Abstract
Methionine sulfoxide reductases (Msrs) are thiol-dependent enzymes which catalyze conversion of methionine sulfoxide to methionine. Three Msr families, MsrA, MsrB, and fRMsr, are known. MsrA and MsrBs are responsible for the reduction of methionine-S-sulfoxide and methionine-R-sulfoxide residues in proteins, respectively, whereas fRMsr reduces free methionine-R-sulfoxide. Besides acting on proteins, MsrA can additionally reduce free methionine-S-sulfoxide. Some MsrAs and MsrBs evolved to utilize catalytic selenocysteine. This includes MsrB1, which is a major MsrB in cytosol and nucleus in mammalian cells. Specialized machinery is used for insertion of selenocysteine into MsrB1 and other selenoproteins at in-frame UGA codons. Selenocysteine offers catalytic advantage to the protein repair function of Msrs, but also makes these proteins dependent on the supply of selenium and requires adjustments in their strategies for regeneration of active enzymes. Msrs have roles in protecting cellular proteins from oxidative stress and through this function they may regulate lifespan in several model organisms.
Keywords: methionine sulfoxide, MsrA, fRMsr, MsrB1, MsrB2, MsrB3, selenoprotein, selenocysteine, aging, antioxidants, ROS
Introduction
Cellular components in aerobic organisms are often confronted with reactive oxygen species (ROS) and reactive nitrogen intermediates (RNI). Molecular oxygen is indispensible for survival of aerobic organisms, which use it for energy pathways in mitochondria and numerous other processes, but the use of oxygen is also associated with the generation of ROS [1]. ROS may damage various biomolecules, such as DNA, proteins, and lipids, and may contribute to the development of cancer, neurodegenerative diseases and other maladies. Oxidative damage to DNA may lead to mutations, which in turn may result in cancer [2, 3]. Lipid oxidation by ROS may change membrane rigidity and permeability. In addition, it may disrupt membrane networks and generate toxic products, such as malondialdehyde, through lipid peroxidation [4, 5]. Oxidized proteins may become nonfunctional due to structural changes and catalytic malfunction. Sulfur-containing amino acids, methionine and cysteine, are the major targets of ROS and are the amino acids most susceptible to oxidation. Oxidized methionine exists in the form of two diastereomers, methionine-S-sulfoxide and methionine-R-sulfoxide [6, 7]. To counteract ROS damage, organisms evolved defense systems, including low molecular weight compounds and antioxidant enzymes that protect against oxidative stress. The latter include glutathione peroxidase (GPx) [8, 9], superoxide dismutase (SOD) [10, 11], catalase [12], thioredoxin reductase (TR) [13], methionine sulfoxide reductase (Msr) [14-17], and several other proteins. Among these antioxidant enzymes, Msrs are prominent because of their roles as repair enzymes and indirect scavengers of ROS. These proteins utilize catalytic redox-active cysteine residues to reverse oxidized methionine back to methionine. Many Msrs use the thioredoxin system in vivo to regenerate the active catalytic cysteine, but other reductants may also be used [16-20]. Because of their protein repair functions, the studies involving Msrs are broadly linked to various research areas such as aging, neurodegenerative disease, cancer, cell signaling, and protein function.
Three types of methionine sulfoxide reductases
The first Msr enzyme, MsrA, was discovered some 30 years ago. It was found that this enzyme can restore the function of oxidized ribosomal protein L12 in E. coli [21]. After this initial finding, Msrs were investigated from a variety of organisms, new forms were discovered, and currently these enzymes can be divided into three large groups according to their substrate specificity and family clustering. MsrA is specific for the reduction of free and protein-based methionine-S-sulfoxide. This protein is the only known enzyme capable of reducing methionine-S-sulfoxide [22-25], but it can also reduce other compounds such as N-acetyl-methionine-S-sulfoxide, dimethyl sulfoxide, ethionine-S-sulfoxide, S-sulindac, S-sulforaphane, etc. [26-28]. It was found that MsrA exist in a selenoprotein form in some lower organisms, such as green algae and some bacteria, wherein it utilizes catalytic selenocysteine in place of cysteine [29, 30].
A second Msr, known as MsrB (described previously as SelR or SelX), is specific for the reduction of protein-based methionine-R-sulfoxide, but also, with low efficiency, of free methionine-R-sulfoxide. For example, catalytic efficiency of E. coli MsrB for reduction of free methionine-R-sulfoxide is 1000-fold lower than that of E. coli MsrA for free methionine-S-sulfoxide. In addition, when the MsrB gene was disrupted in E. coli, specific activity of cell extract for reduction of free methionine-R-sulfoxide was essentially unchanged [31, 32], whereas mutation of another Msr gene (see below) in yeast resulted in reduced activity with this substrate [33]. In human SK-Hep1 cells, MsrBs were also found to be inefficient in reducing free methionine-R-sulfoxide. This study revealed that mammals have important differences in the reduction of free methionine-S-sulfoxide and free methionine-R-sulfoxide [34].
In mammals, there are three MsrB proteins, including MsrB1, MsrB2, and MsrB3. All three contain Zn, coordinated by two CxxC motifs (i.e., two Cys separated by two residues), which stabilizes MsrB structure. Of these three enzymes, MsrB1 is a selenoprotein that contains selenocysteine in the place of the catalytic cysteine residue normally present in other MsrBs [35, 36].
A third Msr type, fRMsr, was only recently discovered [37] and it catalyzes the reduction of free methionine-R-sulfoxide [33, 34, 37]. This enzyme is a GAF-domain-containing protein and is highly specific for its substrate (i.e., it does not act on protein-base sulfoxides) [33, 37, 38]. Interestingly, comparative genomic analysis of fRMsr revealed that its occurrence is limited to unicellular organisms, whereas multicellular organisms, including mammals, lack this protein [33, 37]. This observation suggested that fRMsr was lost during evolution in mammals, which currently lack this function, i.e., they are deficient in the reduction of free methionine-R-sulfoxide (Figure 1).
Figure 1.
Reversible generation and reduction of methionine sulfoxide. MsrA can reduce both free and protein-based methionine-S-sulfoxide. MsrB mainly reduces protein-based methionine-R-sulfoxide and its activity towards free methionine-R-sulfoxide is very weak. fRMsr can only reduce free methionine-R-sulfoxide.
The three Msrs support oxidative protein repair and methionine metabolism functions by reducing free and protein-based methionine-R,S-sulfoxides. The key function of Msr is to repair oxidatively damaged proteins, in which ROS oxidized methionine residues. This oxidation may interfere with protein function, because it leads to an increased negative charge and size, and consequently may result in structural changes and loss of function [6, 7, 39]. In addition, Msrs may work indirectly as ROS scavengers through cyclic oxidation and reduction of methionine and methionine sulfoxide residues. The methionine/Msr system may assist other redox systems, such as thioredoxin and glutathione systems, in maintaining cellular redox homeostasis. This idea is consistent with the data showing that deficiency in various Msrs results in oxidative stress [14, 16, 40, 41]. Methionine sulfoxides and Msrs may also play a role in sensing altered redox status and regulating certain proteins. Recently, it was shown that CaMKII (calcium/calmodulin (Ca2+/CaM)-dependent protein kinase II) was activated by oxidation of its methionine residue in the regulatory domain in the absence of Ca2+/CaM and that this activation could be reversed by MsrA [42]. In addition, human large conductance Ca2+-activated K channels (BKCa or Slo), E. coli Ffh, human potassium channel HERG1, and calmodulin are known to be regulated by oxidation of methionine and repair activity of Msrs [43-45]. Along with these Msr functions, there were several reports showing that MsrA knockout mice exhibit a tip-toe walking behavior consistent with cerebellar dysfunction [46], enhanced neurodegeneration in hippocampus [47], and impaired dopamine regulation [48]. However, the specific link between Msrs and neurodegenerative diseases remains poorly understood.
Localization of mammalian MsrA and MsrBs
Mammals have a single MsrA gene containing a typical mitochondrial signal peptide [49]. However, mouse and rat MsrAs occur not only in mitochondria, but also in cytosol and nucleus. It was suggested that the rate of folding is a crucial factor in the subcellular distribution of MsrA, even though the molecular mechanism and regulation remain unclear [50]. It was also reported that alternative first exon splicing generates an additional MsrA form lacking a mitochondrial signal, which resides in cytosol and nucleus [51-53]. It should be noted, however, that the alternatively spliced MsrA form was not detected in examined mouse tissues, including brain, kidney, and liver, by western blot assays, and only a weak signal was detected at the mRNA level. Studies showed that other alternative forms of MsrA mRNA including an additional exon 2 could be detected in rat aortic vascular smooth muscle cells and aortic tissue preparations [53] and this form was localized exclusively to mitochondria [53, 54].
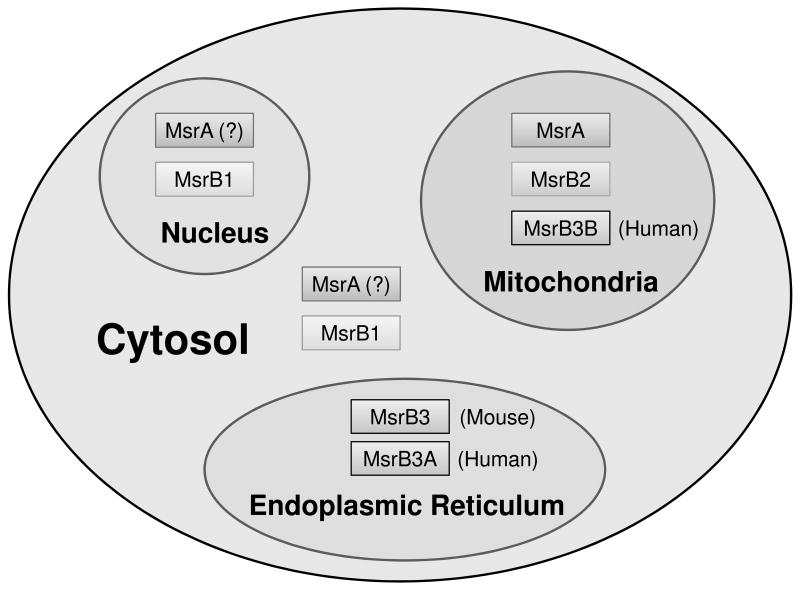
Among three mammalian MsrBs, MsrB1 is present in the cytosol and nucleus and exhibits the highest catalytic activity due to presence of selenocysteine in its active site. Recently, it was found that MsrB1 occurs in two forms, 14 kDa and 5 kDa, in mouse tissues and human HEK 293 cells and that both forms are selenoproteins. The 5 kDa form corresponded to a C-terminal sequence of 14 kDa MsrB1. However, neither function nor regulation of the 5 kDa MsrB1 is known [55, 56]. MsrB2, also known as CBS1, is targeted to mitochondria with the guidance of its N-terminal signal peptide. MsrB2 has cysteine as the catalytic residue. It shows high activity with methionine-R-sulfoxide, but is inhibited by elevated concentrations of the substrate [35, 57, 58]. MsrB3 consists of two forms, MsrB3A and MsrB3B, generated by alternative first exon slicing in humans. MsrB3A is targeted to the endoplasmic reticulum (ER) with the N-terminal ER signal peptide and an ER retention signal at the C-terminus, whereas MsrB3B is targeted to mitochondria by its N-terminal mitochondrial signal peptide. Interestingly, mouse MsrB3 also has the ER and mitochondrial signal peptides located consecutively at the N-terminus, but it is targeted only to the ER due to masking of the mitochondrial signal by the upstream ER signal. Even though a possibility cannot be excluded that small amounts of mouse MsrB3 targeted to mitochondria exist, studies found no evidence of alternative splicing in this gene based on EST, RT-PCR, and western blot analyses [35, 59]. The localization of mammalian MsrA and MsrBs is shown in Figure 2.
Figure 2.
Localization of Msrs in mammals. MsrA and MsrB1 are located in the nucleus and cytosol. MsrA, MsrB2, and MsrB3B are targeted to mitochondria by their mitochondrial targeting signals. Human MsrB3A is located in the ER and has N-terminal ER targeting and C-terminal retention signals, whereas human MsrB3B is a mitochondrial protein. Mouse MsrB3 is located in the ER. There are alternatively spliced variants of MsrA, which are not shown in the figure and localize to cytosol and nucleus, or to mitochondria.
Catalysis by the different Msr types
Mammalian MsrA has three conserved cysteines, which participate in catalysis as catalytic and resolving cysteines. A sulfenic acid intermediate at the catalytic cysteine is generated when this residue attacks the sulfur of methionine-S-sulfoxide and then thiol-disulfide exchange involving two resolving cysteines lead to a disulfide bond on the protein surface, which is finally reduced by thioredoxin or DTT [60]. Recently, it was proposed that yeast fRMsr also has three conserved cysteines that follow a similar sulfenic acid/thiol-disulfide exchange mechanism. In addition, the thioredoxin system can function in recycling fRMsr like it does in the case of MsrA. A difference between MsrA and fRMsr involves accessibility of the substrate. Yeast fRMsr has a narrow binding pocket, which limits access to bulky substrates. This may explain differences in substrate specificity between MsrA and fRMsr, even though the overall catalytic process involving catalytic and resolving cysteines is similar in these enzymes (Figure 3A and B) [33, 37].
Figure 3.
Catalytic mechanisms of Msrs. Catalytic cysteine (CC), resolving cysteine (RC), and/or third cysteine (TC) take part in the catalysis by (A) MsrA, (B) fRMsr, (C) MsrB1, and (D) MsrB2 and MsrB3. MsrA and fRMsr have three conserved cysteines. MsrB1 has one conserved cysteine and one conserved selenocysteine. MsrB2 and MsrB3 have only one redox cysteine and its intermediate sulfenic acid may be directly reduced by thioredoxin or certain compounds.
MsrB1 has one conserved cysteine in the N-terminal portion and the catalytic selenocysteine in the C-terminal region. During catalysis, selenocysteine forms selenenic acid intermediate by attacking the sulfur of methionine-R-sulfoxide and then this intermediate rearranges into selenenylsulfide with the help of the resolving cysteine and is finally reduced by thioredoxin (Figure 3C) [61]. In contrast, MsrB2 and MsrB3 have only one conserved cysteine and appear to have evolved a different catalytic mechanism for the reduction of their sulfenic acid intermediate (i.e., without disulfide formation between catalytic and resolving cysteines), which is reduced directly by thioredoxin (Figure 3D) [61-63]. Although thioredoxin is the most important reductant for Msrs, it was shown that thioredoxin-independent Msr reduction is also possible [19, 20]. For example, thionein, selenocystamine, and CDP32 could be used for the reduction of human MsrB2 and MsrB3, and such thioredoxin-independent reaction may even be more efficient than that involving thioredoxin.
Selenoproteins and catalytic benefits of selenocysteine utilization
Selenoproteins exist in organisms in all three domains of life. Selenium is inserted into these proteins cotranslationally in the form of selenocysteine, in response to UGA codon. Selenocysteine insertion is dependent on the presence of a stem-loop structure, known as the SECIS (selenocysteine insertion sequence) element, in the 3′-UTRs of selenoprotein genes. Selenium is essential in mammals because of its occurrence in selenoproteins (Figure 4). These proteins have various functions and show widely different knockout phenotypes. For example, glutathione peroxidase 4 (GPx4) reduces lipid peroxides and its knockout in mice results in embryonic lethality [64]. Another selenoprotein, Selenoprotein P (SelP), transports selenium from liver to brain, thyroid gland, kidney, testis and likely other organs. Without SelP, a much higher selenium intake is needed to support selenoprotein synthesis [65-67]. Selenoprotein N (SelN) is a protein of unknown function and mutations in this gene are linked to various muscular diseases, such as rigid spine muscular dystrophy [68]. Many of the 25 human selenoproteins contribute to the regulation of redox homeostasis, but the functions of several proteins are not known. Perhaps, the best studied selenoprotein families are glutathione peroxidases and thioredoxin reductases. There are 5 selenoprotein GPxs in humans and 4 in mice, which function as thiol-dependent peroxidases. Thioredoxin reductases 1, 2, and 3 regulate cellular redox homeostasis by controlling the redox state of thioredoxins [69]. SelW and SelK might also play a role in redox regulation. For example, SelW binds glutathione and its overexpression increases resistance to oxidative stress in CHO cells and H1299 cancer cells in a glutathione-dependent manner [70, 71]. Overexpression of SelK reduces intracellular ROS levels and protects cardiomyocytes from oxidative stress-induced toxicity [72].
Figure 4.
Selenoprotein biosynthesis. Selenocysteine is inserted into protein at UGA codons with the help of various factors that form the selenocysteine insertion machinery as shown in the figure. Sec-tRNASec, a tRNA specific for selenocysteine; SBP2, SECIS binding protein 2; EFSec, elongation factor specific for Sec-tRNA.
Like other functionally characterized selenoproteins, MsrB1 is an oxidoreductase. Selenocysteine in this enzyme supports high catalytic efficiency of MsrB1 (>100-fold higher than that of the corresponding cysteine-containing mutant) [35, 61, 73, 74]. This enhancement of activity was also demonstrated for a selenocysteine-containing MsrA from Chlamydomonas [30]. Thus, selenocysteine, strategically positioned in the active site, is a key catalytic residue, whereas its replacement with cysteine leads to a significant drop or loss of catalytic activity. Similar observations were also made with iodothyronine deiodinase in oocytes [75, 76] and clostridial glycine reductase selenoprotein A [75, 77].
However, selenocysteine is not always beneficial under physiological conditions. When the catalytic Cys in MsrB2 and MsrB3 was replaced with selenocysteine, activities of these enzymes were increased up to 100-fold in the presence of DTT as a reducing agent, but these proteins were not active with thioredoxin. One possibility is that a selenenic acid intermediate could not be efficiently reduced by thioredoxin and therefore the enzymes could not be regenerated for the next catalytic cycle, whereas a selenenylsulfide bond, formed between catalytic selenocysteine and resolving cysteine residues, was reducible by thioredoxin. This hypothesis was recently supported by the studies on a natural selenocysteine-containing MsrA and an artificial selenoprotein MsrB from Clostridium sp., an anaerobic bacterium [29]. It was shown that these proteins have catalytic advantage by incorporating selenocysteine as the catalytic residue, but they could not be reduced by thioredoxin, suggesting that the selenenic acid intermediate requires other reductants under physiological conditions.
On the other hand, the use of selenocysteine in MsrB and MsrA is rare when viewed from the evolutionary perspective. The absolute majority of these enzymes are cysteine-containing proteins, and many of these are highly efficient in standard in vitro assays. Interestingly, Msrs are absent in many hyperthermophylic organisms because at higher temperatures, methionine sulfoxide reduction does not require catalysis. By the same token, selenoprotein Msrs might be needed under conditions when their cysteine-containing counterparts are ineffective.
Aging and Msrs
Since the oxidative stress theory of aging was first introduced half a century ago, free radicals and other ROS emerged as important factors that regulate the aging process [1, 78, 79]. Along with the idea of ROS-induced damage, defense mechanisms received much attention, especially in the area of aging research, because efficient protection from oxidative stress and elimination of damage might delay the aging process. It has been shown that various antioxidant enzymes, such as catalase and SOD, could protect from oxidative stress and increase lifespan in some model organisms [80-84]. As an antioxidant enzyme, MsrA has also been examined, and it was found that its overexpression could extend lifespan by 70% in Drosophila [84]. Furthermore, knockout of MsrA in mice or yeast reduced lifespan of these organisms [46, 85]. In contrast to MsrA, the effect of MsrB is less obvious. It was reported that overexpression of MsrB extends lifespan of yeast cells grown under caloric restriction conditions, but not in cells grown in regular medium, i.e., conditions where MsrA could increase lifespan [86].
As mentioned above, MsrA and MsrB have clear differences in their ability for reduction of free methionine sulfoxide as well as in their cellular locations, and these differences might account for the observations in the aging studies. Much of the amino acid metabolism occurs in mitochondria, which is the organelle that generates significant amounts of ROS [87-90]. Since methionine is one of amino acids most susceptible to oxidation by ROS, it is possible that it can serve as a defense system through cyclic oxidation and reduction by mitochondria-targeted Msrs. In addition, methionine is a precursor for other antioxidants and redox compounds, such as cysteine, taurine, and glutathione. Furthermore, S-adenosylmethionine, the principal methyl donor in the cell, is also an intermediate in the methionine metabolism, which may link Msr function to epigenetic regulation [34, 78, 91, 92, 93]. Possible negative effects of excess methionine as a factor that regulates lifespan should be also considered. For example, overflow of methionine may generate excessive amounts of homocysteine [94] (Figure 5). Even though efficient reduction of free methionine sulfoxide might be a factor in protecting cells from ROS and extending lifespan, it should be viewed in the context of overall methionine metabolism under both stress and normal conditions. Finally, since MsrB1 is a selenoprotein, it would be important to examine its effect as well as the effect of dietary selenium on lifespan in mammals.
Figure 5.
Methionine and methionine sulfoxide metabolism in mammals. Protein synthesis, methionine cycle and transsulfuration pathways represent the major uses of free methionine.
Acknowledgments
This study was supported by NIH AG021518 (to V.N.G) and KOSEF R13-2005-005-01004-0 (to H.-Y.K). The NTNU's support (to A.D.) is also acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins A, Harrington V. Repair of oxidative DNA damage: assessing its contribution to cancer prevention. Mutagenesis. 2002;17:489–493. doi: 10.1093/mutage/17.6.489. [DOI] [PubMed] [Google Scholar]
- 3.Loft S, Poulsen HE. Cancer risk and oxidative DNA damage in man. J Mol Med. 1996;74:297–312. doi: 10.1007/BF00207507. [DOI] [PubMed] [Google Scholar]
- 4.Nair U, Bartsch H, Nair J. Lipid peroxidation-induced DNA damage in cancer-prone inflammatory diseases: a review of published adduct types and levels in humans. Free Radic Biol Med. 2007;43:1109–1120. doi: 10.1016/j.freeradbiomed.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 5.Rikans LE, Hornbrook KR. Lipid peroxidation, antioxidant protection and aging. Biochim Biophys Acta. 1997;1362:116–127. doi: 10.1016/s0925-4439(97)00067-7. [DOI] [PubMed] [Google Scholar]
- 6.Stadtman ER. Protein oxidation and aging. Free Radic Res. 2006;40:1250–1258. doi: 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- 7.Shechter Y, Burstein Y, Patchornik A. Selective oxidation of methionine residues in proteins. Biochemistry. 1975;14:4497–4503. doi: 10.1021/bi00691a025. [DOI] [PubMed] [Google Scholar]
- 8.Spector A, Kuszak JR, Ma W, Wang RR. The effect of aging on glutathione peroxidase-i knockout mice-resistance of the lens to oxidative stress. Exp Eye Res. 2001;72:533–545. doi: 10.1006/exer.2001.0980. [DOI] [PubMed] [Google Scholar]
- 9.Barkats M, Millecamps S, Abrioux P, Geoffroy MC, Mallet J. Overexpression of glutathione peroxidase increases the resistance of neuronal cells to Abeta-mediated neurotoxicity. J Neurochem. 2000;75:1438–1446. doi: 10.1046/j.1471-4159.2000.0751438.x. [DOI] [PubMed] [Google Scholar]
- 10.Warner HR. Superoxide dismutase, aging, and degenerative disease. Free Radic Biol Med. 1994;17:249–258. doi: 10.1016/0891-5849(94)90080-9. [DOI] [PubMed] [Google Scholar]
- 11.Orr WC, Sohal RS. Effects of Cu-Zn superoxide dismutase overexpression of life span and resistance to oxidative stress in transgenic Drosophila melanogaster. Arch Biochem Biophys. 1993;301:34–40. doi: 10.1006/abbi.1993.1111. [DOI] [PubMed] [Google Scholar]
- 12.Orr WC, Sohal RS. The effects of catalase gene overexpression on life span and resistance to oxidative stress in transgenic Drosophila melanogaster. Arch Biochem Biophys. 1992;297:35–41. doi: 10.1016/0003-9861(92)90637-c. [DOI] [PubMed] [Google Scholar]
- 13.Arner ES, Holmgren A. The thioredoxin system in cancer. Semin Cancer Biol. 2006;16:420–426. doi: 10.1016/j.semcancer.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Kantorow M, Hawse JR, Cowell TL, Benhamed S, Pizarro GO, Reddy VN, Hejtmancik JF. Methionine sulfoxide reductase A is important for lens cell viability and resistance to oxidative stress. Proc Natl Acad Sci USA. 2004;101:9654–9659. doi: 10.1073/pnas.0403532101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moskovitz J, Flescher E, Berlett BS, Azare J, Poston JM, Stadtman ER. Overexpression of peptide-methionine sulfoxide reductase in Saccharomyces cerevisiae and human T cells provides them with high resistance to oxidative stress. Proc Natl Acad Sci USA. 1998;95:14071–14075. doi: 10.1073/pnas.95.24.14071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stadtman ER. Cyclic oxidation and reduction of methionine residues of proteins in antioxidant defense and cellular regulation. Arch Biochem Biophys. 2004;423:2–5. doi: 10.1016/j.abb.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Stadtman ER, Moskovitz J, Berlett BS, Levine RL. Cyclic oxidation and reduction of protein methionine residues is an important antioxidant mechanism. Mol Cell Biochem. 2002;234-235:3–9. [PubMed] [Google Scholar]
- 18.Brot N, Weissbach L, Werth J, Weissbach H. Enzymatic reduction of protein-bound methionine sulfoxide. Proc Natl Acad Sci USA. 1981;78:2155–2158. doi: 10.1073/pnas.78.4.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding D, Sagher D, Laugier E, Rey P, Weissbach H, Zhang XH. Studies on the reducing systems for plant and animal thioredoxin-independent methionine sulfoxide reductases B. Biochem Biophys Res Commun. 2007;361:629–633. doi: 10.1016/j.bbrc.2007.07.072. [DOI] [PubMed] [Google Scholar]
- 20.Sagher D, Brunell D, Hejtmancik JF, Kantorow M, Brot N, Weissbach H. Thionein can serve as a reducing agent for the methionine sulfoxide reductases. Proc Natl Acad Sci USA. 2006;103:8656–8661. doi: 10.1073/pnas.0602826103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caldwell P, Luk DC, Weissbach H, Brot N. Oxidation of the methionine residues of Escherichia coli ribosomal protein L12 decreases the protein's biological activity. Proc Natl Acad Sci USA. 1978;75:5349–5352. doi: 10.1073/pnas.75.11.5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moskovitz J, Poston JM, Berlett BS, Nosworthy NJ, Szczepanowski R, Stadtman ER. Identification and characterization of a putative active site for peptide methionine sulfoxide reductase (MsrA) and its substrate stereospecificity. J Biol Chem. 2000;275:14167–14172. doi: 10.1074/jbc.275.19.14167. [DOI] [PubMed] [Google Scholar]
- 23.Boschi-Muller S, Gand A, Branlant G. The methionine sulfoxide reductases: Catalysis and substrate specificities. Arch Biochem Biophys. 2008;474:266–273. doi: 10.1016/j.abb.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Sharov VS, Ferrington DA, Squier TC, Schoneich C. Diastereoselective reduction of protein-bound methionine sulfoxide by methionine sulfoxide reductase. FEBS Lett. 1999;455:247–250. doi: 10.1016/s0014-5793(99)00888-1. [DOI] [PubMed] [Google Scholar]
- 25.Lee BC, Lee YK, Lee HJ, Stadtman ER, Lee KH, Chung N. Cloning and characterization of antioxidant enzyme methionine sulfoxide-S-reductase from Caenorhabditis elegans. Arch Biochem Biophys. 2005;434:275–281. doi: 10.1016/j.abb.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 26.Etienne F, Resnick L, Sagher D, Brot N, Weissbach H. Reduction of Sulindac to its active metabolite, sulindac sulfide: assay and role of the methionine sulfoxide reductase system. Biochem Biophys Res Commun. 2003;312:1005–1010. doi: 10.1016/j.bbrc.2003.10.203. [DOI] [PubMed] [Google Scholar]
- 27.Weissbach H, Resnick L, Brot N. Methionine sulfoxide reductases: history and cellular role in protecting against oxidative damage. Biochim Biophys Acta. 2005;1703:203–212. doi: 10.1016/j.bbapap.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Moskovitz J, Weissbach H, Brot N. Cloning the expression of a mammalian gene involved in the reduction of methionine sulfoxide residues in proteins. Proc Natl Acad Sci USA. 1996;93:2095–2099. doi: 10.1073/pnas.93.5.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HY, Zhang Y, Lee BC, Kim JR, Gladyshev VN. The selenoproteome of Clostridium sp. OhILAs: Characterization of anaerobic bacterial selenoprotein methionine sulfoxide reductase A. Proteins. 2008;74:1008–1017. doi: 10.1002/prot.22212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HY, Fomenko DE, Yoon YE, Gladyshev VN. Catalytic advantages provided by selenocysteine in methionine-S-sulfoxide reductases. Biochemistry. 2006;45:13697–13704. doi: 10.1021/bi0611614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimaud R, Ezraty B, Mitchell JK, Lafitte D, Briand C, Derrick PJ, Barras F. Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J Biol Chem. 2001;276:48915–48920. doi: 10.1074/jbc.M105509200. [DOI] [PubMed] [Google Scholar]
- 32.Etienne F, Spector D, Brot N, Weissbach H. A methionine sulfoxide reductase in Escherichia coli that reduces the R enantiomer of methionine sulfoxide. Biochem Biophys Res Commun. 2003;300:378–382. doi: 10.1016/s0006-291x(02)02870-x. [DOI] [PubMed] [Google Scholar]
- 33.Le DT, Lee BC, Marino SM, Zhang Y, Fomenko DE, Kaya A, Hacioglu E, Kwak GH, Koc A, Kim HY, Gladyshev VN. Functional Analysis of Free Methionine-R-sulfoxide Reductase from Saccharomyces cerevisiae. J Biol Chem. 2009;284:4354–4364. doi: 10.1074/jbc.M805891200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee BC, Le DT, Gladyshev VN. Mammals reduce methionine-S-sulfoxide with MsrA and are unable to reduce methionine-R-sulfoxide, and this function can be restored with a yeast reductase. J Biol Chem. 2008;283:28361–28369. doi: 10.1074/jbc.M805059200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim HY, Gladyshev VN. Methionine sulfoxide reduction in mammals: characterization of methionine-R-sulfoxide reductases. Mol Biol Cell. 2004;15:1055–1064. doi: 10.1091/mbc.E03-08-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kryukov GV, Kumar RA, Koc A, Sun Z, Gladyshev VN. Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc Natl Acad Sci USA. 2002;99:4245–4250. doi: 10.1073/pnas.072603099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Z, Johnson LC, Weissbach H, Brot N, Lively MO, Lowther WT. Free methionine-(R)-sulfoxide reductase from Escherichia coli reveals a new GAF domain function. Proc Natl Acad Sci USA. 2007;104:9597–9602. doi: 10.1073/pnas.0703774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho YS, Burden LM, Hurley JH. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 2000;19:5288–5299. doi: 10.1093/emboj/19.20.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stadtman ER, Van Remmen H, Richardson A, Wehr NB, Levine RL. Methionine oxidation and aging. Biochim Biophys Acta. 2005;1703:135–140. doi: 10.1016/j.bbapap.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 40.Cabreiro F, Picot CR, Perichon M, Friguet B, Petropoulos I. Overexpression of methionine sulfoxide reductases A and B2 protects MOLT-4 cells against zinc-induced oxidative stress. Antioxid Redox Signal. 2009;11:215–225. doi: 10.1089/ars.2008.2102. [DOI] [PubMed] [Google Scholar]
- 41.Marchetti MA, Pizarro GO, Sagher D, Deamicis C, Brot N, Hejtmancik JF, Weissbach H, Kantorow M. Methionine sulfoxide reductases B1, B2, and B3 are present in the human lens and confer oxidative stress resistance to lens cells. Invest Ophthalmol Vis Sci. 2005;46:2107–2112. doi: 10.1167/iovs.05-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erickson JR, Joiner ML, Guan X, Kutschke W, Yang J, Oddis CV, Bartlett RK, Lowe JS, O'Donnell SE, Aykin-Burns N, Zimmerman MC, Zimmerman K, Ham AJ, Weiss RM, Spitz DR, Shea MA, Colbran RJ, Mohler PJ, Anderson ME. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell. 2008;133:462–474. doi: 10.1016/j.cell.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su Z, Limberis J, Martin RL, Xu R, Kolbe K, Heinemann SH, Hoshi T, Cox BF, Gintant GA. Functional consequences of methionine oxidation of hERG potassium channels. Biochem Pharmacol. 2007;74:702–711. doi: 10.1016/j.bcp.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ezraty B, Grimaud R, Hassouni MEl, Moinier D, Barras F. Methionine sulfoxide reductases protect Ffh from oxidative damages in Escherichia coli. EMBO J. 2004;23:1868–1877. doi: 10.1038/sj.emboj.7600172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carruthers NJ, Stemmer PM. Methionine oxidation in the calmodulin-binding domain of calcineurin disrupts calmodulin binding and calcineurin activation. Biochemistry. 2008;47:3085–3095. doi: 10.1021/bi702044x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moskovitz J, Bar-Noy S, Williams WM, Requena J, Berlett BS, Stadtman ER. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc Natl Acad Sci USA. 2001;98:12920–12925. doi: 10.1073/pnas.231472998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pal R, Oien DB, Ersen FY, Moskovitz J. Elevated levels of brain-pathologies associated with neurodegenerative diseases in the methionine sulfoxide reductase A knockout mouse. Exp Brain Res. 2007;180:765–774. doi: 10.1007/s00221-007-0903-6. [DOI] [PubMed] [Google Scholar]
- 48.Oien DB, Osterhaus GL, Latif SA, Pinkston JW, Fulks J, Johnson M, Fowler SC, Moskovitz J. MsrA knockout mouse exhibits abnormal behavior and brain dopamine levels. Free Radic Biol Med. 2008;45:193–200. doi: 10.1016/j.freeradbiomed.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vougier S, Mary J, Friguet B. Subcellular localization of methionine sulphoxide reductase A (MsrA): evidence for mitochondrial and cytosolic isoforms in rat liver cells. Biochem J. 2003;373:531–537. doi: 10.1042/BJ20030443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim HY, Gladyshev VN. Role of structural and functional elements of mouse methionine-S-sulfoxide reductase in its subcellular distribution. Biochemistry. 2005;44:8059–8067. doi: 10.1021/bi0501131. [DOI] [PubMed] [Google Scholar]
- 51.Kim HY, Gladyshev VN. Alternative first exon splicing regulates subcellular distribution of methionine sulfoxide reductases. BMC Mol Biol. 2006;7:11. doi: 10.1186/1471-2199-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee JW, Gordiyenko NV, Marchetti M, Tserentsoodol N, Sagher D, Alam S, Weissbach H, Kantorow M, Rodriguez IR. Gene structure, localization and role in oxidative stress of methionine sulfoxide reductase A (MSRA) in the monkey retina. Exp Eye Res. 2006;82:816–827. doi: 10.1016/j.exer.2005.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haenold R, Wassef R, Hansel A, Heinemann SH, Hoshi T. Identification of a new functional splice variant of the enzyme methionine sulphoxide reductase A (MSRA) expressed in rat vascular smooth muscle cells. Free Radic Res. 2007;41:1233–1245. doi: 10.1080/10715760701642096. [DOI] [PubMed] [Google Scholar]
- 54.Haenold R, Wassef R, Brot N, Neugebauer S, Leipold E, Heinemann SH, Hoshi T. Protection of vascular smooth muscle cells by over-expressed methionine sulphoxide reductase A: role of intracellular localization and substrate availability. Free Radic Res. 2008;42:978–988. doi: 10.1080/10715760802566541. [DOI] [PubMed] [Google Scholar]
- 55.Kryukov GV, Kryukov VM, Gladyshev VN. New mammalian selenocysteine-containing proteins identified with an algorithm that searches for selenocysteine insertion sequence elements. J Biol Chem. 1999;274:33888–33897. doi: 10.1074/jbc.274.48.33888. [DOI] [PubMed] [Google Scholar]
- 56.Fomenko DE, Novoselov SV, Natarajan SK, Lee BC, Koc A, Carlson BA, Lee TH, Kim HY, Hatfield DL, Gladyshev VN. MsrB1 (Methionine-R-sulfoxide Reductase 1) Knock-out Mice: Roles of MsrB1 in redox regulation and identification of a novel selenoprotein form. J Biol Chem. 2009;284:5986–5993. doi: 10.1074/jbc.M805770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cabreiro F, Picot CR, Perichon M, Castel J, Friguet B, Petropoulos I. Overexpression of mitochondrial methionine sulfoxide reductase B2 protects leukemia cells from oxidative stress-induced cell death and protein damage. J Biol Chem. 2008;283:16673–16681. doi: 10.1074/jbc.M708580200. [DOI] [PubMed] [Google Scholar]
- 58.Huang W, Escribano J, Sarfarazi M, Coca-Prados M. Identification, expression and chromosome localization of a human gene encoding a novel protein with similarity to the pilB family of transcriptional factors (pilin) and to bacterial peptide methionine sulfoxide reductases. Gene. 1999;233:233–240. doi: 10.1016/s0378-1119(99)00131-6. [DOI] [PubMed] [Google Scholar]
- 59.Kim HY, Gladyshev VN. Characterization of mouse endoplasmic reticulum methionine-R-sulfoxide reductase. Biochem Biophys Res Commun. 2004;320:1277–1283. doi: 10.1016/j.bbrc.2004.06.078. [DOI] [PubMed] [Google Scholar]
- 60.Lowther WT, Brot N, Weissbach H, Honek JF, Matthews BW. Thiol-disulfide exchange is involved in the catalytic mechanism of peptide methionine sulfoxide reductase. Proc Natl Acad Sci USA. 2000;97:6463–6468. doi: 10.1073/pnas.97.12.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim HY, Gladyshev VN. Different catalytic mechanisms in mammalian selenocysteine- and cysteine-containing methionine-R-sulfoxide reductases. PLoS Biol. 2005;3:e375. doi: 10.1371/journal.pbio.0030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee TH, Kim HY. An anaerobic bacterial MsrB model reveals catalytic mechanisms, advantages, and disadvantages provided by selenocysteine and cysteine in reduction of methionine-R-sulfoxide. Arch Biochem Biophys. 2008;478:175–180. doi: 10.1016/j.abb.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 63.Kim HY, Kim JR. Thioredoxin as a reducing agent for mammalian methionine sulfoxide reductases B lacking resolving cysteine. Biochem Biophys Res Commun. 2008;371:490–494. doi: 10.1016/j.bbrc.2008.04.101. [DOI] [PubMed] [Google Scholar]
- 64.Borchert A, Wang CC, Ufer C, Schiebel H, Savaskan NE, Kuhn H. The role of phospholipid hydroperoxide glutathione peroxidase isoforms in murine embryogenesis. J Biol Chem. 2006;281:19655–19664. doi: 10.1074/jbc.M601195200. [DOI] [PubMed] [Google Scholar]
- 65.Hill KE, Zhou J, McMahan WJ, Motley AK, Atkins JF, Gesteland RF, Burk RF. Deletion of selenoprotein P alters distribution of selenium in the mouse. J Biol Chem. 2003;278:13640–13646. doi: 10.1074/jbc.M300755200. [DOI] [PubMed] [Google Scholar]
- 66.Schomburg L, Schweizer U, Holtmann B, Flohe L, Sendtner M, Kohrle J. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem J. 2003;370:397–402. doi: 10.1042/BJ20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schweizer U, Michaelis M, Kohrle J, Schomburg L. Efficient selenium transfer from mother to offspring in selenoprotein-P-deficient mice enables dose-dependent rescue of phenotypes associated with selenium deficiency. Biochem J. 2004;378:21–26. doi: 10.1042/BJ20031795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ferreiro A, Quijano-Roy S, Pichereau C, Moghadaszadeh B, Goemans N, Bonnemann C, Jungbluth H, Straub V, Villanova M, Leroy JP, Romero NB, Martin JJ, Muntoni F, Voit T, Estournet B, Richard P, Fardeau M, Guicheney P. Mutations of the selenoprotein N gene, which is implicated in rigid spine muscular dystrophy, cause the classical phenotype of multiminicore disease: reassessing the nosology of early-onset myopathies. Am J Hum Genet. 2002;71:739–749. doi: 10.1086/342719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 70.Beilstein MA, Vendeland SC, Barofsky E, Jensen ON, Whanger PD. Selenoprotein W of rat muscle binds glutathione and an unknown small molecular weight moiety. J Inorg Biochem. 1996;61:117–124. doi: 10.1016/0162-0134(95)00045-3. [DOI] [PubMed] [Google Scholar]
- 71.Jeong D, Kim TS, Chung YW, Lee BJ, Kim IY. Selenoprotein W is a glutathione-dependent antioxidant in vivo. FEBS Lett. 2002;517:225–228. doi: 10.1016/s0014-5793(02)02628-5. [DOI] [PubMed] [Google Scholar]
- 72.Lu C, Qiu F, Zhou H, Peng Y, Hao W, Xu J, Yuan J, Wang S, Qiang B, Xu C, Peng X. Identification and characterization of selenoprotein K: an antioxidant in cardiomyocytes. FEBS Lett. 2006;580:5189–5197. doi: 10.1016/j.febslet.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 73.Kuiper GG, Klootwijk W, Visser TJ. Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase reduces catalytic efficiency and alters substrate preference. Endocrinology. 2003;144:2505–2513. doi: 10.1210/en.2003-0084. [DOI] [PubMed] [Google Scholar]
- 74.Kim HY, Gladyshev VN. Methionine sulfoxide reductases: selenoprotein forms and roles in antioxidant protein repair in mammals. Biochem J. 2007;407:321–329. doi: 10.1042/BJ20070929. [DOI] [PubMed] [Google Scholar]
- 75.Stadtman TC. Selenocysteine. Annu Rev Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- 76.Berry MJ, Banu L, Larsen PR. Type I iodothyronine deiodinase is a selenocysteine-containing enzyme. Nature. 1991;349:438–440. doi: 10.1038/349438a0. [DOI] [PubMed] [Google Scholar]
- 77.Garcia GE, Stadtman TC. Clostridium sticklandii glycine reductase selenoprotein A gene: cloning, sequencing, and expression in Escherichia coli. J Bacteriol. 1992;174:7080–7089. doi: 10.1128/jb.174.22.7080-7089.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Friguet B, Bulteau AL, Petropoulos I. Mitochondrial protein quality control: implications in ageing. Biotechnol J. 2008;3:757–764. doi: 10.1002/biot.200800041. [DOI] [PubMed] [Google Scholar]
- 79.Murphy MP, Partridge L. Toward a control theory analysis of aging. Annu Rev Biochem. 2008;77:777–798. doi: 10.1146/annurev.biochem.77.070606.101605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chiang AN, Wu HL, Yeh HI, Chu CS, Lin HC, Lee WC. Antioxidant effects of black rice extract through the induction of superoxide dismutase and catalase activities. Lipids. 2006;41:797–803. doi: 10.1007/s11745-006-5033-6. [DOI] [PubMed] [Google Scholar]
- 81.Mollnau H, Wenzel P, Oelze M, Treiber N, Pautz A, Schulz E, Schuhmacher S, Reifenberg K, Stalleicken D, Scharffetter-Kochanek K, Kleinert H, Munzel T, Daiber A. Mitochondrial oxidative stress and nitrate tolerance--comparison of nitroglycerin and pentaerithrityl tetranitrate in Mn-SOD+/- mice. BMC Cardiovasc Disord. 2006;6:44. doi: 10.1186/1471-2261-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Orr WC, Sohal RS. Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science. 1994;263:1128–1130. doi: 10.1126/science.8108730. [DOI] [PubMed] [Google Scholar]
- 83.Radyuk SN, Michalak K, Klichko VI, Benes J, Rebrin I, Sohal RS, Orr WC. Peroxiredoxin 5 confers protection against oxidative stress and apoptosis and also promotes longevity in Drosophila. Biochem J. 2009 doi: 10.1042/BJ20082003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ruan H, Tang XD, Chen ML, Joiner ML, Sun G, Brot N, Weissbach H, Heinemann SH, Iverson L, Wu CF, Hoshi T. High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc Natl Acad Sci USA. 2002;99:2748–2753. doi: 10.1073/pnas.032671199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koc A, Gladyshev VN. Methionine sulfoxide reduction and the aging process. Ann NY Acad Sci. 2007;1100:383–386. doi: 10.1196/annals.1395.042. [DOI] [PubMed] [Google Scholar]
- 86.Koc A, Gasch AP, Rutherford JC, Kim HY, Gladyshev VN. Methionine sulfoxide reductase regulation of yeast lifespan reveals reactive oxygen species-dependent and -independent components of aging. Proc Natl Acad Sci USA. 2004;101:7999–8004. doi: 10.1073/pnas.0307929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zorov DB, Juhaszova M, Sollott SJ. Mitochondrial ROS-induced ROS release: an update and review. Biochim Biophys Acta. 2006;1757:509–517. doi: 10.1016/j.bbabio.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 88.Han D, Williams E, Cadenas E. Mitochondrial respiratory chain-dependent generation of superoxide anion and its release into the intermembrane space. Biochem J. 2001;353:411–416. doi: 10.1042/0264-6021:3530411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Starkov AA. The role of mitochondria in reactive oxygen species metabolism and signaling. Ann NY Acad Sci. 2008;1147:37–52. doi: 10.1196/annals.1427.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moller IM, Kristensen BK. Protein oxidation in plant mitochondria as a stress indicator. Photochem Photobiol Sci. 2004;3:730–735. doi: 10.1039/b315561g. [DOI] [PubMed] [Google Scholar]
- 91.Prudova A, Bauman Z, Braun A, Vitvitsky V, Lu SC, Banerjee R. S-adenosylmethionine stabilizes cystathionine beta-synthase and modulates redox capacity. Proc Natl Acad Sci USA. 2006;103:6489–6494. doi: 10.1073/pnas.0509531103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brosnan JT, Brosnan ME. The sulfur-containing amino acids: an overview. J Nutr. 2006;136:1636S–1640S. doi: 10.1093/jn/136.6.1636S. [DOI] [PubMed] [Google Scholar]
- 93.Waterland RA. Assessing the effects of high methionine intake on DNA methylation. J Nutr. 2006;136:1706S–1710S. doi: 10.1093/jn/136.6.1706S. [DOI] [PubMed] [Google Scholar]
- 94.Perla-Kajan J, Twardowski T, Jakubowski H. Mechanisms of homocysteine toxicity in humans. Amino Acids. 2007;32:561–572. doi: 10.1007/s00726-006-0432-9. [DOI] [PubMed] [Google Scholar]