Abstract
Argonaute (Ago) proteins form the core of RNA-induced silencing complexes (RISCs) and mediate small RNA-guided gene silencing. In RNAi, short interfering RNAs (siRNAs) guide RISCs to complementary target RNAs, leading to cleavage by the endonuclease Ago2. Noncatalytic Ago proteins, however, contribute to RNAi as well but cannot cleave target RNA and often generate off-target effects. Here we show that synthetic siRNA duplexes interact with all Ago proteins, but a functional RISC rapidly assembles only around Ago2. By stabilizing the siRNA duplex, we show that the noncatalytic Ago proteins Ago1, -3, and -4 can be selectively blocked and do not form functional RISCs. In addition, stabilized siRNAs form an Ago2-RISC more efficiently, leading to increased silencing activity. Our data suggest novel parameters for the design of siRNAs with selective activation of the endonuclease Ago2.
Keywords: argonaute proteins, RNAi, gene silencing, siRNAs
INTRODUCTION
In RNAi, short interfering RNAs (siRNAs) guide RNA-induced silencing complexes (RISCs) to complementary RNA molecules, leading to endonucleolytic cleavage of the target RNA (Dorsett and Tuschl 2004; Meister and Tuschl 2004). Depending on the organism, siRNAs can derive from long double-stranded RNA (dsRNA), intramolecular folding structures such as hairpins, or exogenously introduced synthetic siRNA duplexes (Kim et al. 2009). After transfection of synthetic siRNAs into mammalian cells, the two strands are separated and one strand gives rise to the guide strand. The other strand, referred to as the passenger strand, is destabilized and removed from the cell (Meister and Tuschl 2004). The decision of which strand is selected as guide strand and which strand is degraded is based on the thermodynamic stability of the ends of the double-stranded siRNA. According to the asymmetry rule, the strand with the less stably paired 5′ end is finally incorporated into the RISC and becomes the guide strand (Khvorova et al. 2003; Schwarz et al. 2003; Tomari et al. 2004).
Argonaute (Ago) proteins form the core of RISC and directly interact with the siRNA guide strand. In humans, eight different Argonaute genes exist, which can be divided into the Ago (Ago1–4) and the Piwi subfamilies (HIWI1-3, HILI). Piwi proteins are utilized for silencing of mobile genetic elements in the germline and interact with a specific class of small RNAs termed Piwi interacting RNAs (piRNAs). Ago proteins, in contrast, are thought to be ubiquitously expressed and interact with endogenous microRNAs (miRNAs) and siRNAs (Ender and Meister 2010). Biochemical as well as structural studies revealed that some Ago proteins are endonucleases and cleave the target RNA that is complementary to the guide strand (Jinek and Doudna 2009). In humans, for example, only Ago2 is catalytically active, although critical residues are conserved in other Ago proteins as well (Liu et al. 2004; Meister et al. 2004).
The discovery that Ago2 is an endonuclease in mammals raised the questions of whether passenger strands within the siRNA duplexes might be cleaved by Ago2 as well and whether such a cleavage process might influence RISC loading. Studies in different organisms revealed that, indeed, cleavage competent Ago proteins cleave the siRNA passenger strand (Matranga et al. 2005; Miyoshi et al. 2005; Rand et al. 2005; Leuschner et al. 2006; Kim et al. 2007). How noncatalytic Ago proteins (e.g., Ago1, Ago3, and Ago4 in human) are loaded, however, remained unclear. Experiments in Drosophila suggest that bypass mechanisms exist and that noncleaving Ago proteins require reduced stability for efficient loading (Forstemann et al. 2007; Tomari et al. 2007; Kawamata et al. 2009).
Human Ago1–4 mainly interact with endogenous miRNAs, which guide Ago proteins to target mRNAs, leading to repression of gene expression. In mammals, perfectly complementary targets are rather rare, and sequence-specific cleavage, as observed for siRNAs, may have only a limited impact on miRNA function. Instead, mammalian miRNAs guide Ago proteins to only partially complementary sequences on mRNAs, leading to deadenylation-dependent mRNA decay and more rarely to translational silencing without changes in mRNA levels (Eulalio et al. 2008; Filipowicz et al. 2008; Guo et al. 2010). It has been demonstrated that synthetic siRNAs that are transfected into mammalian cells not only bind to Ago2 and lead to mRNA cleavage but also bind to all other Ago proteins (Liu et al. 2004). A consequence of such interactions is that siRNAs function to some extent as miRNAs and guide Ago proteins to partially complementary RNAs, leading to hardly controllable “off-target” effects in RNAi experiments (Jackson et al. 2003, 2006b; Lin et al. 2005; Birmingham et al. 2006; Aleman et al. 2007).
siRNAs are currently used to develop novel RNAi-based drugs (Castanotto and Rossi 2009). It is highly desirable to produce potent siRNAs with minimal off-target effects. We therefore set out to analyze siRNA loading into different human Ago proteins using a biochemical approach. We find that all four Ago proteins initially interact with the siRNA duplex. The catalytically active Ago2 removes the passenger strand immediately, likely by cleaving it. A significant fraction of noncatalytic Ago proteins, however, remains with the duplex over days, and single-stranded guide strands accumulate only slowly. Strikingly, duplex stabilization by locked nucleic acid (LNA) modification trapped noncatalytic Ago proteins with double-stranded siRNAs, while Ago2 even more efficiently removes the modified passenger strand. Moreover, the guide strand of stabilized siRNAs is enriched in Ago2. Our results show that thermodynamic duplex stability is a major determinant of passenger strand displacement in all Ago proteins, leading to decreased off-target effects but increased Ago2-mediated on-target effects.
RESULTS
Cellular Ago protein concentrations
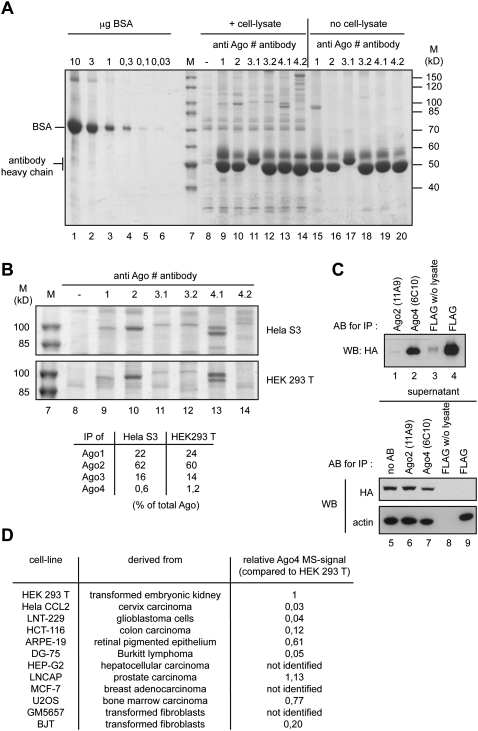
It has been demonstrated before that the noncatalytic Ago proteins contributed to on- and off-target effects in RNAi experiments (Aleman et al. 2007; Wu et al. 2008; Vickers et al. 2009). To better understand the effect of noncatalytic Ago proteins on RNAi, we analyzed siRNA loading into the individual Ago proteins. In order to examine siRNA interactions with Ago proteins, we first measured cellular Ago protein levels by immunoprecipitation (IP) using monoclonal antibodies that discriminated between the human Ago proteins (Fig. 1; Beitzinger et al. 2007; Rudel et al. 2008; Weinmann et al. 2009). Endogenous Ago1–4 were immunoprecipitated from HeLa S3 and HEK 293T cells (Fig. 1A,B), and the isolated proteins were analyzed by mass spectrometry. In Ago1–3 IPs, the bands with a molecular weight of ∼100 kDa were clearly identified as Ago1–3. For Ago4, however, the antibody 6C10 precipitated very little Ago4 and mainly cross-reacts with an unrelated protein. Another anti-Ago4 antibody (3G5), which immunoprecipitates Flag/HA-tagged (FH) Ago4, does not show such cross-reactivities, but endogenous Ago4 was not detectable in the immunoprecipitates, suggesting that Ago4 is not abundant in HEK 293T or HeLa S3 cells. To rule out that the low Ago4 levels in IP experiments are due to low antibody affinity, we immunoprecipitated FH-Ago4 from a stably transfected HeLa S3 cell line (Fig. 1C). Indeed, the antibody 6C10 immunoprecipitated FH-Ago4 efficiently, supporting the idea that Ago4 protein expression is rather low in HEK 293T and HeLa S3 cells.
FIGURE 1.
Estimation of Ago1–4 protein amounts in different human cell lines. (A) Cell lysate was prepared from HeLa S3 cells, and Ago1–4 proteins were immunoprecipitated using monoclonal antibodies against Ago1 (lane 9), Ago2 (lane 10), Ago3 (two different antibodies 3.1 and 3.2; lanes 11,12), and Ago4 (two different antibodies 4.1 and 4.2; lanes 13,14). The samples were analyzed on a SDS-polyacrylamid-gel together with a BSA standard (lanes 1–6) and stained with colloidal Coomassie. In lanes 15–20, lysate was omitted from the IP. M indicates molecular weight marker. (B) Cell lysate was prepared from HEK 293T cells, and Ago1–4 proteins were immunoprecipitated as described in A. The protein bands at a size of ∼100 kD were analyzed by mass spectrometry (MS). The signal intensities of all peptides for each Ago protein were used to compare the relative amounts of Ago1–4 with each other. For Ago3, the intensities from both antibody clones 3.1 and 3.2 were averaged. For Ago4, only signals from antibody clone 4.1 were used (table). (C) Lysate from a cell line stably expressing FLAG-HA (FH)–tagged Ago4 was prepared as described in A. The IP of Ago4 by the anti-Ago4 antibody (lane 2) was compared to the IP of tagged Ago4 with anti-FLAG antibody (lane 4). Anti-Ago2 (11A9) IP from FH-Ago4–expressing cells served as control. The experiment was analyzed by Western blotting using an antibody against the HA tag. In addition, the supernatants of the IP samples were analyzed using the anti-HA-antibody and an anti-beta-actin antibody (lower panel). Anti-Ago2 (11A9) IP from FH-Ago4–expressing cell lysates served as control (lane 1). (D) Ago4 was immunoprecipitated from lysates of different cell lines with antibody 6C10 (4.1 in A). The IPs were analyzed by MS. Sums of the intensities of all peptides identified for Ago4 were compared among the different cell lines as described in B. The peptide intensity sum from HEK 293T was defined as 1 (corresponding to ∼1% of all Ago in HEK 293T).
By using the described IP approach, we set out to measure Ago protein levels in HEK 293T and HeLa S3 cells. Defined amounts of BSA served as a standard, and we estimated the intracellular concentration of Ago2 as ∼100 nM (Fig. 1A). A comparison of the overall peptide signal intensities of each Ago protein in our mass spectrometry experiments revealed that Ago2 represents ∼60%, Ago1 20%–25%, Ago3 ∼15%, and Ago4 ∼1% of the total Ago protein pool in HEK 293T or HeLa S3 cells (Fig. 1B, table). In search for Ago4-expressing cells, we analyzed 13 different cell lines, and from none of them could we immunoprecipitate higher amounts of Ago4 (Fig. 1D), indicating that the contribution of Ago4 to RNAi on- and off-target effects in the analyzed cell lines might be negligible. In summary, we have measured cellular Ago protein levels for the first time and found that Ago2 is the most abundant Ago protein followed by Ago1 and Ago3 in the commonly used cell lines HEK 293T and HeLa S3. In many cell lines, only a little Ago4 protein is found. Ago4 mRNA levels have been quantified before, and mRNA levels significantly differ from the Ago4 protein levels measured here (Meister et al. 2004). Therefore, it is very likely that Ago4 expression is to some extent regulated post-transcriptionally.
siRNA strand incorporation into different Ago proteins
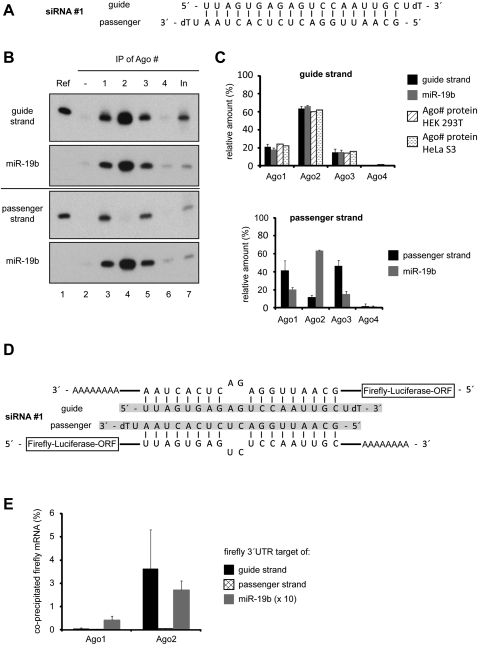
We used our RISC IP approach to analyze siRNA strand loading into the four different human Ago proteins in detail. A thermodynamically asymmetric siRNA (one strand is preferentially incorporated into RISC, siRNA#1) (Fig. 2A) was transfected into HEK 293T cells, and the individual Ago proteins were immunoprecipitated as described above. Small RNAs were extracted and analyzed by Northern blotting using probes against the guide or the passenger strand (Fig. 2B). Since different probes have different binding affinities, we loaded the corresponding synthetic siRNA strand onto each Northern blot for signal normalization. As expected for an asymmetric siRNA, the guide strand is incorporated into the different Ago proteins according to their protein expression levels (Fig. 2B,C, upper panel). The passenger strand, however, is present in Ago1-containing and Ago3-containing, but not in Ago2-containing, RISCs (Fig. 2B,C, lower panel). Similar results were obtained for several other siRNA duplexes (data not shown).
FIGURE 2.
Analysis of siRNA strand loading into different Ago proteins. (A) The thermodynamically asymmetric siRNA#1 used for the following experiments. (B) Cells were lysed 20 h after transfection, and Ago1 (lane 3), Ago2 (lane 4), Ago3 (lane 5), and Ago4 (lane 6) were immunoprecipitated from the lysates. The coimmunoprecipitated siRNA#1 guide strand (upper panel) and the passenger strand (lower panel) were analyzed by Northern blotting. Blots were stripped and reprobed for miR-19b. Ref indicates 3 pmol of the respective siRNA single strand; In, 2% of the input sample. (C) The radioactive signals from Northern blots of three independent experiments as shown in B were quantified using a phosphoimager system, and the relative signal intensities of the siRNA#1 guide strand (upper graph, black columns), the passenger strand (lower graph, black columns), and miR-19b (gray columns) were compared between the four Ago proteins. Standard deviations of the signals are indicated. In addition, the relative protein amounts of the individual Ago proteins in HEK 293T and Hela S3 cells as calculated in Figure 1B are included in the upper graph (diagonally and horizontally hatched columns). (D) Targeting sequences for the guide (upper part) or the passenger strand (lower part) of siRNA#1 were cloned into the 3′ UTR of the firefly luciferase mRNA of a pMir-REPORT vector. The siRNA targeting sequences contain mismatches in the annealing positions 10 and 11 of the respective siRNA strands in order to avoid cleavage by Ago2. As a reference, a construct containing a targeting sequence for miR-19b also with mismatches in annealing positions 10 and 11 of the miRNA was used (data not shown). (E) HEK 293T cells were transfected with siRNA#1 and either a pMir-REPORT vector containing a targeting site for the guide strand (black columns), the passenger strand (cross-hatched columns), or miR-19b (gray columns). Twenty hours post-transfection, cells were lysed and Ago1 or Ago2 immunoprecipitated from the lysates. The coimmunoprecipitated RNA was isolated and transcribed into cDNA, and the relative amount of firefly luciferase mRNA in each sample was analyzed by qPCR. The graph analyzes the relative amounts (percentage) compared with the input of the respective mRNAs coimmunoprecipitated with the Ago-siRNA or Ago-miRNA complexes.
The presence of the passenger strand on Ago1 and Ago3 could be due to unwound double-stranded siRNA or could also be in agreement with the idea that Ago1 and Ago3 do not follow the asymmetry rule and load both strands equally well. Therefore, we analyzed if Ago1/3 carries single- or mainly double-stranded siRNAs in our experiments. We cloned target sites for the passenger strand and the guide strand into the 3′ untranslated region (UTR) of a firefly luciferase reporter. To prevent cleavage by Ago2, the target site is not complementary to the individual siRNA strand at positions 10 and 11 of the siRNA (Fig. 2D). Reporter constructs were cotransfected with siRNA#1 into HEK 293T cells. Ago1 and Ago2 were immunoprecipitated using monoclonal antibodies, and coprecipitated luciferase reporter mRNA was analyzed by quantitative PCR (Fig. 2E). As expected, the anti-Ago2 antibody predominantly coimmunoprecipitated the mRNA targeted by the siRNA guide strand. Antibodies directed against Ago1, however, did not immunoprecipitate significant amounts of the target mRNA, suggesting that Ago1 carries mainly siRNA duplexes, thereby preventing interactions with the corresponding target RNA (Fig. 2E). An artificial target of the endogenous miR-19b, however, was coimmunoprecipitated by the anti-Ago1 and the anti-Ago2 antibody with similar efficiencies. The weaker signal for anti-Ago1 IPs is due to lower Ago1 protein levels (Fig. 2E).
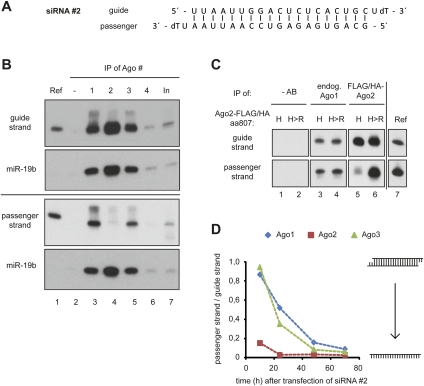
Noncatalytic Ago proteins remove the passenger strand inefficiently
Since it has been suggested that both Ago1 and Ago2 possess cleavage activity toward the passenger strand (Wang et al. 2009), we asked next whether passenger strand cleavage is required for the observed rapid removal of the passenger strand by Ago2. Another asymmetric siRNA (Fig. 3A, siRNA#2) was transfected into HEK 293T cells, and siRNA loading was analyzed as described above. Similar to siRNA#1 (Fig. 2B and 2C), the guide strand of siRNA#2 is loaded into the four Ago proteins according to their protein expression levels (Fig. 3B). The passenger strand is mainly found in Ago1 and Ago3 complexes (Fig. 3B, lanes 3,5). For the analysis of Ago2 cleavage activity, we transfected FH-Ago2 or a catalytically inactive FH-Ago2 mutant (Ago2-H807R) together with siRNA#2 into HEK 293T cells. Both wild-type (wt) FH-Ago2 (H) and FH-Ago2 H807R (H>R) efficiently immunoprecipitated the guide strand (Fig. 3C, lanes 5,6, upper panels). The passenger strand, however, remained only with the catalytically inactive Ago2 H807R mutant, indicating that Ago2's cleavage activity is required for efficient passenger strand displacement. The inactive Ago2 mutant behaves similarly to endogenous Ago1 (Fig. 3C, lanes 6,3,4), and therefore, our data suggest that Ago1 does not possess distinct passenger strand cleavage activity in our experiments.
FIGURE 3.
Noncatalytic Ago proteins remove passenger strands inefficiently. (A) The thermodynamically asymmetric siRNA#2 used for the following experiments. (B) Twenty hours after transfection, cells were lysed, and Ago1–4 (lanes 3–6) isolated, and the coimmunoprecipitated RNA was analyzed by Northern blotting using probes against the guide strand (upper panel) or the passenger strand (lower panel). Ref indicates 3 pmol of the respective siRNA single strand; In, 2% of input sample. (C) Cells were transfected with plasmids encoding wt FH-Ago2 (“H”; lanes 1,3,5) or the endonuclease inactive FH-Ago2 H807R mutant (“H>R”; lanes 2,4,6). Twenty hours after the first transfection, cells were transfected with siRNA#2. Twenty hours after siRNA transfection, cells were lysed; endogenous Ago1 was immunoprecipitated using the anti-Ago1 antibody (lanes 3,4) and FH-Ago2 variants using an anti-FLAG antibody (lanes 5,6). In lanes 1 and 2, antibodies were omitted from the reaction as control. Coimmunoprecipitated RNAs were isolated and analyzed by Northern blotting. Ref indicates 3 pmol of the respective siRNA single strand. (D) The same experimental setup as in B was applied to analyze the kinetics of passenger strand displacement in Ago1–3. Cells were lysed 10 h, 24 h, 48 h, and 70 h after siRNA transfection and further processed as described in B. Probe signals from Northern blots (data not shown) were quantified; passenger strand and guide strand signals were compared by normalization to the reference signals. The ratio of passenger strand to guide strand coimmunoprecipitated with Ago1, Ago2, and Ago3 at the indicated time points is shown in the graph.
Since noncatalytic Ago proteins stably interact with siRNA duplexes 20 h after siRNA transfection, we analyzed the kinetics of Ago1, Ago2, and Ago3 passenger strand removal. The asymmetric siRNA#2 was transfected into HEK 293T cells, and siRNA passenger strand displacement by the different Ago proteins was analyzed over time (Fig. 3D). While Ago2 immediately removes the passenger strand, likely by endonucleolytic cleavage (red line), the passenger strand is only slowly displaced from the noncatalytic Ago proteins Ago1 and Ago3 over a time period of ∼2–3 d (Fig. 3D, blue and green lines). Taken together, our data demonstrate that all human Ago proteins initially interact with synthetic siRNA duplexes, but only Ago2 is capable of rapid removal of the passenger strand.
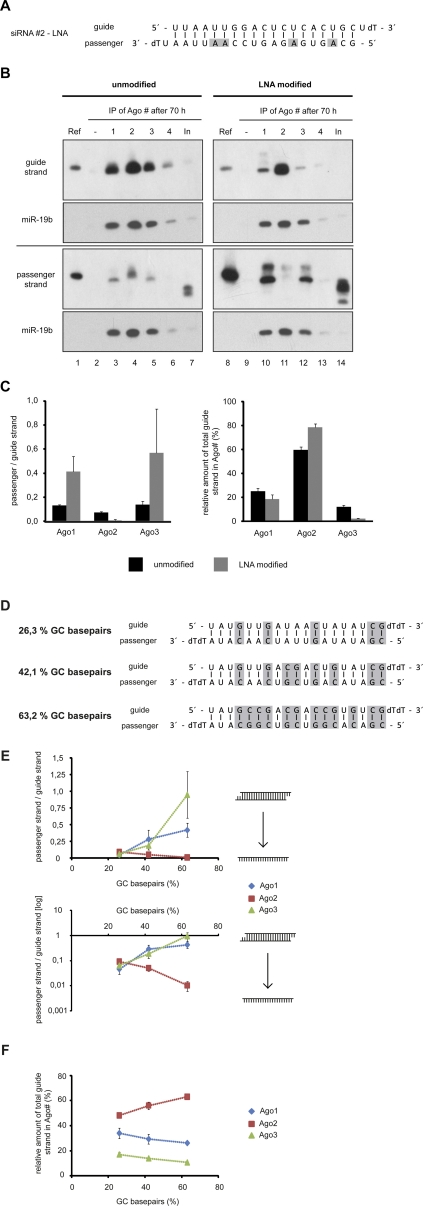
siRNA duplex stabilization traps noncatalytic Ago proteins on double-stranded siRNAs
Our striking results on siRNA strand displacement prompted us to ask whether or not RISC activation of noncatalytic Ago proteins can be inhibited by stabilized siRNA duplexes. Such siRNAs would have minimal off-target effects caused by the noncatalytic Ago proteins. We synthesized a variant of siRNA#2 with four LNA nucleotides in the passenger strand, which form very stable interactions with complementary RNA nucleotides (Fig. 4A). The LNA-modified siRNA as well as the unmodified version were transfected into HEK 293T cells, and strand incorporation as well as displacement was analyzed as described above (Fig. 4B). Ago2 is capable of displacing the passenger strand even in the presence of LNA-modified nucleotides very efficiently. Furthermore, Ago1 and Ago3 are trapped on the modified double-stranded siRNA, and even after longer time periods, the passenger strand is not efficiently removed (Fig. 4B, cf. passenger strand panels). Signal quantification revealed that about three to four times as much of the modified passenger strand remains on Ago1 or Ago3 compared with the unmodified siRNA strand (Fig. 4C). Of note, even less passenger strand is observed on Ago2 when the LNA-modified passenger strand was used (Fig. 4B,C). In addition, we observed a relative enrichment of the guide strand in Ago2 and a decrease of guide strand in noncatalytic Ago proteins when the cells were transfected with the LNA-modified siRNA#2 compared with the unmodified siRNA#2 (Fig. 4B, cf. guide strand panels and C, right graph). Similar results were obtained with alternatively LNA-modified passenger strand variants of siRNA#2 (data not shown). These results suggest that the LNA-stabilized siRNA duplexes not only block siRNA activity mediated by Ago1 and Ago3 but also promote guide strand loading onto Ago2 as well as passenger strand displacement from Ago2 (Fig. 4C).
FIGURE 4.
Thermodynamically stabilized siRNA duplex lead to reduced passenger strand displacement efficiency of noncatalytic Ago proteins. (A) LNA-modified siRNA#2 used for the following experiments. LNA-modified nucleotides are highlighted in gray. (B) Seventy hours after transfection with siRNA#2 or LNA-modified siRNA#2, cells were lysed and Ago1–4 (lanes 3–6,10–13) were immunoprecipitated from the lysates. RNA was isolated and analyzed by Northern blotting. Blots were stripped and reprobed for miR-19b. Ref indicates 3 pmol of the respective siRNA single strand; In, 2% of the input sample. (C) For the left graph, Northern blot signals from four independent experiments as the example shown in B were quantified, and the ratio of passenger strand to guide strand normalized by the reference signals (Ref in B) was plotted for Ago1–3. The right graph shows the relative distribution (percentage) of guide strand to Ago1–3 from the same set of experiments. Black bars indicate siRNA#2; gray bars, the LNA-modified siRNA#2. Error bars, SDs of the signals. (D) Three siRNAs with a similar sequence backbone but increasing GC content used for the following experiments. GC base pairs are indicated in gray. (E) siRNAs as shown in D were transfected into HEK 293T cells; cells were lysed 24 h later and Ago1–3 immunoprecipitated, and the amount of coimmunoprecipitated siRNA passenger and guide strands was quantified via Northern blots (data not shown). The ratio of passenger to guide strand of the siRNAs from three independent experiments is plotted as a function of the GC content on a linear scale (upper graph) and on a logarithmic scale (basis 10; lower graph). (F) The graph shows the relative distribution (percentage) of siRNA guide strand as a function of the siRNA GC content for the experiments described in E. Error bars in E and F, SDs.
In order to analyze whether the observed effects are due to the LNA modification or the thermodynamic stability of the siRNA duplex, we synthesized three different siRNA duplexes with partially similar sequences but increasing GC content and therefore increasing thermodynamic stability (Fig. 4D). The siRNA duplexes were transfected into HEK 293T cells, and the strand incorporation was analyzed by Northern blotting as described above. Quantification of the normalized Northern blot signals revealed that increasing thermodynamic stability leads to the reduced passenger strand displacement of Ago1 and Ago3 (Fig. 4E, upper panel shows a linear scale and the lower panel a logarithmic scale). Consistent with the LNA-modified siRNA duplex, we observe even less passenger strand being present in Ago2 when the siRNA duplex is stabilized by increasing GC contents (Fig. 4E). In addition, the relative amount of guide strand found in Ago2 is also increased when siRNAs with high GC contents were used (Fig. 4F). Taken together, our data indicate that the increased thermodynamic stability of siRNA duplexes leads to reduced passenger strand displacement by Ago1 and Ago3 and, on the other hand, to increased passenger strand displacement and guide strand loading onto Ago2.
Thermodynamically stabilized siRNA duplexes show reduced off-target effects
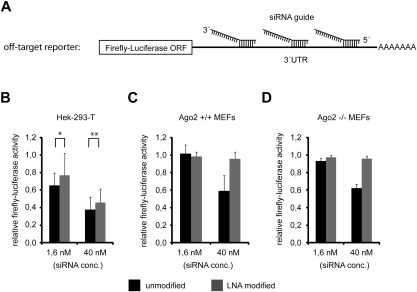
In order to analyze whether the reduced strand displacement of Ago1 and Ago3 leads to reduced off-target effects, we performed luciferase reporter experiments in different cell lines (Fig. 5). Three partially complementary binding sites for the guide strand of the siRNA#2 duplex (Figs. 3A, 4A) were cloned behind a firefly luciferase reporter (Fig. 5A). The reporter plasmid was transfected into HEK 293T cells together with two different concentrations (1.6 or 40 nM) of either the unmodified or the LNA-modified version of siRNA#2 (Fig. 5B). At both concentrations, the LNA-modified siRNA#2 repressed the off-target reporter less efficiently. The effects, however, were rather mild but nevertheless significant.
FIGURE 5.
(A) Scheme of the luciferase based off-target reporter to measure siRNA#2 guide strand off-target activity. The 3′ UTR of the firefly-luciferase contains three targeting-sites complementary to the seed region of siRNA#2 guide strand. Off-target reporter activity in the presence of indicated concentrations of unmodified siRNA#2 (black bars) and LNA-modified siRNA#2 (as shown in Fig. 4A, gray bars) was measured 48 h post-transfection in HEK 293T cells, Ago2+/+ MEFs, and Ago2−/− MEFs (B–D), respectively. For B, P-values were calculated (t-test): *0.109; **0.007.
In order to distinguish between Ago2- and Ago1/3/4-mediated off-target effects, we changed our experimental system and switched to Ago2-deficient mouse embryonic fibroblasts (MEFs). Off-target effects observed in these cells are caused by Ago1, Ago3, and/or Ago4 only. The off-target reporter was transfected together with two concentrations of either the unmodified or the LNA-modified siRNA#2 into wt or Ago2−/− MEFs (Fig. 5C,D). Indeed, Ago1/3/4-mediated off-target effects were not measureable for the LNA-modified version of siRNA#2 compared with the unmodified siRNA#2 at the higher siRNA concentration (40 nM) in Ago2−/− MEFs, although there was a pronounced off-target effect of the unmodified siRNA#2 (Fig. 5D). Similar results were obtained at later time points (Supplemental Fig. 1A). Not much change was observed when low concentrations (1.6 nM) of both variants of siRNA#2 were transfected, most likely due to reduced overall activity. Off-target effects are similar between the wt and Ago2−/− MEFs, suggesting that mainly Ago1/3 and 4 might cause the observed off-target effects in the wt MEFs as well as in the Ago2−/− MEFs, while Ago2 might cause a major portion of the off-target effects observed in HEK 293T cells. In summary, our data suggest that thermodynamically stabilized siRNAs duplexes can almost completely eliminate Ago1/3/4-mediated off-target effects.
Increased on-target effects of thermodynamically stable siRNA duplexes
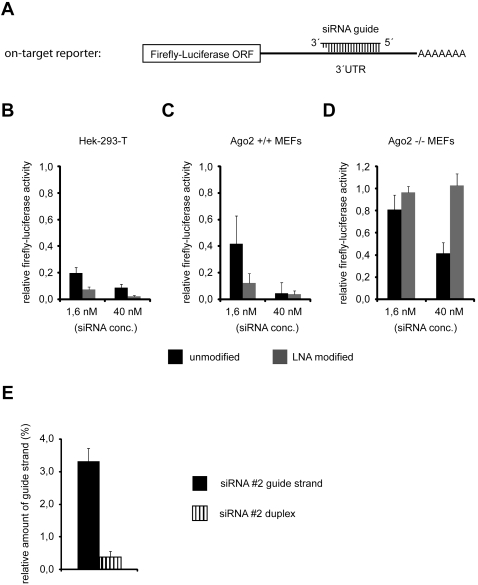
Our experiments showed that the guide strands of LNA-modified siRNAs or siRNAs with a high GC content are more efficiently loaded into Ago2 (see Fig. 4B,C,F). In addition, a lower proportion of passenger strand was found in Ago2 (Fig. 4B,C,E), indicating an enhanced activation of Ago2 containing RISCs. We hypothesized that the use of such stabilized siRNAs might therefore result in better knock-down efficiencies. To test our hypothesis, we generated an “on-target” luciferase reporter (Fig. 6A) containing a perfectly complementary target site for the siRNA#2 guide strand (Figs. 3A, 4A). The reporter construct was cotransfected with two concentrations (1.6 and 40 nM) of unmodified or LNA-stabilized siRNA#2 into HEK 293T cells (Fig. 6B). Strikingly, the LNA-modified siRNA produced a stronger knock down compared with the unmodified siRNA at both siRNA concentrations. In order to test if the increased knock-down efficiency of the LNA-modified siRNA was due to an increased activation of Ago2, we used the Ago2-deficient MEFs described above. While the LNA-modified siRNA generated a stronger knock down in wt MEFs compared with the corresponding unmodified siRNA (Fig. 6C), almost no knock down by the LNA-modified siRNA was observed in the Ago2−/− MEFs; however, there was still pronounced knock-down efficiency of the unmodified siRNA (Fig. 6D). Similar results were obtained at a later time point (72 h post-transfection) (Supplemental Fig. 1B). In summary, the increased on-target activities of the LNA-modified siRNA compared with the unmodified siRNA measured in HEK 293T cells and wt MEFs and, in contrast, the almost complete absence of on-target activity of the LNA-modified siRNA in Ago2−/− MEFs strongly support the hypothesis that thermodynamically stabilized siRNAs indeed selectively activate the endonucleolytic Ago2 and can thereby even increase the overall knock-down efficiency of an siRNA.
FIGURE 6.
The LNA-modified version of siRNA#2 selectively enables Ago2-mediated siRNA activity, resulting in increased on-target activity. (A) Scheme of the luciferase-based reporter to measure siRNA#2 guide strand on-target activity. The 3′ UTR of the firefly-luciferase contains one targeting site with perfect complementarity to siRNA#2 guide strand. Luciferase reporter activity in the presence of indicated concentrations of unmodified siRNA#2 (black bars) or LNA-modified siRNA#2 (as shown in Fig. 4A, gray bars) was measured in HEK 293T cells, Ago2+/+ MEFs, and Ago2−/− MEFs (B–D), respectively. Luciferase reporter activity was measured 48 h after cotransfection of cells with reporter encoding plasmid and siRNA. Average values from at least three independent experiments are shown. Error bars, SDs. (E) siRNA single strand shows higher affinity for Ago1 than corresponding siRNA duplex. P32-labeled 5′-phosphorylated siRNA#2 guide strand or siRNA#2 duplex was incubated for 1 h with purified, recombinant His-Ago1. His-Ago1 was immunoprecipitated with monoclonal anti-Ago1 antibody afterward, and the amount of coimmunoprecipitated radioactive siRNA#2 guide strand was analyzed by scintillation counting. The relative amount (compared to the input) of coimmunoprecipitated siRNA#2 guide strand as a single strand (black bar) or as part of an siRNA#2 duplex (vertically hatched bar) is depicted in the graph. Average values from three independent experiments are shown; error bars, SDs.
What might be the molecular reasons for the increased knock-down efficiencies of thermodynamically stabilized siRNAs? One plausible explanation of the effects could be that Ago2 associates with the double-stranded siRNA, immediately cleaves and removes the passenger strand, and loads the single-stranded guide strand. Thus, Ago2 would rapidly generate stable RISCs. We hypothesize that human Ago proteins possess lower affinity to double-stranded siRNAs than to single-stranded siRNAs, as it has been described for the recombinant Aquifex aeolicus Ago protein (Yuan et al. 2005). Since Ago1/3 and -4 do not cleave the passenger strand, they remain longer with double-stranded siRNAs. Because of the low affinity to double-stranded siRNAs, off rates of the siRNA duplex from the noncatalytic Ago proteins might be high and directly correlate with the thermodynamic stability of the siRNA duplex. Consequently, more siRNA duplexes are available for Ago2, leading to increased RNAi efficiency. Since our hypothesis is based on different Ago affinities for single-stranded or double-stranded siRNAs, we analyzed Ago-siRNA binding (Fig. 6E). Recombinant, purified His-Ago1 was incubated with a radiolabeled guide strand only or double-stranded siRNA#2. After incubation, beads were washed, and the bound RNA was quantified. Indeed, Ago1 binds much stronger to the single-stranded siRNA#2 guide strand compared with its double-stranded version, suggesting that our model explaining increased Ago2 guide strand loading and, as a consequence, increased knock down efficiency of thermodynamically stabilized siRNAs might be correct.
DISCUSSION
RNAi is widely used as a research tool for the sequence-specific knock down of gene expression on the post-transcriptional level. RNAi is also becoming more and more important for medical research and drug development. However, off-target effects are still a major problem for RNAi experiments (Svoboda 2007). Generally, sequence-specific off-target activity is thought to originate from the seed regions (nucleotides 2–8) of the passenger and the guide strand and affects mRNAs containing sequence motifs complementary to passenger and guide strand seed regions (Lin et al. 2005; Birmingham et al. 2006; Jackson et al. 2006b). In order to avoid siRNA passenger strand off-target activity and favor guide strand loading, several studies have analyzed siRNA strand selection and established siRNA design concepts that very efficiently help to avoid loading of the passenger strand into functional RISCs (Khvorova et al. 2003; Schwarz et al. 2003; Bramsen et al. 2007, 2009; Chen et al. 2008).
During investigation of how the individual Ago proteins contribute to siRNA on-target activity, it has been suggested that mainly the endonuclease Ago2 contributes to siRNA on-target activity (Vickers et al. 2007; Diederichs et al. 2008). How the individual Ago proteins contribute to siRNA off-target activity is still under discussion, but several studies have implicated that the contribution of the noncatalytic Ago proteins Ago1/3/4 to siRNA off-target activity is disproportionately high (Aleman et al. 2007; Wu et al. 2008; Vickers et al. 2009). However, with respect to the observation that Ago2 mainly triggers siRNA on-target activity, while noncatalytic Ago1/3/4 might strongly contribute to siRNA off-target activity, a comprehensive study aiming at the identification of siRNA features that are important for Ago protein selection has not been performed. Therefore, we used our monoclonal antibodies against the individual Ago proteins and analyzed Ago-siRNA loading and RISC activation in living cells in molecular detail.
Our data based on coimmunoprecipitation studies of individual Ago proteins from cells transfected with siRNAs that are characterized by different thermodynamic duplex stabilities suggest that increasing the thermodynamic stability of the siRNA duplexes inhibits the activation of noncatalytic Ago proteins. Furthermore, duplex stabilization has no negative effect on Ago2 loading, indicating that such modified siRNAs can indeed be used for RNAi. Minimizing the off-target effects caused by all four Ago proteins is one of the major challenges in the application of RNAi. We find that the LNA-stabilized variant of a siRNA has reduced activity toward an off-target luciferase reporter in HEK 293T cells. This reduction of off-target activity is even more pronounced in wt MEFs. By the use of Ago2-deficient MEFs, we show that siRNA-based Ago1/3/4 off-target activity is almost completely eliminated by our duplex stabilizing siRNA design. Nevertheless, in contrast to previous studies (Aleman et al. 2007; Wu et al. 2008; Vickers et al. 2009), our observations do not necessarily lead to the conclusion that siRNA off-target activity mediated by Ago2 is lower than off-target activity mediated by the noncatalytic Ago1/3/4 proteins. We still observe off-target activity of the LNA-stabilized siRNA in HEK 293T cells (Fig. 5B), where Ago2 contributes ∼60% of the total Ago (Fig. 1A,B). As the LNA-stabilized siRNA has neither measureable on- nor off-target activity in Ago2−/− MEFs (Figs. 6D, 5D, respectively), we conclude that its off-target activity measured in HEK 293T cells is largely caused by Ago2.
In addition to reduced Ago1/3/4-mediated off-target effects, thermodynamically stable siRNAs show increased Ago2-mediated on-target effects. The elevated RNAi efficiency seems to be due to an increased loading of the guide strands into Ago2 compared with Ago1 and Ago3. Since we find that the Ago1 protein possesses higher binding affinity to single-stranded siRNA compared with double-stranded siRNA, stable siRNA duplexes might dissociate more frequently after binding from noncatalytic Ago proteins due to the low duplex unwinding rates on these Ago proteins. Consequently, this could lead to a larger pool of siRNAs that are available for loading into the catalytic Ago protein Ago2, thus leading to more efficient RNAi. In addition, we find an extremely low proportion of passenger strand in Ago2 for thermodynamically stabilized siRNAs, compared with less stable siRNA duplexes. This observation could indicate, that Ago2 might cleave the passenger strands of thermodynamically more stable siRNA duplexes more efficiently, thereby increasing the efficiency of RISC activation. But this hypothesis needs to be addressed in future studies experimentally.
In order to reduce off-target effects caused by the noncatalytic Ago proteins as well as to increase on-target effects by Ago2, we suggest using stabilized siRNAs. Stabilization could either be achieved by simply selecting target sites with higher G/C content, although it remains to be tested how efficient G/C-rich sequences can be targeted by RNAi. Alternatively, passenger strands could be modified using LNAs, 2′-O-methyl or 2′-O-fluoro nucleotides that form base pairs with increased thermodynamic stability. In addition, it could be very interesting combining our approach of siRNA passenger strand modification to generate thermodynamically stabilized siRNA duplexes with guide strand modifications. Very interesting combinations could be siRNA guide strand modifications in the seed region, which have been described to cause reduced siRNA off-target activity, for example, 2′-O-methyl nucleotides (Jackson et al. 2006a) or UNA nucleotides (Bramsen et al. 2010; Laursen et al. 2010; Vaish et al. 2010). Since siRNAs are being used as drugs our siRNA selection parameters may also have an impact on siRNA-based therapy.
MATERIALS AND METHODS
IP of Ago proteins
For IP of human Ago proteins, the following monoclonal antibodies have been used: Ago1, clone 4B8 (Beitzinger et al. 2007); Ago2, clone 11A9 (Rudel et al. 2008); Ago3, clone 5A3 (#3.1) (Weinmann et al. 2009) and clone 4A11 (#3.2; for siRNA and miRNA co-IP clone anti-Ago3 4A11 was used) (this work); and Ago4, clone 6C10 (#4.1) (Weinmann et al. 2009) and clone 3G5 (#4.2) (this work) (for siRNA and miRNA co-IP clone anti-Ago4 6C10 was used). All monoclonal anti-Ago antibodies are available from Ascenion (www.ascenion.de). For IP of Flag-tagged proteins, ANTI-FLAG M2 Agarose (Sigma-Aldrich) was used. For IP of endogenous Ago proteins, antibodies were immobilized on Protein G Sepharose 4 Fast Flow (GE Healthcare). For mRNA coimmunoprecipitation experiments, Dynabeads Protein G (Invitrogen) have been used.
After antibody coupling, column materials were equilibrated with cell lysis buffer, cell lysate was added, and tubes were rotated overnight (analysis of cellular Ago protein amounts) or for 3 h (analysis of coprecipitating RNAs). After incubation with cell lysate, beads were washed twice with lysis buffer and twice with phosphate buffered sodium (PBS) containing 0.01% TritonX100. After the final washing step, the remaining buffer was removed quantitatively. For the analysis of the immunoprecipitated Ago proteins, sample buffer was added to the beads, and proteins were denatured at 65°C for 30 min. For the analysis of coprecipitated RNAs, samples were treated with Proteinase K in 250 μL of Proteinase K buffer (0.3 μg/μL Proteinase K, 300 mM NaCl, 25 mM EDTA, 2% SDS, 200 mM Tris at pH 7.5) and incubated for 15 min at 65°C. RNA was extracted with acidic phenol extraction and precipitated from the aqueous phase by adding 2.5 vol of pure ethanol followed by incubation overnight at −20°C. The RNA from the input samples was extracted with PeqGOLD TriFast (Peqlab) according to the manufacturer's instructions and was precipitated overnight at −20°C.
siRNAs
The siRNAs #1 and #2 were synthesized in house as described before (Chen et al. 2008): siRNA#1, guide strand UUAGUGAGAGUCCAAUUGCUT and passenger strand GCAAUUGGACUCUCACUAAUT; siRNA#2, guide strand UUAAUUGGACUCUCACUGCUT and passenger strand GCAGUGAGAGUCCAAUUAAUT. The siRNAs with increasing GC content used for experiments in Figure 4, D through F, were synthesized externally (Biomers).
Northern blotting
Northern blotting was carried out as described before (Ender et al. 2008). The following probes were used: siRNA#1, guide strand 5′-TTAGTGAGAGTCCAATTGCTT -3′ and passenger strand 5′-GCAATTGGACTCTCACTAATT-3′; siRNA#2, guide strand 5′-TTAATTGGACTCTCACTGCTT-3′ and passenger strand 5′-GCAGTGAGAGTCCAATTAATT-3′; and miR-19b, 5′-TCAGTTTTGCATCCATTTGCACA-3′. Radioactive blots were exposed to BioMax MS films (Kodak) using an intensifying screen (GE Healthcare). For radioactive signal quantifications, blots were exposed to BAS-IP MS 2040 imaging plates (FUJI) and scanned with a FLA-5000 Phosphoimager System (FUJI) or a PMI Phosphoimager System (Biorad).
Plasmids
For pMir-REPORT constructs, sequences complementary to siRNA#1 guide strand and passenger strand and human miR19b were cloned into the 3′ UTR of the firefly luciferase of a pMir-REPORT vector (Ambion). The vector was cut with the restriction enzymes SacI and NaeI. DNA-oligonucleotides encoding the siRNA/miRNA targeting sites were pre-annealed and 5′-phosphorylated with PNK:
pMir-siRNA#1 guide target, 5′-CATCGCCACCTTGTTTAAGCCGCAATTGGAGACTCACTAAATTAGACCTACGCACTCCAGGCC-3′ and
5′-GGCCTGGAGTGCGTAGGTCTAATTTAGTGAGTCTCCAATTGCGGCTTAAACAAGGTGGCGATGAGCT-3′;
pMir-siRNA#1 passenger target, 5′-CATCGCCACCTTGTTTAAGCCTTAGTGAGACACCAATTGCATTAGACCTACGCACTCCAGGCC-3′ and
5′-GGCCTGGAGTGCGTAGGTCTAATGCAATTGGTGTCTCACTAAGGCTTAAACAAGGTGGCGATGAGCT-3′; and
pMir-miR19b target, 5′-CATCGCCACCTTGTTTAAGCCTCAGTTTTGCATCCATTTGCACAATTAGACCTACGCACTCCAGGCC-3′ and
5′-GGCCTGGAGTGCGTAGGTCTAATTGTGCAAATGGATGCAAAACTGAGGCTTAAACAAGGTGGCGATGAGCT-3′.
For luciferase-based reporters, the following pre-annealed DNA-oligonucleotides were cloned into pMir-RL plasmids in a similar way as described for pMir-REPORT cloning above:
pMir-RL-siRNA#2 guide strand on-target reporter, 5′-CATCGCCACCTTGTTTAAGCCGCAGTGAGAGTCCAATTAAATTAGACCTACGCACTCCAGGCC-3′ and
5′-GGCCTGGAGTGCGTAGGTCTAATTTAATTGGACTCTCACTGCGGCTTAAACAAGGTGGCGATGAGCT-3′;
siRNA#2 guide strand off-target reporter, 5′-CATCCAATTACGCCACCTACTGTTTAAGCCCCAATTAATTAGACCTACTAGCACTCCCCAATTATGGCC-3′ and
5′-GGCCATAATTGGGGAGTGCTAGTAGGTCTAATTAATTGGGGCTTAAACAGTAGGTGGCGTAATTGGATGAGCT-3′.
Vp5-FLAG-HA-Ago2 has been described before (Meister et al. 2004). The Vp5-FLAG-HA-Ago2 H807R variant was generated via PCR, using the following primer for mutagenesis: 5′-CCAGCATACTACGCTCGCCTGGTGGCC-3′.
Cell culture
All cell lines mentioned in this publication were cultivated in Dulbecco's modified Eagle's medium (DMEM; PAA) supplemented with 10% fetal bovine serum (GIBCO), 100 U/mL penicillin, and 100 μg/mL streptomycin (GIBCO) at 37°C in 5% CO2 atmosphere. The Ago2−/− MEFs (Liu et al. 2004) as well as the Hela S3 cell line stably expressing FH-tagged Ago4 have been described before (Meister et al. 2004).
Transfection of HEK 293T cells with siRNA and plasmids for Ago-siRNA coimmunoprecipitation
Cells were transfected using calcium phosphate mediated transfection. About 15 h prior to transfection, cells were plated at 20% confluency. For all transfections of either pure siRNA or plasmid to a cell culture dish with a diameter of 15 cm, the following mix was added: 37.5 μL of a 20 μM siRNA stock solution or 10 μg of plasmid DNA was diluted in 645 μL H2O and 91.5 μL 2 M CaCl2. To this mixture was added 750 μL of 2× HEPES-buffered saline (274 mM NaCl, 1.5 mM Na2HPO4, 54.6 mM HEPES-KOH at pH 7.1). For transfections of siRNA and pMir-REPORT plasmid at the same time, the following mix was used: 7.5 μg plasmid and 25 μL of 20 μM of siRNA#1 were diluted in 860 μL H2O and 122 μL 2 M CaCl2. To this mixture was added 1000 μL of 2× HEPES-buffered saline. Transfection mix was incubated for 10 min and was then added drop-wise onto the cells.
Preparation of cell lysates
Cells were washed once with PBS on the cell culture dishes. Afterward cells were scratched from the dishes in the presence of PBS. The cell suspension was transferred into Falcon-tubes, and cells were harvested by centrifugation for 3 min at 1200g. The supernatant was removed quantitatively, and the weight of the cell pellet was determined.
For the IP of Ago proteins in order to determine their cellular amounts, cells were lysed on ice in 5 v/w (mL/g) of the following lysis buffer: 25 mM Tris-HCl (pH 7.5), 2 mM EDTA, 0.6% NP40, 0.6 M KCl. For the IP of Ago proteins in order to coprecipitate and analyze siRNA and miRNA, cells were lysed in the same buffer, but with 0.3% NP40 and 0.3 M KCl; here, 8 v/w (mL/g) has been used for lysis.
Cells were broken by sonification, and the lysate was centrifuged afterward at 17 000g to remove cell debris from the lysate. The lysates were used for IP.
Analysis of protein samples
Samples were separated by 10% denaturating SDS-PAGE. For mass spectrometry analysis, the gels were stained afterward with colloidal Coomassie solution (8.3% phosphoric acid, 8.3% (NH4)2SO4, 20% methanol, 1 g/L Coomassie Brilliant Blue G250; Sigma-Aldrich). Bands of interest were cut out from the gel and analyzed by the mass spectrometry group of the Microchemistry Core Facility, Max-Planck-Institute of Biochemistry (Martinsried, Germany). All intensities of peptides belonging to an Ago in its respective IP sample were added, and the sums of the peptide intensities were compared among the different Ago-IPs in order to roughly calculate the relative amounts of the individual Ago proteins.
For Western blotting after SDS-PAGE, samples were transferred to a nitrocellulose membrane (GE Healthcare) by semidry electroblotting. The membranes were blocked with 5% milk-powder (Roth) in PBS containing 0,05% Tween and were incubated with anti-beta-Actin antibody (ab6276, Biozol) or anti-HA tag antibody (Upstate/Millipore) according to the manufacturers' recommendations.
Real-time PCR-based analysis of coprecipitating reporter-mRNA
HEK 293T were transfected using calcium phosphate, as mentioned above. The cells were harvested 22–24 h after transfection and were lysed as described under “Preparation of Cell Lysates.” After IP of Ago proteins and RNA precipitation, the RNA was washed once with ice-cold 80% ethanol and solved in 25 μL water for 5 min at 70°C. Seven microliters of RNA was subjected to DNase I (Fermentas) digestion for 30 min at 37°C. Two microliters of the RNA was transcribed into cDNA with a First Strand cDNA Synthesis Kit (Fermentas) according to the manufacturer's instructions using Random Hexamer Primer. The resulting cDNA was analyzed by quantitative real-time PCR. It was diluted 1:10; 5 μL per sample of this dilution was mixed with 2 pmol each of forward and reverse primer and 10 μL of 2× MESA Green qPCR MasterMix Plus (Eurogentec) in a total volume of 20 μL. We used the following primers directed against firefly-luciferase mRNA: forward, 5′-GTGTTCGTCTTCGTCCCAGT-3′; reverse, 5′-GCTGGGCGTTAATCAGAGAG-3′. Samples were analyzed on a Biorad MyiQ analyzer.
Luciferase reporter assays
On- or off-target dual luciferase pMir-RL reporter plasmids were cotransfected with siRNA reversely into HEK 293T cells, wt MEFs, or Ago2−/− MEFs (Liu et al. 2004) by the use of Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. siRNA was cotransfected at indicated concentrations together with 10 ng of reporter plasmid into HEK 293T cells or together with 50 ng of reporter plasmid into wt MEFs or Ago2−/− MEFs per well of a 96-well plate. Each sample was transfected as a technical triplicate per experiment. Cells were lysed in Passive Lysis Buffer (Promega) at indicated time points after transfection. Firefly and Renilla luciferase activities were measured essentially as described before (Hock et al. 2007), but as separate samples. Renilla luciferase signals were used to normalize firefly luciferase signals; siRNA effects were compared to the effects of an unrelated control siRNA.
His-Ago1 siRNA binding assay
Human 6xHis-Ago2 was cloned into a modified pFastBac HT A vector (Invitrogen), transformed into DH10MultiBac via the Tn7 recombination site. Recombinant His-Ago1 protein was expressed in SF21 cells using the Baculovirus system. SF21 cells were lysed in lysis buffer (50 mM HEPES at pH 7.5, 500 mM NaCl, 0.1% NP-40) by intensive vortexing. After centrifugation for 30 min at 40000g, the lysate was supplemented with glycerin and imidazol (pH 7.5) to a final concentration of 10% and 10 mM, respectively. His-Ago2 was then purified by Ni2+-affinity chromatography followed by gel-filtration (Superdex200 prep grade, GE-Healthcare). For the Ago1-siRNA binding assay, siRNA#2 guide strand was radioactively labeled by PNK (Fermentas)-mediated 5′-phosphorylation using 32P-γ-ATP (Hartmann Analytics). Furthermore, siRNA#2 guide and passenger strands were phorsphorylated in the presence of cold ATP. Free ATP and PNK were removed from labeling reactions by gel-filtration with MicroSpin G25 columns (GE Healthcare). Concentrations of the purified siRNA strands were measured, and the radioactively labeled and unlabeled siRNA#2 guide strand was mixed in a ratio of 1:10, respectively. Subsequently, half of the mixture was pre-annealed with an equal amount of siRNA#2 passenger strand. The other half was diluted with an equal volume of water.
To test siRNA#2 guide strand and duplex interactions with Ago1, 10 nM of purified His-Ago1 was incubated with 0.5 nM of either siRNA#2 guide strand or duplex in 50 mM HEPES (pH 7.5, 300 mM NaCl, 10% glycerin, 2 mM MgCl2, 0.5 mg/mL BSA) for 90 min at 24°C. As control samples, the siRNA single- and double-strands were incubated in the absence of His-Ago1. Afterward, His-Ago1 was immunoprecipitated from the binding reaction for 60 min by the addition of anti-Ago1 antibody loaded Protein G Sepharose 4 Fast Flow (GE Healthcare). The IP samples were washed two times with 1 mL of binding buffer lacking BSA and another two times with 1 mL PBS containing 0.01% Tween. Thereafter, the samples were Proteinase K digested as described above. Samples were diluted in 5 mL Unisafe I emulsifying scintillator (Zinser Analytic) and measured on a LS6500 scintillation counter (Beckmann).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Corinna Friederich, Sigrun Ammon, and Sabine Rotmüller for excellent technical assistance, as well as Bernd Haas for siRNA synthesis and Gregory Hannon for Ago2−/− MEFs. This study was in part supported by the Deutsche Forschungsgemeinschaft (DFG; FOR855), the Bavarian Genome Research Network (BayGene to G.M.), and Roche Kulmbach GmbH. S.P. receives a fellowship from the Roche Postdoc Fellowship Program.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2348111.
REFERENCES
- Aleman LM, Doench J, Sharp PA 2007. Comparison of siRNA-induced off-target RNA and protein effects. RNA 13: 385–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G 2007. Identification of human microRNA targets from isolated Argonaute protein complexes. RNA Biol 4: 76–84 [DOI] [PubMed] [Google Scholar]
- Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, et al. 2006. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods 3: 199–204 [DOI] [PubMed] [Google Scholar]
- Bramsen JB, Laursen MB, Damgaard CK, Lena SW, Babu BR, Wengel J, Kjems J 2007. Improved silencing properties using small internally segmented interfering RNAs. Nucleic Acids Res 35: 5886–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjaer N, Babu BR, Hojland T, Abramov M, Van Aerschot A, et al. 2009. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res 37: 2867–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramsen JB, Pakula MM, Hansen TB, Bus C, Langkjaer N, Odadzic D, Smicius R, Wengel SL, Chattopadhyaya J, Engels JW, et al. 2010. A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res 38: 5761–5773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D, Rossi JJ 2009. The promises and pitfalls of RNA-interference-based therapeutics. Nature 457: 426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Weinmann L, Gaidatzis D, Pei Y, Zavolan M, Tuschl T, Meister G 2008. Strand-specific 5′-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA 14: 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Jung S, Rothenberg SM, Smolen GA, Mlody BG, Haber DA 2008. Coexpression of Argonaute-2 enhances RNA interference toward perfect match binding sites. Proc Natl Acad Sci 105: 9284–9289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y, Tuschl T 2004. siRNAs: applications in functional genomics and potential as therapeutics. Nat Rev Drug Discov 3: 318–329 [DOI] [PubMed] [Google Scholar]
- Ender C, Meister G 2010. Argonaute proteins at a glance. J Cell Sci 123: 1819–1823 [DOI] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G 2008. A human snoRNA with microRNA-like functions. Mol Cell 32: 519–528 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E 2008. Getting to the root of miRNA-mediated gene silencing. Cell 132: 9–14 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD 2007. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell 130: 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G 2007. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep 8: 1052–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 21: 635–637 [DOI] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. 2006a. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA 12: 1197–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS 2006b. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12: 1179–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Doudna JA 2009. A three-dimensional view of the molecular machinery of RNA interference. Nature 457: 405–412 [DOI] [PubMed] [Google Scholar]
- Kawamata T, Seitz H, Tomari Y 2009. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol 16: 953–960 [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Kim K, Lee YS, Carthew RW 2007. Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes. RNA 13: 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Laursen MB, Pakula MM, Gao S, Fluiter K, Mook OR, Baas F, Langklaer N, Wengel SL, Wengel J, Kjems J, et al. 2010. Utilization of unlocked nucleic acid (UNA) to enhance siRNA performance in vitro and in vivo. Mol Biosyst 6: 862–870 [DOI] [PubMed] [Google Scholar]
- Leuschner PJ, Ameres SL, Kueng S, Martinez J 2006. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep 7: 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Fesik SW, Shen Y 2005. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res 33: 4527–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD 2005. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123: 607–620 [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431: 343–349 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T 2004. Human Argonaute2 Mediates RNA Cleavage Targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC 2005. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev 19: 2837–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X 2005. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123: 621–629 [DOI] [PubMed] [Google Scholar]
- Rudel S, Flatley A, Weinmann L, Kremmer E, Meister G 2008. A multifunctional human Argonaute2-specific monoclonal antibody. RNA 14: 1244–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Svoboda P 2007. Off-targeting and other non-specific effects of RNAi experiments in mammalian cells. Curr Opin Mol Ther 9: 248–257 [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD 2004. A protein sensor for siRNA asymmetry. Science 306: 1377–1380 [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD 2007. Sorting of Drosophila small silencing RNAs. Cell 130: 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaish N, Chen F, Seth S, Fosnaugh K, Liu Y, Adami R, Brown T, Chen Y, Harvie P, Johns R, et al. 2010. Improved specificity of gene silencing by siRNAs containing unlocked nucleobase analogs. Nucleic Acids Res (in press). doi: 10.1093/nar/gkq961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers TA, Lima WF, Nichols JG, Crooke ST 2007. Reduced levels of Ago2 expression result in increased siRNA competition in mammalian cells. Nucleic Acids Res 35: 6598–6610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers TA, Lima WF, Wu H, Nichols JG, Linsley PS, Crooke ST 2009. Off-target and a portion of target-specific siRNA mediated mRNA degradation is Ago2 ‘Slicer’ independent and can be mediated by Ago1. Nucleic Acids Res 37: 6927–6941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Li S, Qi HH, Chowdhury D, Shi Y, Novina CD 2009. Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nat Struct Mol Biol 16: 1259–1266 [DOI] [PubMed] [Google Scholar]
- Weinmann L, Hock J, Ivacevic T, Ohrt T, Mutze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G 2009. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 136: 496–507 [DOI] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG 2008. Importance of translation and nonnucleolytic ago proteins for on-target RNA interference. Curr Biol 18: 1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ 2005. Crystal structure of A. aeolicus Argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell 19: 405–419 [DOI] [PMC free article] [PubMed] [Google Scholar]