SUMMARY
MitoPLD is a member of the phospholipase D superfamily proteins conserved among diverse species. Zucchini, the Drosophila homolog of MitoPLD, has been implicated in primary biogenesis of Piwi-interacting RNAs (piRNAs). By contrast, MitoPLD has been shown to hydrolyze cardiolipin in the outer membrane of mitochondria to generate phosphatidic acid, which is a signaling molecule. To assess whether the mammalian MitoPLD is involved in piRNA biogenesis, we generated MitoPLD mutant mice. The mice display meiotic arrest during spermatogenesis, demethylation and derepression of retrotransposons, and defects in primary piRNA biogenesis. Furthermore, in mutant germ cells, mitochondria and the components of the nuage, a perinuclear structure involved in piRNA biogenesis/function, are mislocalized to regions around the centrosome, suggesting that MitoPLD may be involved in microtubule-dependent localization of mitochondria and these proteins. Our results indicate a conserved role for MitoPLD/Zuc in the piRNA pathway and link mitochondrial membrane metabolism/signaling to small RNA biogenesis.
INTRODUCTION
piRNAs are small RNAs of mostly 24-30 nucleotides (nt) in length that are expressed in germ cells of animals and bound to the PIWI proteins, which represent a subfamily of the Argonaute protein family. The PIWI-piRNA complex recognizes target RNAs dependent on sequence complementarity and cleave the targets through the slicer activity possessed by the PIWI domain. With this activity, the PIWI-piRNA complex represses retrotransposons in germ cells, which preserve the integrity of the genome (Aravin et al., 2007a; Kim et al., 2009). Requirement of the piRNA pathway in repression of retrotransposons has been shown by genetic and molecular studies in mice, zebrafish and flies (Aravin et al., 2007b; Brennecke et al., 2007; Carmell et al., 2007; Houwing et al., 2007; Kuramochi-Miyagawa et al., 2008; Saito et al., 2006; Vagin et al., 2006; Watanabe et al., 2008).
Two pathways for the biogenesis of piRNAs have been discovered—the primary and secondary pathways (Aravin et al., 2007a; Kim et al., 2009). The primary pathway is thought to generate piRNAs (primary piRNAs) from various portions of longer single-stranded piRNA precursors transcribed from their coding genomic regions called piRNA clusters (Aravin et al., 2006; Girard et al., 2006; Grivna et al., 2006; Lau et al., 2006; Watanabe et al., 2006). Retrotransposon sequences are often found within the clusters and serve as sources of piRNAs. However, features that distinguish piRNA precursors from other RNAs remain elusive, and enzymes that are required for the production of primary piRNAs have not been definitively identified. In the secondary pathway, piRNAs (secondary piRNAs) are generated from the 5’ portions of RNA fragments cleaved by existing PIWI-piRNA complexes. The primary and secondary piRNAs guide each other's production in the secondary pathway, many repetitions of which (so-called ping-pong cycle) lead to the accelerated production of these piRNAs (Brennecke et al., 2007; Gunawardane et al., 2007). This secondary pathway is thought to be an adaptive system for genome defense because only the piRNAs from expressed retrotransposons are amplified (Aravin et al., 2007a).
There are three PIWI proteins in mice, named MILI, MIWI and MIWI2, all of which are specifically expressed in germ cells. MILI and MIWI2 begin their expression in the fetal testis, whereas MIWI begin its expression in pachytene spermatocytes in the postnatal testis (Aravin et al., 2008; Deng and Lin, 2002; Kojima et al., 2009; Kuramochi-Miyagawa et al., 2001; Kuramochi-Miyagawa et al., 2008; Reuter et al., 2009; Shoji et al., 2009; Vagin et al., 2009; Wang et al., 2009). Disruptions of the Mili and Miwi2 genes result in meiotic arrest at the zygotene phase due to a burst of expression of L1 retrotransposons (Carmell et al., 2007; Kuramochi-Miyagawa et al., 2004; Kuramochi-Miyagawa et al., 2008). In addition, the Mili mutant displays a reduced mitotic rate of spermatogonia (Unhavaithaya et al., 2009). By contrast, Miwi null mice show spermatogenesis arrest at the early round spermatid stage (Deng and Lin, 2002). All three mouse PIWI proteins are localized to perinuclear electron-dense structures called the nuage, which is presumed to have a role in RNA metabolism and storage (Aravin et al., 2008; Aravin et al., 2009; Chuma et al., 2009). The nuage is also called intermitochondrial cement, pi-body or piP-body, depending on their localization, morphology and/or biochemical properties.
In addition to the PIWI proteins, Zucchini (Zuc) has also been implicated in piRNA biogenesis. The zuc gene was first identified in a screen for female-sterile in Drosophila (Schupbach and Wieschaus, 1991). The zuc mutants show defects in midoogenesis and share phenotypic features with mutants of aubergine that encodes a Piwi family protein. The mutations in zuc cause defects in piRNA biogenesis and derepression of retrotransposons (Pane et al., 2007). Among the Drosophila mutants that show defects in piRNA production, the zuc mutants are unique in that they show defects not only in germ cells but also in somatic cells of the ovary (Malone et al., 2009; Saito et al., 2009). Since only the primary pathway is active in somatic cells (Li et al., 2009; Malone et al., 2009), Zuc is thought to be involved in this pathway. Zuc belongs to the phospholipase D superfamily and is highly conserved among animal species. Because bacterial Nuc protein, another member of the phospholipase D superfamily, has an endonuclease activity (Pohlman et al., 1993; Zhao et al., 1997), Zuc is thought to be a candidate enzyme that is involved in biogenesis/maturation of piRNAs (Haase et al., 2010; Olivieri et al., 2010; Pane et al., 2007; Saito et al., 2009; Saito et al., 2010).
The mammalian homolog of Zuc is known as MitoPLD, which has an activity to hydrolyze cardiolipin, a mitochondrion-enriched lipid molecule. The hydrolytic reaction generates phosphatidic acid (PA) that can act as a signaling molecule (Choi et al., 2006; Huang and Frohman, 2009; Stace and Ktistakis, 2006), and overexpression of MitoPLD in cultured cells results in mitochondrial fusion (Choi et al., 2006). However, it is unclear if the function of mammalian MitoPLD in mitochondria is related to the function of Drosophila Zuc in the piRNA pathway. To elucidate whether mammalian MitoPLD is involved in the piRNA pathway, we have generated MitoPLD mutant mice. Our results demonstrate that MitoPLD has a conserved role in piRNA biogenesis.
RESULTS
Mouse MitoPLD Is Expressed in the Male and Female Germline
The Drosophila Zuc protein has a phospholipase D domain and is characterized by a single H(X)K(X4)D catalytic motif. Its orthologs are found in vertebrates including zebrafish, frogs, mice, rats and humans. MitoPLD (also known as PLD6) is the only ortholog of Drosophila Zuc in humans, which has been previously shown to have a role in mitochondrial fusion in HeLa cells (Choi et al., 2006). To determine which tissues express the MitoPLD gene, we first examined publicly available expressed sequence tag (EST) data. In zebrafish, frogs and mice, MitoPLD ESTs were predominantly found in reproductive tissues such as the testis and ovary. In humans, MitoPLD ESTs were not restricted to the testis and ovary, and were broadly distributed in many tissues (Figure S1A).
To confirm the tissue distribution deduced from the EST data, RT-PCR was performed using RNAs isolated from several mouse tissues and oocytes (Figure S1B). Consistent with the EST data, MitoPLD was predominantly expressed in the testis and growing oocytes. The expression in the testis was analyzed embryonic day 16.5 (E16.5) to the adult stage and MitoPLD mRNA was detected throughout this period. Mice have a single MitoPLD locus on chromosome 11, which consists of two exons that span 3.5 kb (Figure S1C). The predicted mRNA size is ~1,750 nt excluding the polyA tail, and Northern blotting indeed detected a band of 1,800-2,000 nt in the testis (Figure S1D). Regarding the protein products, a 147 amino acid (a.a.) protein is predicted in the UCSC and NCBI databases, whereas a 221 a.a. protein is predicted in the Ensemble database (Figure S1C). The size difference arises from a splicing junction variation: in the UCSC and NCBI databases, a 4-nt insertion is present at a splicing junction, which causes a shift in the reading frame (Figure S1E). Indeed, we identified both types of transcripts in the mouse testicular ESTs and confirmed their presence by sequencing the RT-PCR products from the testis (five of which was the Ensemble type and three of which was the UCSC/NCBI type). However, the shorter protein is predicted to lack the H(X)K(X4)D motif essential for the enzymatic function (Choi et al., 2006) and is not found in the human EST database. Thus, we inferred that only the longer protein is active and evolutionarily conserved. We detected a band of an expected molecular weight for the longer protein in the mouse testis from E16.5 to the adult stage by Western blotting (Figure S1F).
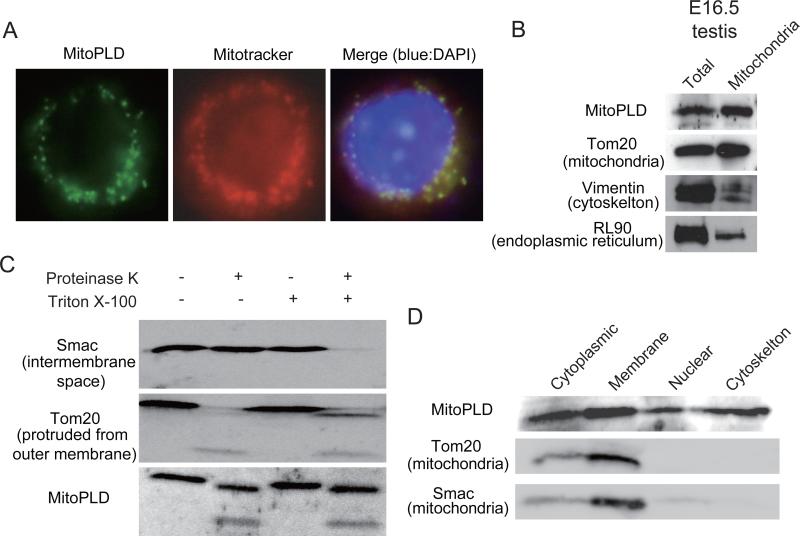
Human MitoPLD was previously shown to be localized to mitochondrial outer membrane when ectopically expressed in cultured cells (Choi et al., 2006). Its N-terminal region has a putative mitochondrial localization signal that encompasses a transmembrane helix domain. This feature is also found in the N-terminal regions of mouse and rat MitoPLDs (data not shown). A mitochondrial localization signal is also found in the orthologs from frogs and flies. Consistent with the presence of the mitochondrial localization signal, an ectopically expressed mouse MitoPLD was localized to mitochondria in NIH3T3 cells (Figure 1A). We collected mitochondria from fetal and adult mouse testicular cells by centrifugation and confirmed the presence of MitoPLD by Western blotting (Figure 1B and data not shown). To characterize the localization of MitoPLD further, the testicular mitochondria were treated by a protease. Irrespective of presence or absence of the detergent that disrupts membrane structure, digestion of MitoPLD occurred at a similar rate (Figure 1C), suggesting that this protein is present on the outer surface of mitochondria. However, a subcellular fractionation of the testicular cells showed that MitoPLD is also found in other fractions (Figure 1D).
Figure 1. MitoPLD is localized to the mitochondrial outer membrane.
A. Mouse MitoPLD ectopically expressed in NIH3T3 cells is localized to mitochondria. Immunostaining was performed with an antibody raised against a C-terminal peptide (left). Mitotracker labels mitochondria (center). A merged view with DAPI staining (right).
B. MitoPLD is found in mitochondria of testicular cells. Mitochondria were purified by centrifugation from E16.5 testes. A total testis lysate and a mitochondrial lysate were subjected to Western blotting with antibodies against a middle portion of MitoPLD, Tom20, Vimentin and RL90 are markers for mitochondria, cytoskeleton and endoplasmic reticulum, respectively.
C. MitoPLD is localized on the outer membrane of mitochondria. Mitochondria were treated with proteinase K in the presence or absence of Triton X-100. Triton X-100 disrupts the membrane structure of mitochondria. Smac is localized in the inter-membrane space and thus is digested only when Triton X-100 is present. Tom20 is protruded from the outer membrane and therefore digested by proteinase K regardless of the presence/absence of Triton-X-100.
D. MitoPLD is also found in other subcellular fractions. Testicular cells were subjected to subcellular fractionation, and Western blotting was performed with antibodies against MitoPLD, Tom20 and Smac.
MitoPLD Mutants Show Meiotic Defects during Spermatogenesis
To know the role of the mammalian MitoPLD in the piRNA pathway, we generated MitoPLD mutant mice. The first exon, which encodes the mitochondrial localization signal encompassing the transmembrane domain and the N-terminal part of the conserved PLD domain, was replaced with a Pgk-Neo cassette flanked by loxP sites (Figure S2A). Correct targeting was confirmed by Southern blotting (Figure S2B). After chimera formation and successful germline transmission, the Pgk-Neo cassette was removed by crossing with CAG-Cre mice. The resulting MitoPLD+/- males and females were crossed, and viable MitoPLD+/+, +/- and -/- mice were obtained at a ratio expected for Mendelian inheritance. Western blotting confirmed the loss of the protein in the MitoPLD-/- testes (Figure S2C).
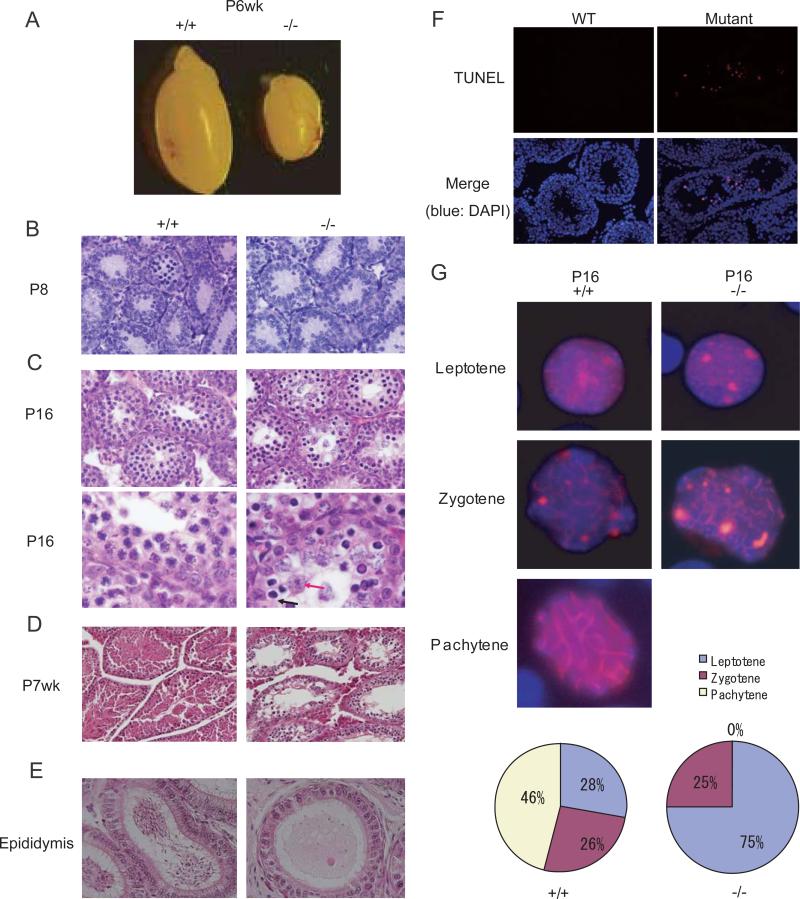
The MitoPLD-/- females showed no discernible phenotype and were fertile. However, none of the MitoPLD-/- males produced progeny, despite the presence of vaginal plugs in the crossed females. Notably, the testes from the MitoPLD-/- males were smaller than those of the MitoPLD+/- and wildtype males (Figure 2A). In normal development, fetal testes at E13-E19.5 contain germ cells termed prospermatogonia. At postnatal day 2-3 (P2-P3), the prospermatogonia start to develop to undifferentiated spermatogonia, which either self-renew or further develop to produce differentiated spermatogonia. In the first wave of germ cell differentiation after birth, the spermatogonia enter meiosis at about P8. No discernible abnormality was found in the MitoPLD-/- testes at P8 (Figure 2B). When differentiation reaches the mid-pachytene spermatocyte stage at P16, atypical spermatocytes started to appear (Figure 2C). Two types of abnormal spermatocytes were noted: one with a condensed nucleus (Figure 2C: black arrow) and the other with a swollen nucleus (Figure 2C: red arrow). Similar defects were previously observed in Miwi2 and Mael mutants (Carmell et al., 2007; Soper et al., 2008). Spermatogenesis is spatially regulated in the seminiferous tubules and spermatogonia are normally found at the basal layer of the seminiferous epithelium (Bellve et al., 1977; de Rooij and Grootegoed, 1998). As differentiation proceeds, germ cells move towards the luminal side. In wildtype testes at postnatal week 7 (P7wk), both round and elongated spermatids are present in the luminal side (Figure 2D). By contrast, no spermatids were observed in the MitoPLD-/- testes, and no spermatozoon in the epididymis (Figure 2D and 2E). A TUNEL assay revealed apoptosis of spermatocytes in MitoPLD-/- tubules (Figure 2F).
Figure 2. Meiotic defects in MitoPLD mutant testes.
A. Testes from MitoPLD-/- mice were significantly smaller than those from +/+ control mice at P6wk.
B. Sections from MitoPLD-/- testes show no discernable defect at P8.
C. MitoPLD-/- testes show spermatocytes with abnormal morphology at P16. In higher magnification views (bottom), MitoPLD-/- testes have spermatocytes with a condensed nucleus (blue arrow) and those with a swollen nucleus (red arrow).
D. MitoPLD-/- testes lack round and elongated spermatids at P7wk.
E. No spermatozoa are observed in MitoPLD-/- epididymides at P8wk.
F. TUNEL assays identify apoptotic cells in MitoPLD-/- testes at P6wk.
G. Spermatocytes are arrested at the zygotene stage in MitoPLD-/- testes. Nuclei of spermatocytes from P16 testes were immunostained with an SYCP3 antibody (red) and counterstained with DAPI (blue). Percentage of spermatocytes at each stage of the meiotic prophase is shown at the bottom.
To define the stage of meiotic arrest more precisely, we stained testicular cells from P16 testes using an antibody against synaptonemal complex protein 3 (SYCP3), which is a component of the axial/lateral elements of the synaptonemal complex (Dobson et al., 1994). We observed some MitoPLD-/- spermatocytes of the zygotene stage, as judged by the formation of elongated axial/lateral elements (Figure 2G). However, we never observed pachytene spermatocytes in MitoPLD-/- cell preparations, as judged by the lack of cells with SYCP3 staining along the length of the synapsed chromosomes (Figure 2G).
Retrotransposon Burst in Mutant Testes
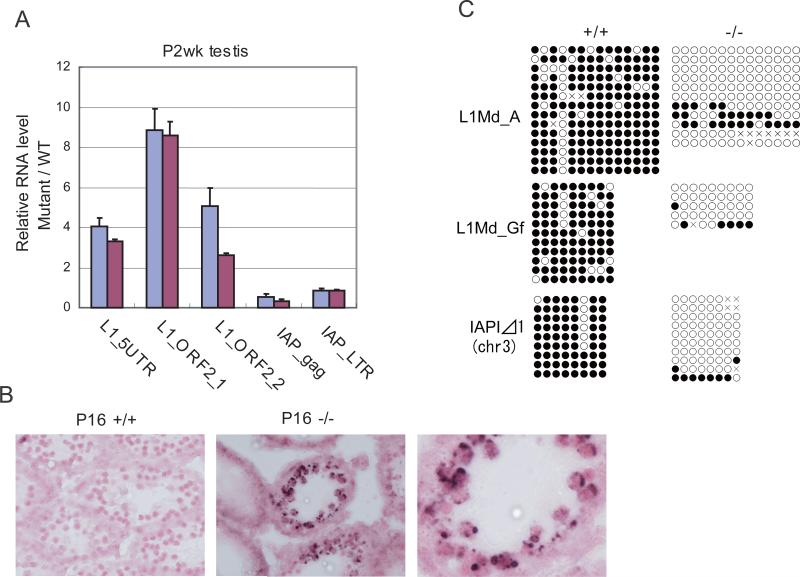
The spermatogenesis arrest phenotype observed in MitoPLD-/- mice was essentially the same as that observed in Mili, Miwi2, Tdrd9, Gasz, Mael and Mov10l1 mutant mice (Carmell et al., 2007; Frost et al., 2010; Kuramochi-Miyagawa et al., 2008; Ma et al., 2009; Shoji et al., 2009; Soper et al., 2008; Zheng et al., 2010). The phenotype of these mutants was previously attributed to an inappropriate activation of L1 retrotransposons in spermatocytes. We examined the levels of L1 and IAP retrotransposon transcripts in MitoPLD-/- testes at P14 and observed a 3- to 9-fold increase in the L1 RNA expression (Figure 3A). However, the IAP RNA level was modestly decreased. In situ hybridization using a probe that detects the L1 RNA showed an accumulation of signals in cytoplasmic regions around the nucleus of MitoPLD-/- spermatocytes (Figure 3B). The region most likely correspond to the cytoplasmic territories previously observed in Mael and Tdrd9 mutants, which are surrounded by a membrane and filled with L1 ribonucleoproteins (Shoji et al., 2009; Soper et al., 2008).
Figure 3. Retrotransposons are derepressed in MitoPLD mutant testes.
A. Quantitative PCR analysis of L1Md and IAP retrotransposon RNAs in P2wk testes. Testes from two MitoPLD-/- mice were analyzed (red and blue bars). The level of Gapdh mRNA was used to normalize the data from mutants and wildtype mice. The relative RNA levels are shown. Error bars represent S. D. (n = 3).
B. In situ hybridization analysis of P16 testes with a probe that detects L1Md RNA. Lump-like signals of L1Md RNA are observed around nuclei of spermatocytes only in MitoPLD-/- testes. Nuclei were stained with Nuclear Fast Red.
C. DNA methylation analysis of retrotransposon sequences in P5-7 spermatogonia by bisulfite sequencing.
Retrotransposons are normally stably repressed by CpG methylation. The CpG methylation is once erased in primordial germ cells and re-established during gametogenesis. In the male germline, de novo CpG methylation occur between E12.5 and E18.5, when MILI and MIWI2 are expressed (Girard and Hannon, 2008; Reik, 2007; Sasaki and Matsui, 2008; Schaefer et al., 2007). We isolated neonatal spermatogonia from MitoPLD-/- testes and analyzed the DNA methylation status of L1 sequences by bisulfite sequencing. L1 retrotransposons of A-type and F-type, the latter of which is further subdivided into Tf-type and Gf-type, are known to be active in mice (Goodier et al., 2001). We examined the methylation status of the 5’ repeat of A-type and Gf-type L1s, which could be easily distinguished by the sequence difference. In MitoPLD-/- spermatogonia, both A-type and Gf-type L1s were hypomethylated, suggesting that the observed derepression of L1 retrotransposons is, at least partly, a consequence of increased transcription (Figure 3C). We could not find a difference in DNA methylation in the bulk of IAP1 (a subtype of IAP) sequences between wildtype and MitoPLD-/- spermatogonia (data not shown). However, using a primer pair that detects a specific IAP1 copy on chromosome 3, we found that the copy was hypomethylated in MitoPLD-/- spermatogonia (Figure 3C). The same copy was previously shown to be hypomethylated in Mili mutants (Kuramochi-Miyagawa et al., 2008).
piRNA Biogenesis Is Affected in Mutant Testes
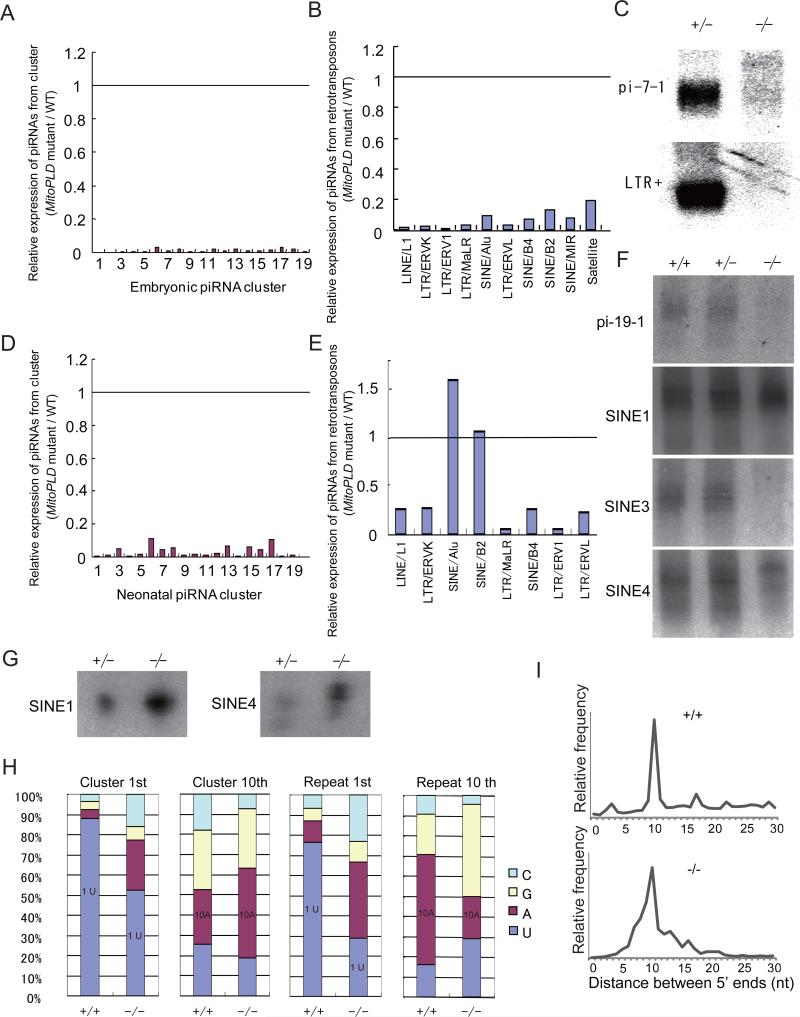
To evaluate the impact of the MitoPLD mutation on piRNA biogenesis, we deep-sequenced 20-33 nt total small RNAs obtained from E16.5 MitoPLD-/- testes. As the levels of some miRNAs examined by Northern blotting were not changed between wildtype and MitoPLD-/- testes (Figure S3A), we used the total miRNA level for normalization (Table S1). It was previously shown that most piRNAs are mapped to retrotransposon sequences and the remaining ones to defined unique sequence regions on the genome (piRNA clusters) (Aravin et al., 2007a; Girard and Hannon, 2008; Kim et al., 2009). In E16.5 MitoPLD-/- testes, small RNAs mapped to the known piRNA clusters were almost absent (200-fold decrease compared with wildtype) (Figure 4A and Table S1), and small RNAs from retrotransposons were decreased by 40-fold (Table S1 and Figure 4B). Furthermore, small RNAs from retrotransposons lost the features of piRNAs in MitoPLD-/- testes. Namely, they showed a broad size distribution extending beyond 25-28 nt (Figure S3B) and lost the enrichment for uridine at the first position or for adenine at the 10th position (Figure S3C). Consistent with these results, we did not detect piRNAs from the clusters or retrotransposons by Northern blotting in MitoPLD-/- testes (Figure 4C). These results suggest that piRNA biogenesis is severely affected in MitoPLD-/- testes at the fetal stage.
Figure 4. Defects in primary piRNA biogenesis in MitoPLD mutant testes.
A. Expression of unique sequence piRNAs in MitoPLD-/- testes relative to MitoPLD+/+ testes at E16.5. The data for the piRNAs from the top 19 piRNA clusters producing the largest numbers (amounts) of piRNA in wildtype testes are shown. The positions of these clusters are shown in Supplemental Information. The piRNA counts were normalized by the miRNA counts.
B. Expression of piRNAs from retrotransposons in MitoPLD-/- testes relative to MitoPLD+/+ testes at E16.5. The data for the piRNAs from the retrotransposons of top 10 piRNA sources are shown. The retrotransposons are ordered from left to right according to the number of piRNAs they produce in wildtype. The total miRNA counts were used for normalization.
C. Northern blotting analysis of piRNAs in MitoPLD+/- and -/- testes at E16.5. pi-7-1 is a piRNA generated from the largest embryonic piRNA cluster on chr7. LTR+ piRNA has a sequence of IAP1 retrotransposon.
D. Expression of unique sequence piRNAs in MitoPLD-/- testes relative to MitoPLD+/+ testes at P10. See (A).
E. Expression of piRNAs from retrotransposons in MitoPLD-/- testes relative to MitoPLD+/+ testes at P10. The data for the top 8 sources of piRNAs in wildtype are shown. Retrotransposons are ordered from left to right according to the number of piRNAs that they produce. See (B).
F. Northern blotting analysis of piRNAs in MitoPLD+/+, +/- and -/- testes at P10. pi-19-1 is a unique piRNA from a piRNA cluster. pi-SINE1, 3 and 4 have sequences of SINE B1.
G. The piRNAs from SINE B1 are bound to MILI. The MILI complex was immunoprecipitated from P10 testes and the bound RNAs were probed with oligonulceotides for pi-SINE1 and pi-SINE4 in (F). RNA from five pairs of testes was loaded in each lane.
H. piRNAs from MitoPLD-/- testes tend to lose the characteristics of the primary piRNA (1st U) but gain that of the secondary piRNA (complementarity between 1st base and 10th base). The nucleotide compositions of the 1st and 10th nucleotide are shown for piRNAs from clusters and repeat from P10 testes.
I. The distance between the 5’ ends of complementary piRNAs. piRNAs from P10 testes that matched the consensus sequence of IAP1 retrotransposon with up to 3 mismatches were used. A peak at 10 nt in MitoPLD-/- testes suggests that the ping-pong cycle (the secondary pathway) is still active.
To determine whether piRNA biogenesis is also affected in postnatal stages, we sequenced small RNAs isolated from P10 MitoPLD-/- testes and compared the results with the previous data from wildtype testes at the same stage (Aravin et al., 2008). The effect of the MitoPLD mutation was milder at P10 than at E16.5. Whereas postnatal prepachytene piRNAs from the clusters showed a 30-fold decrease in expression, those from retrotransposons showed an only 2.1-fold decrease (Table S1 and Figure 4D and 4E). When small RNAs from individual retrotransposon species were examined, the effect of the mutation was considerably different among the retrotransposons (Figure 4E). Whereas small RNAs from L1s were decreased 4-fold, small RNAs from SINE/B1 elements were increased 1.6-fold. Consistent with the deep-sequencing results, we detected small RNAs from SINE/B1s in MitoPLD-/- by Northern blotting (Figure 4F). Interestingly, there was a shift in prepachytene piRNA size from 27 nt to 28 nt (Figure S3D). Of the three mouse PIWI family proteins, MILI is the only one expressed at P10 (Kuramochi-Miyagawa et al., 2001; Kuramochi-Miyagawa et al., 2008). We immunoprecipitated MILI-piRNA complexes from P10 MitoPLD-/- testes and bound RNAs were analyzed by Northern blotting. The study detected small RNAs from SINE/B1s (Figure 4G), indicating that they are indeed piRNAs. Together, these results suggest that the piRNA profile is dramatically altered in MitoPLD-/- testes.
Secondary piRNA Biogenesis Is Active in Mutant Testes
In the primary pathway, piRNAs are generated from precursor RNAs transcribed from the piRNA clusters (Aravin et al., 2006; Girard et al., 2006; Grivna et al., 2006; Lau et al., 2006; Watanabe et al., 2006). In the secondary pathway, the ping-pong cycle, which assures adaptive retrotransposon silencing, generates secondary piRNAs from the end of RNAs cleaved by the PIWI-piRNA complexes (Brennecke et al., 2007; Gunawardane et al., 2007). Consequently, piRNAs from the clusters are mostly primary piRNAs whereas those from retrotransposons are more enriched in secondary piRNAs (Figure 4H). The more profound defects observed in piRNAs from the clusters than those from retrotransposons in postnatal MitoPLD-/- testes suggest that it is primarily the primary biogenesis that was affected. Consistent with this idea, the content of uridine at the first position, a feature of primary piRNAs, was diminished in piRNAs from both the clusters and retrotransposons in P10 MitoPLD-/- testes (Figure 4H). Because PIWI-mediated cleavage of the target RNA occurs at a position between the 10th and 11th nucleotide of the guide piRNA, a resulting piRNA pair has a 10-nt overlap between the 5’ ends. We measured the distance between the 5’ ends of complementary piRNAs from IAP1 retrotransposons and observed a peak at 10 nt (Figure 4I), suggesting that the secondary pathway is active. The 10th nucleotide of secondary piRNAs is enriched for adenine. Despite the decreased uridine content at the first position, piRNAs from the clusters showed an increased content of adenine at the 10th position in MitoPLD-/- testes (Figure 4H). This suggests that, although the overall piRNA production is low, some piRNAs from the clusters are produced through the secondary pathway in MitoPLD-/- testes. In piRNAs from retrotransposons, we observed increased contents of adenine and cytosine at the first position and uridine and guanine at the 10th position in MitoPLD-/- testes (Figure 4H), further suggesting the active operation of the secondary pathway.
Mislocalization of the piRNA Pathway Components around the Centrosome
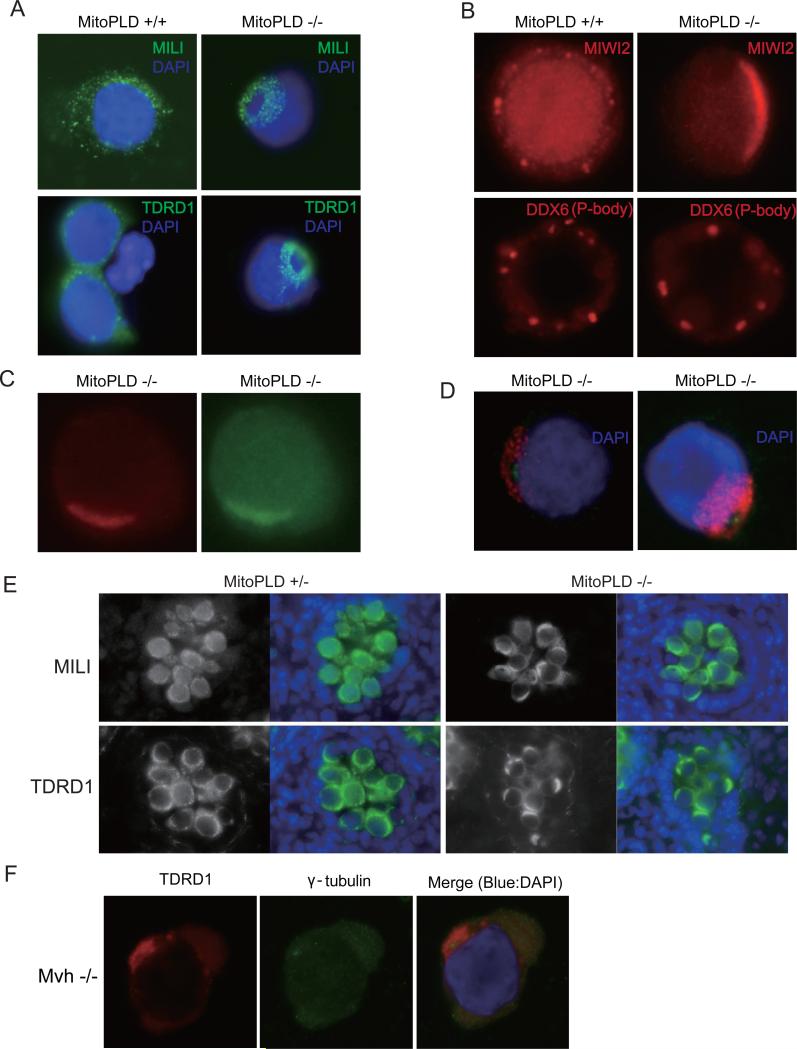
The components of the piRNA pathways are localized to two distinct granules called the pi-body and piP-body (collectively represent the nuage) in the fetal testis (Aravin et al., 2007a; Aravin et al., 2009). The pi-body, which is also called the inter-mitochondrial cement, contains MILI and TDRD1, while the piP-body, which is also called the MIWI2 body, contains MIWI2, TDRD9 and MAEL. In the piP-bodies, the components of the P-body, which is involved in RNA degradation/translational control including the miRNA- and siRNA-mediated pathways (Parker and Sheth, 2007), are also localized. We analyzed the localization of the piRNA pathway components in E16.5 MitoPLD-/- prospermatogonia by immunostaining. The pi-body proteins MILI and TDRD1 were mislocalized and showed a donut-shaped staining pattern in a region adjacent to the nucleus in individual cells (Figure 5A). We also observed mislocalization of the piP-body protein MIWI2 to the region where MILI was localized (Figure 5B and 5C). However, we did not observe mislocalization of DDX6 (Figure 5B), a P-body protein normally found in the piP-bodies (Aravin et al., 2009). Remarkably, at the center of the donut-shaped staining for MILI and TDRD1, a signal for gamma-tubulin, a marker for the centrosome (Jeng and Stearns, 1999), was observed (Figure 5D). Thus, the localization of multiple components of the p-bodies and piP-bodies, which may be dependent on the function or organization of the microtubules, is misregulated in MitoPLD-/- prospermatogonia.
Figure 5. MitoPLD is required for nuage localization.
A. Mislocalization of the pi-body components in MitoPLD-/- testes. Prospermatogonia from E16.5 testes were immunostained with MILI or TDRD1 antibodies in the presence of 0.1 % Triton X-100.
B. Mislocalization of the piP-body components in MitoPLD-/- testes. Prospermatogonia from E16.5 testes were immunostained with MIWI2 or DDX6 antibodies in the absence of the detergent.
C. Colocalization of MILI and MIWI2 in MitoPLD-/- testes. Prospermatogonia from E16.5 testes were double stained in the absence of detergent. The difference in MILI staining pattern between (A) and (C) (doughnut-shaped staining and crescent-like staining) results from the presence or absence of Triton X-100.
D. MILI and TDRD1 are mislocalized around the centrosome in MitoPLD-/- testes. Prospermatogonia from E16.5 testes were double-stained with gamma-tubulin, a marker for the centrosome, and MILI (left) or TDRD1 (right) anitibodies.
E. Mislocalization of nuage components in MitoPLD and Mvh mutants. Immunostaining of MILI and TDRD1 show crescent-like staining in MitoPLD-/- testes at E16.5.
F. Immunostaining patterns of TDRD1 and gamma-tubulin are essentially the same in Mvh mutant and MitoPLD mutant prospermatogonia at E16.5.
In cryosections of E16.5 MitoPLD-/- testes, immunostaining for MILI and TDRD1 showed crescent-shaped localization patterns around the prospermatogonial nuclei (Figure 5E). This pattern is very similar to the one previously observed in prospermatogonia and spermatogonia of Mvh mutants (Chuma et al., 2006; Kuramochi-Miyagawa et al., 2010). We then examined the Mvh-/- prospermatogonia by co-staining for TDRD1 and gamma-tubulin. The TDRD1 signals were predominantly localized around the centrosome (Figure 5F), although a few signals were observed away from centrosome (data not shown). These results indicate that MitoPLD and Mvh might have a role in the same context.
Mislocalization of Mitochondria
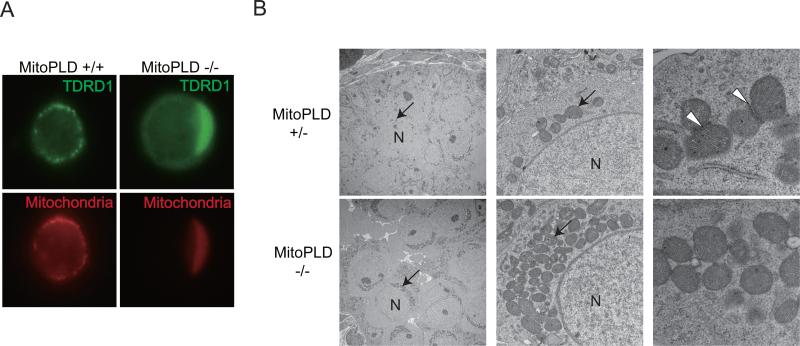
Since MILI and TDRD1 are normally localized to the intermitochondrial cement (pi-body) (Aravin et al., 2009; Chuma et al., 2006; Vagin et al., 2009), we examined the localization of mitochondria in MitoPLD-/- prospermatogonia. We found that mitochondria were also mislocalized to the region where TDRD1 accumulated (Figure 6A), suggesting that they were also localized around the centrosome. By contrast, the localizations of ER and Golgi apparatus were not affected (data not shown). Electron microscopic studies showed that mitochondria were clustered at a particular region adjacent to the nucleus in MitoPLD-/- prospermatogonia (Figure 6B, left and center). Furthermore, the electron-dense pi-bodies disappeared in MitoPLD-/- prospermatogonia (Figure 6B, right), suggesting that not only the localization but also the formation of the pi-body was impaired.
Figure 6. MitoPLD is required for mitochondrial localization.
A. Mitochondria are mislocalized around the centrosome and colocalize with TDRD1 in MitoPLD-/- testes. Prospermatogonia from E16.5 testes were double-stained with MitoTracker and TDRD1 antibody.
B. Conglomeration of mitochondria around the nucleus and dininished pi-body formation in MitoPLD-/- prospermatogonia observed by electron microscopy. Sections from E16.5 testes were studied. Mitochondria are indicated by arrows and pi-bodies in the control testes are indicated by arrowheads. Nuclei are marked “N” in the left and middle panels.
DISCUSSION
To ask whether mammalian MitoPLD is involved in the piRNA biogenesis pathway, we have generated and analyzed mouse MitoPLD mutants. The MitoPLD-/- mice showed spermatogenesis arrest at the zygotene stage of meiosis. This phenotype was accompanied by a burst of L1 retrotransposon expression with demethylation of their genomic sequences in the mutant testes. A similar demethylation of the L1 sequences was previously observed in Mili and Miwi2 mutant testes (Aravin et al., 2007b; Carmell et al., 2007; Kuramochi-Miyagawa et al., 2008), and thus the PIWI-piRNA complexes have been proposed to have a role in de novo methylation of this retrotransposon. Our findings are consistent with this notion. The piRNA biogenesis is thought to occur through two pathways, namely the primary pathway and the secondary (so-called ping-pong) pathway. In MitoPLD mutant testes, the primary pathway is heavily affected, whereas the secondary pathway seems to be still active. A defect in the primary pathway has also been reported in zuc mutants of Drosophila (Haase et al., 2010; Malone et al., 2009; Olivieri et al., 2010; Saito et al., 2009; Saito et al., 2010). Thus, MitoPLD/Zuc has a conserved role in piRNA biogenesis.
Biogenesis of piRNAs involves generation of their 5’ and 3’ ends through cleavage of the precursor RNAs. In the secondary pathway, the 5’ end of piRNAs is generated presumably through the nuclease activity of PIWI proteins (Brennecke et al., 2007; Gunawardane et al., 2007). However, the enzymes involved in the generation of the 5’ end in the primary pathway and the 3’ end in both pathways are unknown. As bacterial Nuc, a homolog of Drosophila Zuc, has a DNase activity (Pohlman et al., 1993), Zuc has been thought to be a candidate for the RNase involved in piRNA biogenesis (Pane et al., 2007). In the MitoPLD mutants, although piRNAs were severely affected, at least some primary piRNAs and a significant amount of secondary piRNAs are still observed. In addition, we have so far detected no discernible RNase activity using the recombinant MitoPLD protein that we produced (Figure S3E and S3F). These observations do not favor MitoPLD as an RNase involved in piRNA biogenesis.
Then how might MitoPLD be involved in the piRNA pathway? A previous report (Choi et al., 2006) and the present work showed that MitoPLD is localized on the surface of the mitochondrial outer membrane. The accompanying paper by Huang et al. and recent paper by Saito et al. also show the localization of Drosophila Zuc in mitochondria (Saito et al., 2010). Taken together, it is interesting to note that two subcellular structures involved in the piRNA pathway, the pi-body and piP-body collectively representing the nuage, are normally found in regions close to mitochondria (Aravin et al., 2009; Chuma et al., 2009). The pi-body is observed between aggregated mitochondria as a cementing material. In the MitoPLD-/- prospermatogonia, mitochondria and some components of the pi-body and motichondria themselves were mislocalized and found primarily around the centrosome. Considering that the centrosome is the main microtubule organizing center, it is possible that the proper localization of the pi-body and mitochondria is regulated in a microtubule-dependent manner, and this may be essential for piRNA biogenesis. Furthermore, our electron microscopic studies showed that MitoPLD is required for not only the localization but also the proper assembly of the pi-body. In addition to the pi-body components, MIWI2, which is a component of piP-body, was also mislocalized around the centrosome in our MitoPLD mutants. By contrast, DDX6, which is a component of both piP-body and P-body, was not colocalized with MIWI2, suggesting a failure of formation of proper piP-body in MitoPLD -/- spermatogonia.
Biochemically, MitoPLD hydrolyzes cardiolipin, a mitochondrion-specific lipid molecule, to generate PA on the surface of mitochondria in cultured cell (Choi et al., 2006). In an accompanying paper, Huang et al. shows that, in the absence of lipin1-- a PA-metabolizing enzyme, the abundance of the nuage is increased and localization of TDRD1 is altered. This suggests that it is PA that regulates the process. Then, how does PA signaling affect the piRNA pathway? One possible mechanism is that PA produced on the surface of mitochondria may work as a signaling molecule (Huang and Frohman, 2009) to activate/recruit molecules that are required for the localization of mitochondria and assembly/localization of the nuage. In fact, PA and other phospholipids are known to bind directly to proteins related to trafficking, including Kinesin (Klopfenstein et al., 2002; Manifava et al., 2001). Mislocalization of the nuage components may lead to misassembly and/or dysfunction, and this may be the cause of greatly reduced primary piRNA biogenesis observed in the MitoPLD mutants. The mislocalized nuage in the MitoPLD mutants could further affect the recognition/transport of the precursor RNAs of primary piRNAs. Some RNA binding proteins are known to recognize specific mRNAs and localize them to specific cellular location by using microtubule dependent motor proteins (St Johnston, 2005). Therefore, it is tempting to speculate that the microtubule dependent transport of precursor RNAs to the nuage may be required for the generation of piRNAs.
In summary, our results unequivocally demonstrate a conserved role for MitoPLD/Zuc in the piRNA biogenesis pathway and link lipid metabolism/signaling on the mitochondrial membrane to small RNA biogenesis. Further studies are required to reveal the precise mechanism of this highly conserved biological system for genome defense in germ cells.
EXPERIMENTAL PROCEDURES
Generation of MitoPLD Mutant Mice
Genomic fragments for construction of the targeting vector were obtained by PCR amplification using KOD plus high-fidelity polymerase. PCR primer sequences are found in Supplemental Information. The targeting construct was generated by ligation of these fragments to the lox-neo DTA vector. The targeting vector was linearlized using SacII and introduced into R1 ES cells by electroporation. Approximately 1,500 colonies were picked up and analyzed by PCR to identify homologous recombination events. Three colonies were positive and further confirmed by Southern blotting. All three ES cell lines contributed to the germline in chimeras and transmitted the targeted allele to their progeny. Heterozygous mice were crossed to CAG-Cre mice and the Neo cassette was removed. The homozygous mice from the three different ES cell lines showed the same phenotype. The results presented herein were obtained from mice derived by crossing to C57BL/6J for 3–5 generations. The genotyping was performed by PCR.
Antibodies
Rabbits were immunized with MitoPLD peptides corresponding to a.a. positions 137-151 (RKAGIQVRHDQDLGY) and 207-221 (DPTKYSFFPQKHRGH). Antibodies were affinity purified from the antisera by using affinity columns to which the peptides had been coupled. The SYCP3 and TDRD1 antibodies are described previously (Chuma and Nakatsuji, 2001; Chuma et al., 2003). The MIWI2 antibody is a gift from Javier Martinez. Other antibodies were from commercial suppliers: MILI, gamma-tubulin and calnexin (Abcam); DDX6 (Bethyl Laboratories); Tom20 (Santa Cruz); Smac (ProSci).
Transfection of NIH3T3 Cells
A full-length MitoPLD cDNA was inserted downstream of the CMV promoter of the plasmid vector pDsRed1-N1 (Clontech). The plasmid was transfected into NIH3T3 cells. The cells were cultured in the presence of MitoTracker CMTMRos (Invitrogen).
Collection of Testicular Cells and Mitochondria
Testes from E16.5 C57BL/6J male fetuses were treated with 0.25% trypsin /1 mM EDTA. Testicular cells were suspended in DMEM supplemented with 10% FCS. After filtration through a cell strainer (40 μm, BD Falcon), germ cells were collected by centrifugation. Mitochondria from testicular cells were collected by sucrose gradient centrifugation. Details are found in Supplementary Information.
FACS Isolation of Spermatogonia and Bisulfite Analysis
Spermatogonia were isolated by fluorescence activated cell sorting (FACS) using Ep-CAM immunofluorescence (Anderson et al., 1999). Postnatal testes at P5-7 were treated with 0.25% trypsin/1 mM EDTA for 10 min, and cells were suspended in DMEM supplemented with 10% FCS. After filtration using a cell strainer (40 μm, BD Falcon), cells were collected by centrifuge. Testicular cells were incubated with rat monoclonal anti-Ep-CAM antibody (G8.8, Santa Cruz) for 30 minutes on ice, and then incubated with Alexa Fluor 488 anti-rat IgG (Invitrogen) for 30 minutes on ice. Positive cells were collected by FACS and their genomic DNA was isolated. Bisulfite treatment of DNA was performed as described (Hirasawa et al., 2008). In brief, DNA was denatured with NaOH and then treated with 9 M sodium bisulfite from BisulFAST kit (TOYOBO). DNA was collected using a microcolumn, desulphonated and eluted using EZ DNA methylation kit (Zymo Research). The primer sequences used are listed below.
Immunocytochemistry
Cells were fixed in 2% parafolmaldehyde for 10 min on ice and attached to glass slides using cytospin. After washing with phosphate buffered saline (PBS), cells were incubated with 1% goat serum/PBS for 30 min in the presence or absence of 0.1 % Triton X-100. Slides were incubated with the primary antibodies diluted as indicated for 30 minutes: SYCP3 1/500, MILI 1/500, TDRD1 1/500, MIWI2 1/250, DDX6 1/250 and gamma-tubulin 1/100. After washing, the slides were incubated with Alexa fluor conjugated secondary antibodies (Invitrogen).
Histological Analysis and Apoptosis Assay
For paraffin sections, testes and epididymis were fixed in Bouin's fixative at 4°C overnight. After washing in PBS, the specimens were dehydrated and embedded in paraffin. Paraffin sections were stained using hematoxylin and eosin. For cryosections, testes were fixed in 2% parafolmaldehyde at 4°C for 3-5 hr. After washing in PBS, the specimens were incubated in 20% sucrose/PBS overnight on a rotating platform. The specimens were then embedded in O.C.T. compound and frozen. Apoptosis was analyzed by TUNEL labeling in cryosections using In Situ Cell Death Detection Kit (Roche).
Small RNA Sequencing
To prepare small RNA libraries, 4 pairs of P10 testes and 10 pairs of E16.5 testes were collected and total RNAs were extracted using ISOGEN (Nippon Gene). Small RNAs ranging in size from 15 to 40 nt were gel purified and used for library construction with Digital Gene Expression for Small RNA Sample Prep Kit (Illumina). The clones were sequenced using Genome Analyzer II (Illumina). After removing the linker sequences, 20-33 nt (E16.5) or 24-33 nt (P10) sequences were extracted. The sequences were mapped to the genome (mm9) using SeqMap (Jiang and Wong, 2008), and only perfectly matched sequences were extracted. Details are found in Supplementary Information.
mRNA Northern Blotting
For Northern blotting analysis of mRNAs, probes were synthesized by random priming. Hybridization was performed in 5xSSPE, 50% formamide, 5xDenhalt's, 0.5% SDS, and 50 μg/ml salmon sperm DNA at 42°C. The membrane was washed three times with a 0.2xSSC/0.1%SDS solution at 68°C.
Small RNA Northern Blotting
Hybridization was performed in 0.2 M NaHPO4 (pH 7.2), 1 mM EDTA, 1% BSA and 7% SDS at 42°C overnight. Other procedures were performed as described previously (Watanabe et al., 2007).
Immunoprecipitation of MILI-piRNA Complex
The MILI complex was immunoprecipitated from a lysate prepared from 10 pairs of P10 testes using the anti-MILI antibody (Kuramochi-Miyagawa et al., 2008). The testes were homogenized in a lysis buffer (20 mM HEPES pH 7.3, 150 mM NaCl, 2.5 mM MgCl2, 0.1% NP-40, 1X Roche-Complete). The extract was sonicated, and then centrifuged at 20,000 g for 30 minutes. Cleared extract was incubated with anti-Mili antibody for 12 h at 4 °C. Using protein G Sepharose, Mili-piRNA-antibody complexes were collected, and then washed five times using wash buffer (20 mM HEPES pH 7.3, 320 mM NaCl, 2.5 mM MgCl2, 0.1% NP-40, 1X Roche-Complete) for 15 min.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Javier Martinez and Dubravka Pezic for the MIWI2 antibody; Takashi Tanaka for information about the DDX6 antibody; Seok-Yong Choi and Vamsi Gangaraju for advice of MitoPLD protein purification; Alexei Aravin and Gregory J. Hannon for small RNA sequence information; Kenji Ichiyanagi, Hiroyasu Furuumi, Yasuhiro Tsutsui, Ryosuke Fujita and Tokumasa Horiike for scientific and technical advice; Naoko Iida and Hiroko Horiike for technical assistance. The small RNA sequences are registered in NCBI Gene Expression Omnibus with the accession number GSE20327. T.W. is a JSPS Postdoctoral Fellow for Research Abroad. This work was supported in part by Grants-in-Aid for Scientific Research on Priority Area from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to H.S and by NIH R01HD42012 to H.L..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anderson R, Schaible K, Heasman J, Wylie C. Expression of the homophilic adhesion molecule, Ep-CAM, in the mammalian germ line. J Reprod Fertil. 1999;116:379–384. doi: 10.1530/jrf.0.1160379. [DOI] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007a;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007b;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 2009;5:e1000764. doi: 10.1371/journal.pgen.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellve AR, Cavicchia JC, Millette CF, O'Brien DA, Bhatnagar YM, Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Choi SY, Huang P, Jenkins GM, Chan DC, Schiller J, Frohman MA. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat Cell Biol. 2006;8:1255–1262. doi: 10.1038/ncb1487. [DOI] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Kitamura K, Kasai S, Fujioka M, Hiyoshi M, Takamune K, Noce T, Nakatsuji N. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci U S A. 2006;103:15894–15899. doi: 10.1073/pnas.0601878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Tanaka T, Nakatsuji N. Ultrastructural characterization of spermatogenesis and its evolutionary conservation in the germline: germinal granules in mammals. Mol Cell Endocrinol. 2009;306:17–23. doi: 10.1016/j.mce.2008.11.009. [DOI] [PubMed] [Google Scholar]
- de Rooij DG, Grootegoed JA. Spermatogonial stem cells. Curr Opin Cell Biol. 1998;10:694–701. doi: 10.1016/s0955-0674(98)80109-9. [DOI] [PubMed] [Google Scholar]
- Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- Dobson MJ, Pearlman RE, Karaiskakis A, Spyropoulos B, Moens PB. Synaptonemal complex proteins: occurrence, epitope mapping and chromosome disjunction. J Cell Sci. 1994;107(Pt 10):2749–2760. doi: 10.1242/jcs.107.10.2749. [DOI] [PubMed] [Google Scholar]
- Frost RJ, Hamra FK, Richardson JA, Qi X, Bassel-Duby R, Olson EN. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1007158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18:136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Du K, Kazazian HH., Jr. A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 2001;11:1677–1685. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, Pappin DJ, Chen C, Gordon A, Hannon GJ. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev. 2010 doi: 10.1101/gad.1968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22:1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Huang H, Frohman MA. Lipid signaling on the mitochondrial surface. Biochim Biophys Acta. 2009;1791:839–844. doi: 10.1016/j.bbalip.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng R, Stearns T. Gamma-tubulin complexes: size does matter. Trends Cell Biol. 1999;9:339–342. doi: 10.1016/s0962-8924(99)01621-9. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wong WH. SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics. 2008;24:2395–2396. doi: 10.1093/bioinformatics/btn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Klopfenstein DR, Tomishige M, Stuurman N, Vale RD. Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell. 2002;109:347–358. doi: 10.1016/s0092-8674(02)00708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K, Kuramochi-Miyagawa S, Chuma S, Tanaka T, Nakatsuji N, Kimura T, Nakano T. Associations between PIWI proteins and TDRD1/MTR-1 are critical for integrated subcellular localization in murine male germ cells. Genes Cells. 2009;14:1155–1165. doi: 10.1111/j.1365-2443.2009.01342.x. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Yomogida K, Kuroiwa A, Tadokoro Y, Fujita Y, Sato M, Matsuda Y, Nakano T. Two mouse piwi-related genes: miwi and mili. Mech Dev. 2001;108:121–133. doi: 10.1016/s0925-4773(01)00499-3. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 2010;24:887–892. doi: 10.1101/gad.1902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Buchold GM, Greenbaum MP, Roy A, Burns KH, Zhu H, Han DY, Harris RA, Coarfa C, Gunaratne PH, et al. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet. 2009;5:e1000635. doi: 10.1371/journal.pgen.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manifava M, Thuring JW, Lim ZY, Packman L, Holmes AB, Ktistakis NT. Differential binding of traffic-related proteins to phosphatidic acid- or phosphatidylinositol (4,5)- bisphosphate-coupled affinity reagents. J Biol Chem. 2001;276:8987–8994. doi: 10.1074/jbc.M010308200. [DOI] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010;29:3301–3317. doi: 10.1038/emboj.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schupbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Pohlman RF, Liu F, Wang L, More MI, Winans SC. Genetic and biochemical analysis of an endonuclease encoded by the IncN plasmid pKM101. Nucleic Acids Res. 1993;21:4867–4872. doi: 10.1093/nar/21.21.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol. 2009;16:639–646. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010 doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- Schaefer CB, Ooi SK, Bestor TH, Bourc'his D. Epigenetic decisions in mammalian germ cells. Science. 2007;316:398–399. doi: 10.1126/science.1137544. [DOI] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17:775–787. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Soper SF, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15:285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D. Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- Stace CL, Ktistakis NT. Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim Biophys Acta. 2006;1761:913–926. doi: 10.1016/j.bbalip.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, Nakano T, Lin H. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J Biol Chem. 2009;284:6507–6519. doi: 10.1074/jbc.M809104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Saxe JP, Tanaka T, Chuma S, Lin H. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr Biol. 2009;19:640–644. doi: 10.1016/j.cub.2009.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Sasaki H, Minami N, Imai H. Analysis of small RNA profiles during development. Methods Enzymol. 2007;427:155–169. doi: 10.1016/S0076-6879(07)27009-0. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Stuckey JA, Lohse DL, Dixon JE. Expression, characterization, and crystallization of a member of the novel phospholipase D family of phosphodiesterases. Protein Sci. 1997;6:2655–2658. doi: 10.1002/pro.5560061221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, McLaughlin KJ, Stark A, Sachidanandam R, Pillai RS, Wang PJ. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1003953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.