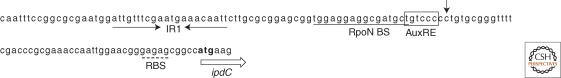Abstract
Microbial synthesis of the phytohormone auxin has been known for a long time. This property is best documented for bacteria that interact with plants because bacterial auxin can cause interference with the many plant developmental processes regulated by auxin. Auxin biosynthesis in bacteria can occur via multiple pathways as has been observed in plants. There is also increasing evidence that indole-3-acetic acid (IAA), the major naturally occurring auxin, is a signaling molecule in microorganisms because IAA affects gene expression in some microorganisms. Therefore, IAA can act as a reciprocal signaling molecule in microbe-plant interactions. Interest in microbial synthesis of auxin is also increasing in yet another recently discovered property of auxin in Arabidopsis. Down-regulation of auxin signaling is part of the plant defense system against phytopathogenic bacteria. Exogenous application of auxin, e.g., produced by the pathogen, enhances susceptibility to the bacterial pathogen.
Pathogenic bacteria can interfere with plant processes by producing the phytohormone auxin, which also regulates bacterial gene expression. Plants respond by down-regulating auxin signaling as a defense mechanism.
The phytohormone auxin (from the Greek “auxein,” meaning to grow) regulates a whole repertoire of plant developmental processes, as documented in previous articles on this topic. Perhaps less well known is the fact that some microorganisms also produce auxin (Costacurta and Vanderleyden 1995; Patten and Glick 1996). In their interaction with plants, these microorganisms can interfere with plant development by disturbing the auxin balance in plants. This is best documented for phytopathogenic bacteria like Agrobacterium spp. and Pseudomonas savastanoi pv. savastanoi, causing tumors and galls, respectively (Jameson 2000; Mole et al. 2007), and plant growth promoting rhizobacteria (PGPR) such as Azospirillum spp. that impact on plant root development (Persello-Cartieaux et al. 2003; Spaepen et al. 2007a). The term rhizobacteria refers to the fact that their numbers are highly enriched in the rhizosphere, i.e., the narrow band of soil that surrounds the root (Hiltner 1904; Smalla et al. 2006; van Loon 2007). Of more recent date is the observation that auxin (indole-3-acetic acid or IAA) is a signaling molecule in some microorganisms (Spaepen et al. 2007a). Bringing these data together, it follows that auxin can have a major impact in microorganism-plant interactions. This is the main theme addressed in this article. Finally, the recent finding that auxin signaling in plants is also part of the Arabidopsis defense response against a leaf pathogen (Navarro et al. 2006) is discussed in relation to bacterial IAA synthesis.
BIOSYNTHESIS PATHWAYS IN BACTERIA
Production of IAA has been reported for many bacteria. It is even assumed that over 80% of the bacteria isolated from the rhizosphere are capable to synthesize IAA (Patten and Glick 1996; Khalid et al. 2004). The main precursor for the synthesis of IAA is tryptophan. Addition of tryptophan to culture media results in all cases in higher IAA production. Biosynthesis of tryptophan starts from the metabolic node chorismate in a five-step reaction encoded by the trp genes. The branch point compound chorismate is synthesized starting from phosphoenolpyruvate and erythrose 4-phosphate in the shikimate pathway, a common pathway for the biosynthesis of aromatic amino acids and many secondary metabolites (Dosselaere and Vanderleyden 2001; Merino et al. 2008).
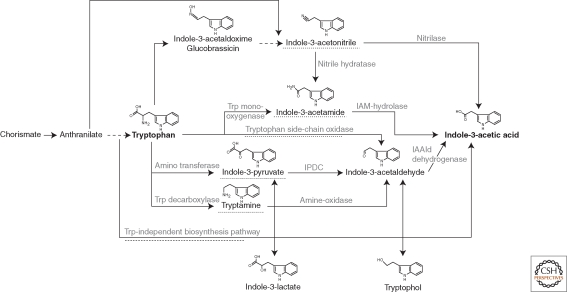
Starting with tryptophan, at least five different pathways have been described for the synthesis of IAA, and most pathways show similarity to those described in plants, although some intermediates can differ (Patten and Glick 1996; Woodward and Bartel 2005; Spaepen et al. 2007a). The IAA biosynthetic pathways have been identified using several genetic and biochemical methods. However, not all genes, enzymes encoded, or intermediates have been characterized. Especially, the use of feeding experiments with labeled precursors in combination with the identification of intermediates has been a major source for the identification of biosynthetic pathways. The pathways described in literature are illustrated in Figure 1 and are mostly named according to a key intermediate of the given pathway. In several studied microorganisms, redundancy of IAA biosynthetic pathways is observed, meaning that multiple pathways are present and active in one single microorganism. This observation is mainly based on knockout studies in which some IAA production could still be observed after inactivation of a single pathway. In the following sections, an overview of the IAA biosynthetic pathways in bacteria is given.
Figure 1.
Overview of IAA biosynthetic pathways in bacteria. Dashed lines refer to the name of pathways, mostly named after an intermediate. IAAld, indole-3-acetaldehyde; IAM, indole-3-acetamide; IPDC, indole-3-pyruvate decarboxylase; Trp, tryptophan. Adapted from Spaepen et al. (2007).
Indole-3-Acetamide Pathway
As the indole-3-acetamide (IAM) pathway is present in several pathogens and can contribute to the virulence of these strains by producing high amounts of IAA (Jameson 2000; Persello-Cartieaux et al. 2003), the genes/proteins and their regulation involved in this pathway are well characterized. The pathway consists of two distinct steps. In the first step tryptophan monooxygenase (encoded by iaaM gene) converts tryptophan to IAM; in the second step IAM is hydrolyzed to IAA and ammonia by an IAM hydrolase (encoded by iaaH gene). Both genes have been characterized from many phytopathogens such as Agrobacterium tumefaciens, P. savastanoi, Pseudomonas syringae, Pantoea agglomerans, but also in symbiotic nitrogen-fixing bacteria belonging to Rhizobium and Bradyrhizobium species (Sekine et al. 1989; Clark et al. 1993; Morris 1995; Theunis et al. 2004). In A. tumefaciens, these genes are located in the T-DNA region of the pTi plasmid together with the ipt gene (coding for dimethylallyl-pyrophosphate:AMP dimethylallyltransferase), involved in cytokinin production (for reviews see Hooykaas and Beijersbergen 1994; Costacurta and Vanderleyden 1995). A. tumefaciens can infect dicotyledonous plants at wound sites by transferring the T-DNA inside the plant cells (genetic colonization), where it is stably integrated into the genome. Several genes encoded on the T-DNA are expressed in the plant nucleus, causing an imbalance in the hormonal status of the plant cells and thereby inducing crown gall tumors (for reviews see Zambryski et al. 1989; Zambryski 1992; Hooykaas and Beijersbergen 1994). In the plant pathogen P. savastanoi pv. savastanoi, which produces high amounts of IAA in culture medium (Glickmann et al. 1998; Manulis et al. 1998; Buell et al. 2003), the iaaM and iaaH genes are located on plasmid pIAA1 (Comai and Kosuge 1982). Both genes are clustered in the iaa operon, and loss of the pIAA1 plasmid results in the loss of IAA production in free living cultures. Strains without the pIAA1 plasmid no longer cause gall formation on host plants such as olive and oleander.
In the symbiotic bacteria Rhizobium fredii and Bradyrhizobium japonicum, the production of IAA via the IAM pathway was suggested, as bacterial cultures can convert an IAM analog. A gene (bam) with sequence similarity with iaaH could be isolated from B. japonicum (Sekine et al. 1988; Sekine et al. 1989). Later, the presence of the IAM pathway was also confirmed for Rhizobium sp. strain NGR234 by LC-MS detection of intermediates (Theunis et al. 2004).
Indole-3-Pyruvate Pathway
The indole-3-pyruvate (IPA) pathway is a major auxin pathway in plants. The IPA pathway could be shown in many bacteria such as phytopathogens (P. agglomerans), plant beneficial bacteria (Azospirillum, Bacillus, Bradyrhizobium, Enterobacter cloacae, Paenibacillus, Pseudomonas, and Rhizobium), and even cyanobacteria. The pathway consists of three steps. In a first step the precursor tryptophan is transaminated to IPA by an aminotransferase. Aminotransferase activity has been shown in different IAA-producing bacteria such as Azospirillum brasilense and Azospirillum lipoferum (Ruckdäschel et al. 1988; Baca et al. 1994). In the rate-limiting step IPA is decarboxylated to indole-3-acetaldehyde (IAAld), which is then oxidized to IAA by a dehydrogenase. The decarboxylation step is catalyzed by the key enzyme indole-3-pyruvate decarboxylase (encoded by ipdC gene). Based on phylogenetic and biochemical analysis, this group of enzymes could be divided into two subgroups, indicating a distinct origin of the encoding genes (Spaepen et al. 2007b). Several members have been isolated and characterized from diverse bacteria such as A. brasilense, A. lipoferum, E. cloacae, Pseudomonas putida, and P. agglomerans (Koga et al. 1991; Costacurta et al. 1994; Brandl and Lindow 1996; Patten and Glick 2002b). Inactivation of these genes results in impaired IAA production (sometimes >90%), indicating the importance of the encoded enzymes and consequently the IPA pathway in the IAA biosynthesis (Prinsen et al. 1993; Costacurta et al. 1994). Biochemically, the indole-3-pyruvate decarboxylase is best characterized in E. cloacae (Schütz et al. 2003a; Schütz et al. 2005) and A. brasilense. In A. brasilense, the decarboxylase can use both indole-3-pyruvate and phenylpyruvate as substrates (Spaepen et al. 2007b). In addition, the crystal structure of both indole-3-pyruvate decarboxylases has been determined (Schütz et al. 2003b; Versées et al. 2007a; Versées et al. 2007b).
Other Pathways Starting from Tryptophan
In the tryptamine (both TAM and TRM are used as abbreviations) pathway, tryptophan is first decarboxylated to tryptamine by a tryptophan decarboxylase and tryptamine is, in contrast to the TAM pathway in plants, directly converted to IAAld by an amine oxidase. In Bacillus cereus, tryptophan decarboxylase activity could be identified (Perley and Stowe 1966), and in Azospirillum exogenously supplied tryptamine is converted to IAA (Hartmann et al. 1983).
The indole-3-acetonitrile (IAN) pathway in plants has been suggested and studied for many years, but the exact role of the involved players is still under debate. In bacteria, nitrilases with specificity for IAN have been detected (Nagasawa et al. 1990; Kobayashi et al. 1993; Howden et al. 2009). In several Agrobacterium and Rhizobium spp., nitrile hydratase and amidase activity could be measured suggesting the conversion of IAN to IAA via IAM. In addition, the nitrile hydratase enzyme from A. tumefaciens could be purified and characterized (Kobayashi et al. 1995).
The tryptophan side-chain oxidase (TSO) pathway has only been shown in Pseudomonas fluorescens CHA0, and in this pathway tryptophan is directly converted to IAAld bypassing IPA. In the last step IAAld is oxidized to IAA as in the IPA pathway (Oberhänsli et al. 1991).
Tryptophan-Independent Pathway
Analysis of mutants in the tryptophan biosynthetic pathway showed that plants are able to synthesize IAA without the tryptophan precursor, and the branch point for this tryptophan-independent pathway arises at indole-3-glycerol phosphate or indole (Last et al. 1991; Normanly et al. 1993). However, no genes or proteins involved in this pathway could be identified yet in plants (Woodward and Bartel 2005). In bacteria, a tryptophan-independent pathway was suggested to occur in A. brasilense by feeding experiments with labeled tryptophan (Prinsen et al. 1993). However, also in this case, no firm genetic or biochemical evidence has been provided to support this pathway.
IAA Conjugation and Degradation
Some intermediates of the IAA biosynthetic pathways can be reversibly converted into storage compounds (e.g., reduction of IAAld to indole-3-lactic acid), although the exact function of these compounds is not known. In plants much of the IAA molecules are found covalently linked to sugars or amino acids, known as IAA-conjugates (Woodward and Bartel 2005). These compounds have diverse roles, such as protection against degradation (although in some cases conjugation is also suggested as a first step in degradation), storage, and transport, and are important to maintain the IAA homeostasis in the cells (Cohen and Bandurski 1982; Seidel et al. 2006). In bacteria the presence of IAA-conjugates has only been shown for P. savastanoi pv. savastanoi. In addition, a gene, involved in the conjugation process and located on the pIAA1 plasmid, was characterized as an indole-3-acetic acid-lysine synthetase (iaaL gene). This enzyme catalyzes the formation of an amide bond between the carboxyl group of IAA and the epsilon amino group of lysine in an ATP-dependent way (Glass and Kosuge 1986; Glass and Kosuge 1988). The importance of this process was shown by plant experiments with an iaaL knockout mutant. This mutant is unable to accumulate IAA conjugates in the bacterial culture medium and to form typical gall formation, indicating an impaired virulence. It was hypothesized that IAA-conjugates are produced by P. savastanoi to protect IAA against degradation by plant enzymes and are hydrolyzed inside plant cells by plant hydrolases into biologically active IAA, promoting gall formation (Glass and Kosuge 1988).
In plants, the degradation of IAA can occur via decarboxylative catabolism of IAA (modification of both side-chain and indole ring), via nondecarboxylative oxidation of the indole ring or via auxin conjugates that can be degraded. IAA degradation has also been shown in many bacteria, and several intermediates were identified (for an overview of the suggested bacterial degradation pathways, see Leveau and Gerards 2008). Recently, an iac locus involved in the degradation of IAA was characterized in P. putida, but further analysis is needed to propose the exact degradation pathway (Leveau and Lindow 2005; Leveau and Gerards 2008).
REGULATION OF BACTERIAL IAA BIOSYNTHESIS
Whereas reports on IAA biosynthesis in bacteria are numerous, regulation of IAA biosynthesis is only documented in a few bacteria. The main precursor for IAA production, tryptophan, is an important modulator in IAA biosynthesis. The addition of tryptophan to the culture medium enhances IAA production in all studied bacteria.
The best documented case whereby the effect of environmental factors is linked, although only partially, to genetic elements is A. brasilense, a member of the group of plant growth promoting rhizobacteria (PGPR). The key gene in IAA production of A. brasilense Sp245, ipdC, is strictly regulated. The IAA production in A. brasilense Sp245 increases during growth with the highest production in the stationary phase. The increase in IAA production is preceded by an increase of ipdC gene expression. Expression of the ipdC gene is further enhanced by the addition of IAA to the culture medium, indicating positive feed-back regulation by the end-product IAA of the IPA pathway. Other auxins such as naphtalene acetic acid and phenylacetic acid also induce ipdC gene expression (Vande Broek et al. 1999; Somers et al. 2005). The ipdC gene of A. brasilense Sp245 was the first bacterial gene described to be regulated by auxins. In silico analysis of the Sp245 ipdC promoter region revealed several putative cis-regulatory elements, such as an auxin response element (AuxRE) as defined in plant genes, a RpoN binding site, and an inverted repeat (see Fig. 2 for an overview of the regulatory elements in the ipdC promoter of A. brasilense) (Vande Broek et al. 2005). The RpoN binding site has been shown to partially control ipdC gene expression because expression of ipdC in a rpoN mutant is 50% reduced (Gysegom 2005). No other trans-controlling elements affecting ipdC gene expression have been identified so far. The inverted repeat is required for IAA inducibility of the ipdC promoter. Expression of the Sp245 ipdC gene is further enhanced by carbon limitation and acidic pH (Ona et al. 2003; Ona et al. 2005). Although it is tempting to speculate that these environmental conditions mimic the plant rhizosphere, the natural habitat of A. brasilense, it should be noted that not all A. brasilense strains show a similar ipdC expression profile (Rothballer et al. 2005; Malhotra and Srivastava 2008).
Figure 2.
Promoter region of the A. brasilense Sp245 ipdC gene. The start of the ipdC open reading frame is indicated with an open arrow, and the ATG start codon is in boldface. The ribosome binding site is underlined in dots. The transcription initiation site is indicated by an arrow. The RpoN-dependent promoter sequence is underlined, and the auxin-responsive element AuxRE is boxed. The inverted repeat is indicated by opposite arrows, and this repeat is responsible for IAA inducibility of the ipdC gene.
In P. putida GR12-2 and P. agglomerans, ipdC expression is regulated by RpoS (Brandl et al. 2001; Patten and Glick 2002a). RpoS is known to regulate transcription of genes in response to stress conditions and starvation. In Pseudomonas chlororaphis, the GacS/GacA two-component system acts as a negative regulator of the tryptophan-dependent IAA biosynthesis (Haas and Keel 2003; Kang et al. 2006). The GacS/GacA system controls the expression of genes, of which many are induced during late logarithmic growth phase.
In E. cloacae UW5, a putative TyrR box, similar to the recognition sequence of TyrR, was found in the promoter region of the ipdC gene. TyrR is a regulatory protein involved in transport and metabolism of aromatic amino acids. TyrR is directly involved in the induction of ipdC gene expression in E. cloacae in response to tryptophan, phenylalanine, and tyrosine, suggesting a broader function of indole-3-pyruvate decarboxylases (Ryu and Patten 2008).
In Xanthomonas axonopodis, IAA production is increased in the presence of leaf extract from the host plant Citrus sinensis (Costacurta et al. 1998). In P. agglomerans, ipdC gene expression is dramatically increased when the bacterium is grown on plant surfaces, but the specific compounds involved are not known (Brandl and Lindow 1997).
In Ralstonia solanacearum, there is a link between the type III secretion system (TTSS) and IAA synthesis. The host-responsive regulator, HrpG, of the TTSS activation cascade not only controls genes of the TTSS but also genes not belonging to the TTSS pathway including other virulence determinants and genes involved in adaptation to life in the host, such as phytohormone production. Auxin synthesis is reduced in an hrpG mutant (Valls et al. 2006).
In Erwinia chrysanthemi, known to produce IAA via the IAM pathway, it was found that in an iaaM mutant, expression of TTSS genes and the gene encoding a pectate lyase is lower as compared to the wild type (Yang et al. 2007).
In P. agglomerans pv gypsophilae, the interaction of the TTSS, quorum sensing and IAA biosynthesis in relation to plant interaction (epiphytic fitness and virulence) was investigated. It was found that IAA (and cytokinins) are involved in the regulation of the quorum sensing system and TTSS regulatory genes (Chalupowicz et al. 2009). Interestingly, as this bacterium possesses both the IAM and IPA pathway for IAA biosynthesis, effects were found opposite for mutants in the IPA and the IAM pathways, respectively.
In Rhizobium sp. strain NGR234, flavonoid signal molecules can induce IAA production, and the transcriptional regulators NodD1 and NodD2 in combination with nod-box NB15 upstream the IAA biosynthesis operon are involved in the induction (Theunis et al. 2004).
AUXIN AS A MICROBIAL SIGNAL MOLECULE
Changes in bacterial gene expression under the influence of IAA were first described for the ipdC gene of A. brasilense (see earlier discussion, positive feedback regulation of IAA synthesis). In the phytopathogen A. tumefaciens, it was shown that IAA is a signal molecule because vir gene expression is inhibited by IAA (by competing with the phenolic plant signal for interaction with the VirA/G system). This control mechanism is proposed as a signal for A. tumefaciens indicating that plant transformation (transfer and expression of T-DNA) was successful (Liu and Nester 2006). In a follow-up study, microarray analyses revealed that addition of IAA represses the vir regulon and chv genes of A. tumefaciens, and that most of these changes are also induced by the addition of salicylic acid. Expression of a gene encoding an ABC-type transporter, probably involved in the uptake of IAA, was found to be specifically up-regulated by IAA (Yuan et al. 2008).
In the yeast Saccharomyces cerevisiae, the addition of IAA to the culture medium stimulates adhesion and filamentation (inducing invasive growth), and a cell-surface protein encoded by FLO11 is involved in these changes (as supported by the increase of FLO11 transcription). At higher IAA concentrations, cell growth is arrested. Transcript profiling revealed a YAP-1 (fungal-specific transcriptional activator involved in pleiotropic drug resistance) binding site in the promoter of IAA-regulated genes; interestingly, a yap1-1 mutant was found hypersensitive to IAA, suggesting that YAP-1 is a key mediator in the auxin response in yeast. It is postulated that IAA at plant wound sites can serve as a cue for yeast and fungi to sense and adapt to the plant environment (Prusty et al. 2004). In Escherichia coli, it was found that IAA-induced cells are more resistant to different stress agents, probably by the enhanced formation of trehalose, lipopolysaccharides, exopolysaccharides, and biofilm. In the presence of IAA, expression of genes encoding cell envelope components and proteins involved in the adaption to adverse conditions are induced (Bianco et al. 2006a). Further expression studies revealed that genes involved in the tricarboxylic acid cycle, glyoxylate shunt, and amino acid biosynthesis are up-regulated in the presence of IAA. In addition, increased activity of several enzymes in the central metabolism was shown (Bianco et al. 2006b).
The question can be addressed whether IAA as an effector molecule for bacterial gene expression is related to indole as a signaling molecule. Indole is well known as a communication signal in E. coli (Lee and Lee 2010) and is studied in relation to quorum sensing (Walters and Sperandio 2006; Lee et al. 2007). Indole controls biofilm formation in E. coli and Vibrio cholerae (Di Martino et al. 2003; Mueller et al. 2009). In a recent study (Lee et al. 2010), it was shown that indole-3-acetonitrile (an auxin) decreased biofilm formation of E. coli 0157:H7 and Pseudomonas auruginosa virulence. This could be linked with specific effects of IAN on gene expression. However, in cases in which IAA has been found as an effector in bacterial gene expression, no such effect was observed with indole, indicating that auxin effects on bacterial gene expression are distinct from the well-known indole signaling.
AUXIN AS AN EFFECTOR MOLECULE IN BACTERIA PLANT INTERACTIONS
Phytopathogens
As already mentioned before, A. tumefaciens transfers T-DNA, encoding genes for auxin and cytokinin biosynthesis (among others), on plant infection into the plant genome. The genes encoding enzymes for auxin and cytokinine biosynthesis are under the control of strong constitutive eukaryotic promoters, resulting in a high accumulation of phytohormones. Depending on the Agrobacterium strain, and consequently on the exact nature of the genes encoding enzymes for phytohormones, T-DNA transfer results either in undifferentiated plant tissue (tumors) or hairy roots (Zambryski 1992; Zupan et al. 2000).
The production of phytohormones (mainly auxins and cytokinins) in free-living cultures is a hallmark for many phytophatogenic gall-forming bacteria such as P. agglomerans, P. savastanoi pv. savastanoi, P. syringae pv. syringae, R. solanacearum and Rhodococcus fascians (Morris 1995; Vandeputte et al. 2005; Valls et al. 2006). Auxin biosynthesis genes are located either on the chromosome or on a plasmid. This location can modulate the IAA level as plasmids are mostly present in multiple copies. This difference is exemplified by the difference in IAA production between P. savastanoi pv. savastanoi and P. syringae pv. syringae. In the former strain, the genes are located on a plasmid, whereas in the latter strain IAA biosynthetic genes are located on the chromosomal DNA, resulting in a lower IAA production. The IAA production in P. syringae pv. syringae could be increased fourfold by introducing a low-copy plasmid, carrying the IAA biosynthetic operon (Mazzola and White 1994; Patten and Glick 1996; Brandl and Lindow 1996). As already mentioned above, the IAM pathway is mainly linked to phytopathology, whereas the IPA pathway is involved in epiphytic and rhizosphere fitness. In the gall-forming bacterium P. agglomerans pv. gypsophilae, both the IAM and the IPA pathway are present. Reduced gall size was observed by inactivation of the IAM pathway (as for many gall-forming bacteria), whereas only a minor reduction in pathogenicity could be observed on inactivation of the IPA pathway. The opposite effect was observed when looking at the epiphytic fitness. In addition, the difference in function of the two pathways could be linked to a difference in regulation of the pathways: the ipdC gene is up-regulated during colonization of the plant, whereas the iaaM gene is up-regulated during the later phases of the interaction with the plant such as gall formation. It was therefore concluded that the IAM pathway is mainly linked to phytopathology, whereas the IPA pathway is involved in epiphytic fitness (Manulis et al. 1998).
Symbiosis
Auxin is involved in many processes of nodule formation by rhizobia in legume plants, such as founder cell specification (auxin transport inhibition mainly by flavonoids), nodule initiation and differentiation (auxin accumulation), vascular bundle formation, and nodule numbers (long distance auxin transport). Because many rhizobia are capable of producing IAA via different pathways, it is assumed that bacterially produced auxin can alter the auxin balance inside the plant. In addition, rhizobia can also indirectly influence the auxin homeostasis by interfering with plant auxin transport (Mathesius 2008).
In general, root nodules contain more auxin than non-nodulated roots (Badenochjones et al. 1983; Ghosh and Basu 2006). Nodules induced with low IAA-producing Rhizobium mutants contain less IAA than nodules initiated by the wild-type strain (Theunis 2005). Vice versa, bacteroids of a B. japonicum mutant, overproducing IAA, contain higher amounts of IAA (Hunter 1989). Also in Rhizobium leguminosarum bv. viciae, the introduction and expression of the IAM biosynthetic pathway resulted in Vicia hirsuta root nodules containing up to 60-fold more IAA than nodules invoked by the wild-type strain. Nitrogen fixation capacity (bacterial conversion of atmospheric N2 to ammonium) in the former nodules was also increased (Camerini et al. 2008). A Sinorhizobium meliloti strain overexpression of the same IAM biosynthetic pathway showed increased tolerance to several stresses, and Medicago truncatula plants inoculated with this strain have a higher IAA content in nodules and roots and are better resistant to salt stress (Bianco and Defez 2009). In addition, inoculation of M. truncatula with the IAA-overproducing strain resulted in better plant growth under phosphorus deficiency because of the release of organic acids by the bacterium (Bianco and Defez 2010). In co-inoculation studies with Azospirillum and Rhizobium, earlier and faster nodulation and higher crop yields are observed. In addition, nodule numbers and biological nitrogen fixation are increased. Using an Azospirillum ipdC mutant, producing 10% of IAA produced by the wild-type strain, the increase in nodulation and nitrogen fixation was not observed, indicating that bacterial IAA production is important in symbiosis (Remans et al. 2008). An extensive overlap of changes in protein level could be observed in M. truncatula in response to auxin treatment and S. meliloti inoculation, probably because of regulation of these proteins by auxin during the early stages of nodulation (van Noorden et al. 2007).
Plant Growth Promoting Rhizobacteria
Upon inoculation of plants with PGPR, a change in root architecture is observed, mainly as an increase in root hairs and lateral roots and shortening of the root length. In general, the total root surface increases, leading to an enhanced mineral uptake from the soil and root exudation (further stimulating root colonization by PGPR). Already for decades, these morphological changes of the root have been attributed to bacterial auxin production, and studies with mutants altered in IAA production confirm its important role. The plant growth promoting effect of bacteria, for which it has been shown that auxin is involved, is known as phytostimulation. However, because most IAA knock-out mutants are still able to promote some plant growth, IAA biosynthesis alone is not responsible for the overall observed effect (Xie et al. 1996; Dobbelaere et al. 1999; Dobbelaere et al. 2003). In the “additive hypothesis,” it was suggested that multiple mechanisms, such as dinitrogen fixation, phosphate solubilization, and 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity, together with IAA biosynthesis, are responsible for the observed plant growth promotion and yield increase (Bashan and Holguin 1997). In addition, some plant growth-promoting traits do not work independently of each other as exemplified by IAA biosynthesis and ACC deaminase activity. Some bacteria can promote plant growth via the action of the ACC deaminase enzyme. This enzyme converts ACC (the precursor of the stress hormone ethylene) in α-ketobutyrate and ammonia, lowering the ethylene level (inhibitor of root growth) inside the plant because of lack of precursor (Glick et al. 1998; Glick 2005). For a more detailed overview of plant growth-promoting rhizobacteria, we refer to other reviews (Steenhoudt and Vanderleyden 2000; Lugtenberg and Kamilova 2009).
AUXIN SIGNALING IN PLANT DEFENSE RESPONSE
Recently, a new intriguing role of auxin has been shown. Auxin physiology is a key factor in bacterial phytopathogenesis. On recognition of a biotrophic leaf pathogen by the plant, auxin signaling in Arabidopsis is down-regulated, thereby decreasing susceptibility for the pathogen (Navarro et al. 2006). This can be counteracted by the pathogen through the activity of an Hrp system (Chen et al. 2007). Both studies illustrate very nicely a prominent role for host auxin signaling in a particular aspect, effector-triggered susceptibility, pathogen associated molecular pattern-triggered immunity and effector-triggered susceptibility, respectively, of the four-phased model of the plant immune system (Jones and Dangl 2006). Furthermore, both studies showed that exogenous supply of auxin to the host promotes susceptibility to the bacterial pathogen and disease symptom development. As many bacterial pathogens are known to produce IAA (see above), it can be speculated that this property is part of the strategy used by the pathogen to circumvent the plant defense system. The same could apply for auxin producing plant growth-promoting rhizobacteria. However, it remains to be proven that IAA produced by these bacteria indeed contributes to better bacteria-plant-root interaction.
Nevertheless, biotrophic phytopathogens and plant beneficial bacteria are coming closer to each other when taking an auxin perspective. Clearly, answering questions in one direction (phytopathology) raises new fascinating questions in another direction (phytostimulation).
CONCLUDING REMARKS AND PERSPECTIVES
Recent landmark discoveries in plant auxin, in terms of the auxin receptor, auxin transporters, integration of auxin signaling with other signaling pathways (see other articles on this topic), in combination with previous scattered reports on the role of auxin in microbe-plant interactions and microbial synthesis of auxins, respectively, have given a tremendous boost to auxin research in bacteria interacting with plants. This is most visible in research of plant growth promoting rhizobacteria in which reported cases of phytostimulation are vastly increasing. This is well in line with the increasing interest in roots as central in efforts of scientists to produce crops with a better yield.
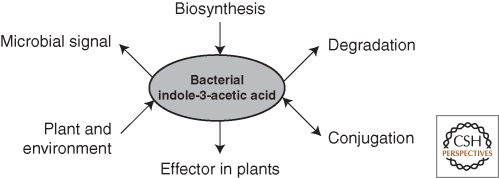
Whereas efforts to fully characterize the multiple microbial biosynthetic pathways of auxin should continue, and this is likely to be the case because similar efforts are being made by plant physiologists and biochemists, also the regulation of microbial IAA biosynthesis should be studied. This will help us to understand the role of IAA in microbial physiology. Until a number of years ago, IAA produced by microorganisms was merely considered as a secondary metabolite, possibly as the result of a detoxification process when tryptophan starts to accumulate in cells. However, the observation that multiple IAA biosynthesis pathways exist in IAA producing bacteria, like it is found for plants, already suggests additional or other roles for IAA in bacteria. The finding that IAA is used as a signal for gene regulation in some bacteria, both in IAA-producers and nonproducers, and Saccharomyces cerevisiae, further supports the idea of IAA being part of genetic networks in some microorganisms. When these microorganisms interact with plants as part of their ecological habitat, it becomes obvious that a reciprocal IAA-mediated signaling process in microbe-plant interactions is likely to occur (Lambrecht et al. 2000). Our further understanding of bacteria-plant interactions, be it pathogenic or beneficial, needs detailed studies that examine hormonal dynamics throughout the course of the interaction (see Fig. 3 for the different processes involved in IAA dynamics from the microbial point of view). With the observation that some rhizosphere bacteria are able to degrade IAA, it is clear that future research on plant roots for better plant growth and yield whereby IAA signaling is considered as a target need to take into account soil conditions, in particular the microbial activities in the rhizosphere.
Figure 3.
IAA dynamics from the bacterial point of view. IAA in plant-microbe interactions is part of a complex network. Bacteria are able to synthesize, conjugate, and degrade IAA, regulating the IAA homeostasis in their cells. Bacterial IAA production can be influenced by different plant and environmental cues as detailed in the text. Bacterial IAA is known as an effector molecule in plant-microbe interactions, both in pathogenesis and phytostimulation. Auxins are also used by bacteria as a signal molecule to steer microbial behavior.
ACKNOWLEDGMENTS
S.S. is a recipient of a postdoctoral fellowship grant from the Research Foundation-Flanders (FWO Vlaanderen).
Footnotes
Editors: Mark Estelle, Dolf Weijers, Ottoline Leyser, and Karin Ljung
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
- Baca BE, Soto-Urzua L, Xochinua-Corona YG, Cuervo-Garcia A 1994. Charaterization of two aromatic amino acid aminotransferases and production of indoleacetic acid in Azospirillum strains. Soil Biol Biochem 26: 57–63 [Google Scholar]
- Badenochjones J, Rolfe BG, Letham DS 1983. Phytohormones, Rhizobium mutants, and nodulation in legumes. 3. Auxin metabolism in effective and ineffective pea root nodules. Plant Physiol 73: 347–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashan Y, Holguin G 1997. Azospirillum-plant relationships: Environmental and physiological advances (1990–1996). Can J Microbiol 43: 103–121 [DOI] [PubMed] [Google Scholar]
- Bianco C, Defez R 2009. Medicago truncatula improves salt tolerance when nodulated by an indole-3-acetic acid-overproducing Sinorhizobium meliloti strain. J Exp Bot 60: 3097–3107 [DOI] [PubMed] [Google Scholar]
- Bianco C, Defez R 2010. Improvement of phosphate solubilization and Medicago plant yield by an indole-3-acetic acid-overproducing strain of Sinorhizobium meliloti. Appl Env Microbiol 76: 4626–4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco C, Imperlini E, Calogero R, Senatore B, Amoresano A, Carpentieri A, Pucci P, Defez R 2006a. Indole-3-acetic acid improves Escherichia coli‘s defences to stress. Arch Microbiol 185: 373–382 [DOI] [PubMed] [Google Scholar]
- Bianco C, Imperlini E, Calogero R, Senatore B, Pucci P, Defez R 2006b. Indole-3-acetic acid regulates the central metabolic pathways in Escherichia coli. Microbiology-Sgm 152: 2421–2431 [DOI] [PubMed] [Google Scholar]
- Brandl MT, Lindow SE 1996. Cloning and characterization of a locus encoding an indolepyruvate decarboxylase involved in indole-3-acetic acid synthesis in Erwinia herbicola. Appl Env Microbiol 62: 4121–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl MT, Lindow SE 1997. Environmental signals modulate the expression of an indole-3-acetic acid biosynthetic gene in Erwinia herbicola. Mol Plant-Microbe Interactions 10: 499–505 [DOI] [PubMed] [Google Scholar]
- Brandl MT, Quinones B, Lindow SE 2001. Heterogeneous transcription of an indoleacetic acid biosynthetic gene in Erwinia herbicola on plant surfaces. Proc Natl Acad Sci 98: 3454–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF, et al. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv tomato DC3000. Proc Natl Acad Sci 100: 10181–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerini S, Senatore B, Lonardo E, Imperlini E, Bianco C, Moschetti G, Rotino GL, Campion B, Defez R 2008. Introduction of a novel pathway for IAA biosynthesis to rhizobia alters vetch root nodule development. Arch Microbiol 190: 67–77 [DOI] [PubMed] [Google Scholar]
- Chalupowicz L, Barash I, Panijel M, Sessa G, Manulis-Sasson S 2009. Regulatory interactions between quorum-sensing, auxin, cytokinin, and the Hrp regulon in relation to gall formation and epiphytic fitness of Pantoea agglomerans pv gypsophilae. Mol Plant-Microbe Interact 22: 849–856 [DOI] [PubMed] [Google Scholar]
- Chen ZY, Agnew JL, Cohen JD, He P, Shan LB, Sheen J, Kunkel BN 2007. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci 104: 20131–20136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark E, Manulis S, Ophir Y, Barash I, Gafni Y 1993. Cloning and characterization of iaaM and iaaH from Erwinia herbicola pathovar gypsophilae. Phytopathology 83: 234–240 [Google Scholar]
- Cohen JD, Bandurski RS 1982. Chemistry and physiology of the bound auxins. Ann Rev Plant Physiol Plant Mol Biol 33: 403–430 [Google Scholar]
- Comai L, Kosuge T 1982. Cloning and characterization of iaaM, a virulence determinant of Pseudomonas savastanoi. J Bacteriol 149: 40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costacurta A, Vanderleyden J 1995. Synthesis of phytohormones by plant-associated bacteria. Crit Rev Microbiol 21: 1–18 [DOI] [PubMed] [Google Scholar]
- Costacurta A, Keijers V, Vanderleyden J 1994. Molecular cloning and sequence analysis of an Azospirillum brasilense indole-3-pyruvate decarboxylase gene. Mol Gen Gen 243: 463–472 [DOI] [PubMed] [Google Scholar]
- Costacurta A, Mazzafera P, Rosato YB 1998. Indole-3-acetic acid biosynthesis by Xanthomonas axonopodis pv. citri is increased in the presence of plant leaf extracts. FEMS Microbiol Lett 159: 215–220 [Google Scholar]
- Di Martino P, Fursy R, Bret L, Sundararaju B, Phillips RS 2003. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can J Microbiol 49: 443–449 [DOI] [PubMed] [Google Scholar]
- Dobbelaere S, Vanderleyden J, Okon Y 2003. Plant growth-promoting effects of diazotrophs in the rhizosphere. Crit Rev Plant Sci 22: 107–149 [Google Scholar]
- Dobbelaere S, Croonenborghs A, Thys A, Vande Broek A, Vanderleyden J 1999. Phytostimulatory effect of Azospirillum brasilense wild type and mutant strains altered in IAA production on wheat. Plant and Soil 212: 155–164 [Google Scholar]
- Dosselaere F, Vanderleyden J 2001. A metabolic node in action: Chorismate-utilizing enzymes in microorganisms. Crit Rev Microbiol 27: 75–131 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Basu PS 2006. Production and metabolism of indole acetic acid in roots and root nodules of Phaseolus mungo. Microbiol Res 161: 362–366 [DOI] [PubMed] [Google Scholar]
- Glass NL, Kosuge T 1986. Cloning of the gene for indoleacetic acid-lysine synthetase from Pseudomonas syringae subsp savastanoi. J Bacteriol 166: 598–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass NL, Kosuge T 1988. Role of indoleacetic acid lysine synthetase in regulation of indoleacetic acid pool size and virulence of Pseudomonas syringae subsp savastanoi. J Bacteriol 170: 2367–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BR 2005. Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol Lett 251: 1–7 [DOI] [PubMed] [Google Scholar]
- Glick BR, Penrose DM, Li JP 1998. A model for the lowering of plant ethylene concentrations by plant growth-promoting bacteria. J Theoretical Biol 190: 63–68 [DOI] [PubMed] [Google Scholar]
- Glickmann E, Gardan L, Jacquet S, Hussain S, Elasri M, Petit A, Dessaux Y 1998. Auxin production is a common feature of most pathovars of Pseudomonas syringae. Mol Plant-Microbe Interact 11: 156–162 [DOI] [PubMed] [Google Scholar]
- Gysegom P 2005. Study of the transcriptional regulation of a key gene in indole-3-acetic acid biosynthesis in Azospirillum brasilense. Ph.D thesis K.U. Leuven [Google Scholar]
- Haas D, Keel C 2003. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Ann Rev Phytopathol 41: 117–153 [DOI] [PubMed] [Google Scholar]
- Hartmann A, Singh M, Klingmüller W 1983. Isolation and characterization of Azospirillum mutants excreting high amounts of indoleacetic acid. Can J Microbiol 29: 916–923 [Google Scholar]
- Hiltner L 1904. Über neuere Erfahrungen und Probleme auf dem Gebiete der Bodenbakteriologie unter besonderer Berücksichtigung der Gründüngung und Brache. Arbeiten der Deutsche Landwirtschaftliche Gesellschaft 98: 59–78 [Google Scholar]
- Hooykaas PJJ, Beijersbergen AGM 1994. The virulence system of Agrobacterium tumefaciens. Ann Rev Phytopathol 32: 157–179 [Google Scholar]
- Howden AJM, Rico A, Mentlak T, Miguet L, Preston GM 2009. Pseudomonas syringae pv. syringae B728a hydrolyses indole-3-acetonitrile to the plant hormone indole-3-acetic acid. Mol Plant Pathol 10: 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter WJ 1989. Indole-3-acetic acid production by bacteroids from soybean root nodules. Physiol Plant 76: 31–36 [Google Scholar]
- Jameson PE 2000. Cytokinins and auxins in plant-pathogen interactions - An overview. Plant Growth Reg 32: 369–380 [Google Scholar]
- Jones JDG, Dangl JL 2006. The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kang BR, Yang KY, Cho BH, Han TH, Kim IS, Lee MC, Anderson AJ, Kim YC 2006. Production of indole-3-acetic acid in the plant-beneficial strain Pseudomonas chlororaphis O6 is negatively regulated by the global sensor kinase GacS. Current Microbiol 52: 473–476 [DOI] [PubMed] [Google Scholar]
- Khalid A, Tahir S, Arshad M, Zahir ZA 2004. Relative efficiency of rhizobacteria for auxin biosynthesis in rhizosphere and non-rhizosphere soils. Aus J Soil Res 42: 921–926 [Google Scholar]
- Kobayashi M, Izui H, Nagasawa T, Yamada H 1993. Nitrilase in biosynthesis of the plant hormone indole-3-acetic acid from indole-3-acetonitrile - Cloning of the Alcaligenes gene and site-directed mutagenesis of cysteine residues. Proc Natl Acad Sci 90: 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Suzuki T, Fujita T, Masuda M, Shimizu S 1995. Occurrence of enzymes involved in biosynthesis of indole-3-acetic acid from indole-3-acetonitrile in plant-associated bacteria, Agrobacterium and Rhizobium. Proc Natl Acad Sci 92: 714–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga J, Adachi T, Hidaka H 1991. Molecular cloning of the gene for indolepyruvate decarboxylase from Enterobacter cloacae. Mol Gen Gen 226: 10–16 [DOI] [PubMed] [Google Scholar]
- Lambrecht M, Okon Y, Vande Broek A, Vanderleyden J 2000. Indole-3-acetic acid: a reciprocal signalling molecule in bacteria-plant interactions. Trends in Microbiol 8: 298–300 [DOI] [PubMed] [Google Scholar]
- Last RL, Bissinger PH, Mahoney DJ, Radwanski ER, Fink GR 1991. Tryptophan mutants in Arabidopsis—the consequences of duplicated tryptophan synthase β genes. Plant Cell 3: 345–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Lee J 2010. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev 34: 426–444 [DOI] [PubMed] [Google Scholar]
- Lee JH, Cho MH, Lee J 2010. 3-Indolylacetonitrile decreases Escherichia coli O157:H7 biofilm formation and Pseudomonas aeruginosa virulence. Environ Microbiol 10.1111/j.1462-2920.2010.02308.x [DOI] [PubMed] [Google Scholar]
- Lee JT, Jayaraman A, Wood TK 2007. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol 7: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveau JHJ, Gerards S 2008. Discovery of a bacterial gene cluster for catabolism of the plant hormone indole 3-acetic acid. FEMS Microbiol Ecol 65: 238–250 [DOI] [PubMed] [Google Scholar]
- Leveau JH, Lindow SE 2005. Utilization of the plant hormone indole-3-acetic acid for growth by Pseudomonas putida strain 1290. Appl Env Microbiol 71: 2365–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Nester EW 2006. Indoleacetic acid, a product of transferred DNA, inhibits vir gene expression and growth of Agrobacterium tumefaciens C58. Proc Natl Acad Sci 103: 4658–4662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B, Kamilova F 2009. Plant-growth-promoting rhizobacteria. Ann Rev Microbiol 63: 541–556 [DOI] [PubMed] [Google Scholar]
- Malhotra M, Srivastava S 2008. Organization of the ipdC region regulates IAA levels in different Azospirillum brasilense strains: molecular and functional analysis of ipdC in strain SM. Env Microbiol 10: 1365–1373 [DOI] [PubMed] [Google Scholar]
- Manulis S, Haviv-Chesner A, Brandl MT, Lindow SE, Barash I 1998. Differential involvement of indole-3-acetic acid biosynthetic pathways in pathogenicity and epiphytic fitness of Erwinia herbicola pv. gypsophilae. Mol Plant-Microbe Interact 11: 634–642 [DOI] [PubMed] [Google Scholar]
- Mathesius U 2008. Auxin: at the root of nodule development? Funct Plant Biol 35: 651–668 [DOI] [PubMed] [Google Scholar]
- Mazzola M, White FF 1994. A mutation in the indole-3-acetic-acid biosynthesis pathway of Pseudomonas syringae pv syringae affects growth in phaseolus vulgaris and syringomycin production. J Bacteriol 176: 1374–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino E, Jensen RA, Yanofsky C 2008. Evolution of bacterial trp operons and their regulation. Curr Opinion Microbiol 11: 78–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mole BM, Baltrus DA, Dangl JL, Grant SR 2007. Global virulence regulation networks in phytopathogenic bacteria. Trends Microbiol 15: 363–371 [DOI] [PubMed] [Google Scholar]
- Morris RO 1995. Genes specifying auxin and cytokinin biosynthesis in prokaryotes . In Plant hormones (ed. Davies PJ) pp. 318–339 Kluwer Academic, Dordrecht, The Netherlands [Google Scholar]
- Mueller RS, Beyhan S, Saini SG, Yildiz FH, Bartlett DH 2009. Indole acts as an extracellular cue regulating gene expression in Vibrio cholerae. J Bacteriol 191: 3504–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa T, Mauger J, Yamada H 1990. A novel nitrilase, arylacetonitrilase, of Alcaligenes faecalis JM3 – Purification and characterization. Eur J Biochem 194: 765–772 [DOI] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JDG 2006. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439 [DOI] [PubMed] [Google Scholar]
- Normanly J, Cohen JD, Fink GR 1993. Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc Natl Acad Sci 90: 10355–10359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhänsli T, Défago G, Haas D 1991. Indole-3-acetic-acid (IAA) synthesis in the biocontrol strain CHA0 of Pseudomonas fluorescens—Role of tryptophan side-chain oxidase. J Gen Microbiol 137: 2273–2279 [DOI] [PubMed] [Google Scholar]
- Ona O, Smets I, Gysegom P, Bernaerts K, Van Impe J, Prinsen E, Vanderleyden J 2003. The effect of pH on indole-3-acetic acid (IAA) biosynthesis of Azospirillum brasilense Sp7. Symbiosis 35: 199–208 [Google Scholar]
- Ona O, Van Impe J, Prinsen E, Vanderleyden J 2005. Growth and indole-3-acetic acid biosynthesis of Azospirillum brasilense Sp245 is environmentally controlled. FEMS Microbiol Lett 246: 125–132 [DOI] [PubMed] [Google Scholar]
- Patten CL, Glick BR 1996. Bacterial biosynthesis of indole-3-acetic acid. Canadian J Microbiol 42: 207–220 [DOI] [PubMed] [Google Scholar]
- Patten CL, Glick BR 2002a. Regulation of indoleacetic acid production in Pseudomonas putida GR12-2 by tryptophan and the stationary-phase sigma factor RpoS. Canadian J Microbiol 48: 635–642 [DOI] [PubMed] [Google Scholar]
- Patten CL, Glick BR 2002b. Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl Environ Microbiol 68: 3795–3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perley JW, Stowe BB 1966. On the ability of Taphrina deformans to produce indoleacetic acid from tryptophan by way of tryptamine. Plant Physiol 41: 234–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persello-Cartieaux F, Nussaume L, Robaglia C 2003. Tales from the underground: molecular plant-rhizobacteria interactions. Plant Cell Environ 26: 189–199 [Google Scholar]
- Prinsen E, Costacurta A, Michiels K, Vanderleyden J, Van Onckelen H 1993. Azospirillum brasilense indole-3-acetic acid biosynthesis: evidence for a non-tryptophan dependent pathway. Mol Plant-Microbe Interact 6: 609–615 [Google Scholar]
- Prusty R, Grisafi P, Fink GR 2004. The plant hormone indoleacetic acid induces invasive growth in Saccharomyces cerevisiae. Proc Natl Acad Sci 101: 4153–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans R, Ramaekers L, Schelkens S, Hernandez G, Garcia A, Reyes JL, Mendez N, Toscano V, Mulling M, Galvez L, Vanderleyden J 2008. Effect of Rhizobium-Azospirillum coinoculation on nitrogen fixation and yield of two contrasting Phaseolus vulgaris L. genotypes cultivated across different environments in Cuba. Plant Soil 312: 25–37 [Google Scholar]
- Rothballer M, Schmid M, Fekete A, Hartmann A 2005. Comparative in situ analysis of ipdC-gfpmut3 promoter fusions of Azospirillum brasilense strains Sp7 and Sp245. Environ Microbiol 7: 1839–1846 [DOI] [PubMed] [Google Scholar]
- Ruckdäschel E, Kittell BL, Helinski DR, Klingmüller W 1988. Aromatic amino acid aminotransferases of Azospirillum lipoferum and their possible involvement in IAA biosynthesis. In Azospirillum IV: Genetics, physiology, ecology (ed. Klingmüller W) Springer-Verlag, Berlin Heidelberg [Google Scholar]
- Ryu RJ, Patten CL 2008. Aromatic amino acid-dependent expression of indole-3-pyruvate decarboxylase is regulated by TyrR in Enterobacter cloacae UW5. J Bacteriol 190: 7200–7208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz A, Golbik R, König S, Hübner G, Tittmann K 2005. Intermediates and transition states in thiamin diphosphate-dependent decarboxylases. A kinetic and NMR study on wild-type indolepyruvate decarboxylase and variants using indolepyruvate, benzoylformate, and pyruvate as substrates. Biochemistry 44: 6164–6179 [DOI] [PubMed] [Google Scholar]
- Schütz A, Golbik R, Tittmann K, Svergun DI, Koch MH, Hübner G, König S 2003a. Studies on structure-function relationships of indolepyruvate decarboxylase from Enterobacter cloacae, a key enzyme of the indole acetic acid pathway. Eur J Biochem 270: 2322–2331 [DOI] [PubMed] [Google Scholar]
- Schütz A, Sandalova T, Ricagno S, Hübner G, König S, Schneider G 2003b. Crystal structure of thiamin diphosphate-dependent indolepyruvate decarboxylase from Enterobacter cloacae, an enzyme involved in the biosynthesis of the plant hormone indole-3-acetic acid. Eur J Biochem 270: 2312–2321 [DOI] [PubMed] [Google Scholar]
- Seidel C, Walz A, Park S, Cohen JD, Ludwig-Muller J 2006. Indole-3-acetic acid protein conjugates: Novel players in auxin homeostasis. Plant Biol 8: 340–345 [DOI] [PubMed] [Google Scholar]
- Sekine M, Ichikawa T, Kuga N, Kobayashi M, Sakurai A, Syono K 1988. Detection of the IAA biosynthetic pathway from tryptophan via indole-3-acetamide in Bradyrhizobium spp. Plant Cell Physiol 29: 867–874 [Google Scholar]
- Sekine M, Watanabe K, Syono K 1989. Molecular cloning of a gene for indole-3-acetamide hydrolase from Bradyrhizobium japonicum. J Bacteriol 171: 1718–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalla K, Sessitsch A, Hartmann A 2006. The Rhizosphere: ‘soil compartment influenced by the root’. FEMS Microbiol Ecol 56: 165. [DOI] [PubMed] [Google Scholar]
- Somers E, Ptacek D, Gysegom P, Srinivasan M, Vanderleyden J 2005. Azospirillum brasilense produces the auxin-like phenylacetic acid by using the key enzyme for indole-3-acetic acid biosynthesis. Appl Environ Microbiol 71: 1803–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J, Remans R 2007a. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev 31: 425–448 [DOI] [PubMed] [Google Scholar]
- Spaepen S, Versées W, Gocke D, Pohl M, Steyaert J, Vanderleyden J 2007b. Characterization of phenylpyruvate decarboxylase, involved in auxin production of Azospirillum brasilense. J Bacteriol 189: 7626–7633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenhoudt O, Vanderleyden J 2000. Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects. FEMS Microbiol Rev 24: 487–506 [DOI] [PubMed] [Google Scholar]
- Theunis M 2005. IAA biosynthesis in rhizobia its potential role in symbiosis. Universiteit Antwerpen; Ph.D thesis [Google Scholar]
- Theunis M, Kobayashi H, Broughton WJ, Prinsen E 2004. Flavonoids, NodD1, NodD2, and nod-box NB15 modulate expression of the y4wEFG locus that is required for indole-3-acetic acid synthesis in Rhizobium sp. strain NGR234. Mol Plant-Microbe Interact 17: 1153–1161 [DOI] [PubMed] [Google Scholar]
- Valls M, Genin S, Boucher C 2006. Integrated regulation of the type III secretion system and other virulence determinants in Ralstonia solanacearum. PLOS 2: 798–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC 2007. Plant responses to plant growth-promoting rhizobacteria. Eur J Plant Pathol 119: 243–254 [Google Scholar]
- van Noorden GE, Kerim T, Goffard N, Wiblin R, Pellerone FI, Rolfe BG, Mathesius U 2007. Overlap of proteome changes in Medicago truncatula in response to auxin and Sinorhizobium meliloti. Plant Physiol 144: 1115–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Broek A, Gysegom P, Ona O, Hendrickx N, Prinsen E, Van Impe J, Vanderleyden J 2005. Transcriptional analysis of the Azospirillum brasilense indole-3-pyruvate decarboxylase gene and identification of a cis-acting sequence involved in auxin responsive expression. Mol Plant-Microbe Interact 18: 311–323 [DOI] [PubMed] [Google Scholar]
- Vande Broek A, Lambrecht M, Eggermont K, Vanderleyden J 1999. Auxins upregulate expression of the indole-3-pyruvate decarboxylase gene in Azospirillum brasilense. J Bacteriol 181: 1338–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeputte O, Oden S, Mol A, Vereecke D, Goethals K, El Jaziri M, Prinsen E 2005. Biosynthesis of auxin by the gram-positive phytopathogen Rhodococcus fascians is controlled by compounds specific to infected plant tissues. Appl Environ Microbiol 71: 1169–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versées W, Spaepen S, Vanderleyden J, Steyaert J 2007a. The crystal structure of phenylpyruvate decarboxylase from Azospirillum brasilense at 1.5 angstrom resolution—Implications for its catalytic and regulatory mechanism. FEBS J 274: 2363–2375 [DOI] [PubMed] [Google Scholar]
- Versées W, Spaepen S, Wood MDH, Leeper FJ, Vanderleyden J, Steyaert J 2007b. Molecular mechanism of allosteric substrate activation in a thiamine diphosphate-dependent decarboxylase. J Biol Chem 282: 35269–35278 [DOI] [PubMed] [Google Scholar]
- Walters M, Sperandio V 2006. Quorum sensing in Escherichia coli and Salmonella. Int J Med Microbiol 296: 125–131 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B 2005. Auxin: Regulation, action, and interaction. Ann Bot 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Pasternak JJ, Glick BR 1996. Isolation and characterization of mutants of the plant growth-promoting rhizobacterium Pseudomonas putida CR12-2 that overproduce indoleacetic acid. Curr Microbiol 32: 67–71 [Google Scholar]
- Yang S, Zhang Q, Guo J, Charkowski AO, Glick BR, Ibekwe AM, Cooksey DA, Yang CH 2007. Global effect of indole-3-acetic acid biosynthesis on multiple virulence factors of Erwinia chrysanthemi 3937. Appl Environ Microbiol 73: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ZC, Liu P, Saenkham P, Kerr K, Nester EW 2008. Transcriptome profiling and functional analysis of Agrobacterium tumefaciens reveals a general conserved response to acidic conditions (pH 5.5) and a complex acid-mediated signaling involved in Agrobacterium-plant interactions. J Bacteriol 190: 494–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambryski PC 1992. Chronicles from the Agrobacterium—plant cell DNA transfer story. Ann Rev Plant Physiol Plant Mol Biol 43: 465–490 [Google Scholar]
- Zambryski P, Tempe J, Schell J 1989. Transfer and function of T-DNA genes from Agrobacterium Ti-plasmid and Ri-plasmid in plants. Cell 56: 193–201 [DOI] [PubMed] [Google Scholar]
- Zupan J, Muth TR, Draper O, Zambryski P 2000. The transfer of DNA from Agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J 23: 11–28 [DOI] [PubMed] [Google Scholar]