene expression by small RNA. Curr Opin Microbiol 12: 674–682 [DOI] [PubMed] [Google Scholar]
- Furchak JRW, Yang P, Jennings C, Walter NG, Kennedy RT 2008. Assay for glucosamine 6-phosphate using a ligand activated ribozyme with fluorescence resonance energy transfer or CE-laser-induced fluorescence detection. Anal Chem 80: 8195–8201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. 2009. Rfam: updates to the RNA families database. Nucleic Acids Res 37: D136–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görke B, Vogel J 2008. Noncoding RNA control of the making and breaking of sugars. Genes Dev 22: 2914–2925 [DOI] [PubMed] [Google Scholar]
- Harmon-Smith M, Celia L, Chertkov O, Lapidus A, Copeland A, Glavina del Rio T, Nolan M, Lucas S, Tice H, Cheng J-F, et al. 2010. Complete genome sequence of Sebaldella termitidis type strain (NCTC 11300T). Stand Genomic Sci 2: 220–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix DK, Brenner SE, Holbrook SR 2005. RNA structural motifs: building blocks of a molecular biomolecule. Q Rev Biophys 38: 221–243 [DOI] [PubMed] [Google Scholar]
- Hu P, Leighton T, Ishkhanova G, Kustu S 1999. Sensing of nitrogen limitation by Bacillus subtilis: comparison to enteric bacteria. J Bacteriol 181: 5042–5050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins C, Kedar V, Fuerst JA 2002. Gene discovery within the planctomycete division of the domain Bacteria using sequence tags from genomic DNA libraries. Genome Biol 3: R31.1–R31.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JN, Roth A, Breaker RR 2007. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2′-deoxyguanosine. Proc Natl Acad Sci 104: 16092–16097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klawuhn K, Jansen JA, Soucheck J, Soukup GA, Soukup JK 2010. Analysis of metal ion dependence in glmS ribozyme self-cleavage and coenzyme binding. ChemBioChem 11: 2567–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Ferré-D'Amaré AR 2006. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science 313: 1752–1756 [DOI] [PubMed] [Google Scholar]
- Klein DJ, Been MD, Ferré-D'Amaré AR 2007. Essential role of an active-site guanine in glmS ribozyme catalysis. J Am Chem Soc 129: 14858–14859 [DOI] [PubMed] [Google Scholar]
- Lim J, Grove BC, Roth A, Breaker RR 2006. Characteristics of ligand recognition by a glmS self-cleaving ribozyme. Angew Chem Int Ed 45: 6689–6693 [DOI] [PubMed] [Google Scholar]
- Link KH, Guo L, Breaker RR 2006. Examination of the structural and functional versatility of glmS ribozymes by using in vitro selection. Nucleic Acids Res 34: 4968–4975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan MT, Martinko JM, Dunlap PV, Clark DP 2008. Brock Biology of Microorganisms, 12th edition Benjamin-Cummings, Reading, MA [Google Scholar]
- McCarthy TJ, Plog MA, Floy SA, Jansen JA, Soukup JK, Soukup GA 2005. Ligand requirements for glmS ribozyme self-cleavage. Chem Biol 12: 1221–1226 [DOI] [PubMed] [Google Scholar]
- Meyer M, Ames TD, Smith DP, Weinberg Z, Schwalbach MS, Giovannoni SJ, Breaker RR 2009. Identification of candidate structured RNAs in the marine organism ‘Candidatus Pelagibacter ubique.’ BMC Genomics 10: 268 doi: 10.1186/1471-2164-10-268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewski S 2002. Glucosamine-6-phosphate synthase—the multi-facets enzyme. Biochim Biophys Acta 1597: 173–192 [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR 2009. Infernal 1.0: inference of RNA alignments. Bioinformatics 25: 1335–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault J, Weinberg Z, Roth A, Popescu O, Chartrand P, Ferbeyre G, Breaker RR 2011. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput Biol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poiata E, Meyer MM, Ames TD, Breaker RR 2009. A variant riboswitch aptamer class for S-adenosylmethionine common in marine bacteria. RNA 15: 2046–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Breaker RR 2009. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem 78: 305–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Nahvi A, Lee M, Jona I, Breaker RR 2006. Characteristics of the glmS ribozyme suggest only structural roles for divalent metal ions. RNA 12: 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A 2009. The long and the short of riboswitches. Curr Opin Struct Biol 19: 251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin DY, Tourova TP, Mußmann M, Muyzer G 2008. Dethiobacter alkaliphilus gen. nov. sp. nov., and Desulfurivibrio alkaliphilus gen. nov. sp. nov.: two novel representatives of reductive sulfur cycle from soda lakes. Extremophiles 12: 431–439 [DOI] [PubMed] [Google Scholar]
- Soukup G 2006. Core requirements for glmS ribozyme self-cleavage reveal a putative pseudoknot structure. Nucleic Acids Res 34: 968–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinsley RA, Furchak JRW, Walter NG 2007. Trans-acting glmS catalytic riboswitch: Locked and loaded. RNA 13: 468–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JX, Breaker RR 2008. Riboswitches that sense S-adenosylmethionine and S-adenosylhomocysteine. Biochem Cell Biol 86: 157–168 [DOI] [PubMed] [Google Scholar]
- Weinberg Z, Breaker RR 2011. R2R - software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics 12: 3 doi: 10.1186/1471-2105-12-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Regulski EE, Hammond MC, Barrick JE, Yao Z, Ruzzo WL, Breaker RR 2008. The aptamer core of SAM-IV riboswitches mimics the ligand-binding site of SAM-I riboswitches. RNA 14: 822–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson SR, Been MD 2005. A pseudoknot in the 3′ non-core region of the glmS ribozyme enhances self-cleavage activity. RNA 11: 1788–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR 2004. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428: 281–286 [DOI] [PubMed] [Google Scholar]
- Xin Y, Hamelberg D 2010. Deciphering the role of glucosamine-6-phosphate in the riboswitch action of glmS ribozyme. RNA 16: 2455–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]