Abstract
CELF (CUG-BP and ETR-3-like factors) proteins are structurally related RNA-binding proteins involved in various aspects of RNA processing including splicing and mRNA stability. The first member of the family, CELF1/CUG-BP1, was identified through its role in myotonic dystrophy, type 1. Several recent studies have uncovered the recurrent implication, to various extents, of CELF proteins or of the functionally related muscleblindlike 1 protein in a number of neurological conditions. This is particularly clear for inherited neurodegenerative disorders caused by expansions of translated or untranslated triplet repeats in the causative gene. Here we review the role played by CELF proteins, at least as modifiers of the pathological phenotype, in a number of neurological diseases. The involvement of CELF proteins suggest that individual pathogenic pathways in a number of neurological conditions overlap at the level of RNA processing.
Key words: CELF proteins, BRUNOL proteins, triplet repeat disorders, neurodegeneration, splicing, RNA processing
Introduction
An increasing number of neurological disorders are being linked to impairment of RNA processing.1–5 Impaired RNA processing can be caused by mutations in cis-acting elements or can result from abnormal activity of RNA-binding proteins. A prominent example of the former is provided by dominant mutations in the MAPT gene, encoding the microtubule-associated protein, tau, in frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17). The RNA- and DNA-binding protein, TDP-43, accumulates in characteristic cytoplasmic inclusions in amyotrophic lateral sclerosis (ALS), and mutations in the TARDBP gene, encoding TDP-43, as well as in the FUS gene, encoding another RNA-binding protein, FUS, have been linked to familial forms of ALS.6 While not a primary cause, aberrant RNA processing has been shown to be part of altered molecular pathways involved in many neurological disorders. Here we review the role played by a group of RNA-binding proteins, the CELF proteins, at least as modifiers of the pathological phenotype, in a number of neurological conditions, particularly in neurodegenerative disorders caused by triplet repeat expansions.
The CELF Family of RNA-Binding Proteins
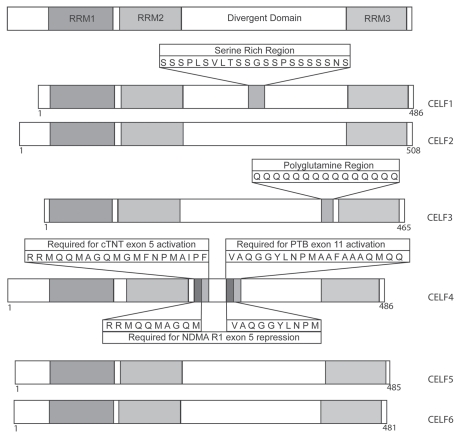
CELF proteins (CUG-BP and ETR-3-like factors) proteins are a family of structurally related RNA-binding proteins involved in various aspects of RNA processing. The first member of the CELF family, CUG-BP1, was identified in investigating the molecular mechanisms of myotonic dystrophy. Myotonic dystrophy, type 1 (DM1) is an inherited progressive muscle wasting disease caused by a CTG repeat expansion from 50 to over 1,000 in the 3′-untranslated region (UTR) of the DM protein kinase (DMPK) gene. The molecular mechanisms of DM1 have been reviewed extensively elsewhere.7–9 A search for proteins binding to repeated CUG sequences identified a ∼50 kDa protein, CUG-BP1, binding to single-stranded CUG repeated sequence.10,11 A comprehensive search of human expressed sequence tag databases for proteins structurally related to CUG-BP1 identified a family of six proteins.12,13 One such protein is ETR-3 (ELAV-type RNA-binding protein 3), related to Drosophila embryonic lethal abnormal vision protein (ELAV). Members of this family are now referred to as CELF (CUG-BP and ETR-3-like factors) proteins. Independently, a similar search for mammalian homologues of the Xenopus laevis BrunoL-1 protein identified the same family of RNA-binding proteins, thus also referred to as Bruno-like (BRUNOL) family.14 In Drosophila, the Bruno protein represses the translation of the oskar mRNA, required in early embryonic development. The structure of the six known members of the CELF family is compared in Figure 1. CELF proteins have been referred to by their CELF or BRUNOL nomenclature, or by specific names (Table 1). The CELF nomenclature will be used throughout this review.
Figure 1.
Comparison of the structure of the six members of the human CELF protein family.12–14 CELF proteins have a similar structure comprising three conserved RR Ms, two in the N-terminus, RR M1 and RR M2 (left as shown on top bar), and a third, RR M3 (right as shown on top bar) in the C-terminus. A non conserved domain, the divergent domain, separates RR M3 from RR M2. Notable differences between the divergent domains of the CELF proteins are a serine-rich region in CELF1 (identified by primary sequence comparison), a polyglutamine stretch in CELF3,12,26 and specific regions in the divergent domain of CELF4 which modulate its splicing activity on different pre-mRNA targets.71
Table 1.
CELF proteins and their alternative nomenclature
| Protein | BRUNOL nomenclature | Alternative name |
| CELF1 | BRUNOL2 | CUG-BP1 |
| NAB50 | ||
| NAPOR | ||
| EDEN-BP | ||
| CELF2 | BRUNOL3 | ETR-3 |
| CUG-BP2 | ||
| NAPOR-2 | ||
| CELF3 | BRUNOL1 | TNRC4 |
| ETR-1 | ||
| CAGH4 | ||
| ER DA4 | ||
| MGC57297 | ||
| CELF4 | BRUNOL4 | |
| CELF5 | BRUNOL5 | |
| CELF6 | BRUNOL6 |
CELF proteins have three conserved RNA recognition motifs (RRM), RRM1 and RRM2 in the N-terminus of the protein and RRM3 in the C-terminus. RRM3 is separated from RRM1 and RRM2 by a non-conserved domain, or divergent domain, specific for each member of the family.12 Each RRM comprises two ribonucleoprotein motifs, (RNP1 and RNP2) responsible for RNA binding; RNP2 is highly conserved among CELF proteins.12,14 Unique among other CELF proteins is CELF3, that contains a 15–18-repeat polyglutamine (polyQ) sequence in its divergent domain. CELF proteins usually bind to single stranded CUG/GUG clusters in introns or in 3′-UTRs of mRNAs.15,16 CELF2 has been implicated in cytoplasmic functions, such as translational control and mRNA stability17 as well as RNA editing.18 CELF1 regulates alternative splicing of several transcripts known to be misregulated in DM1 including cardiac troponin T, the insulin receptor, and the muscle-specific chloride channel.7
In DM1, the CUG expansion in the 3′-UTR of the DMPK mRNA activates protein kinase C (PKC), that phosphorylates CELF1. Phosphorylation of CELF1 stabilizes the protein and results in its upregulation.19 Long CUG repeats in DM1 form an imperfect double stranded hairpin structure that sequesters another RNA-binding protein, Muscleblind-like 1 (MBNL1), an homolog of Drosophila muscleblind.20 MBNL1 co-localizes with CUG RNA nuclear foci in muscle from DM1 patients.21 Sequestration of MBNL1 by long CUG repeats decreases its activity as a splicing regulator.22 CELF1 and MBNL1 regulate a number of common splicing events, but in opposite directions.23 Abnormal splicing observed in DM1 is likely to be the result of the combined effects of loss of MBNL1 activity and upregulation of CELF1 activity.
The expression pattern of CELF proteins across adult and foetal human tissues has been examined by Northern blotting, dot-blot analysis or high-density microarrays.12–14,24 These studies revealed that all CELF proteins were expressed at similar levels in all adult brain areas examined and throughout development. CELF3 and CELF5 are preferentially expressed in the brain and some ubiquitous proteins (e.g., CELF2 and 4) are expressed at a high level in the brain. CELF3 appears early in development and is expressed in brain and testis only.12,13,25 The CELF3 transcript is translated into a bona fide ∼50 kDa protein in the brain.26 In the mouse, CELF4 is brain-specific and is expressed in neurons at a particularly high level in the pyramidal neurons of the CA2 and CA3 regions.27 Although little information is available to date, neuronal CELF proteins appear to be important in maintaining neuronal function. In Caenorhabditis elegans, mutations in unc-75, the orthologue of CELF3, CELF4 and CELF5 result in sprouting defects in specific subpopulations of motor neurons accompanied by a locomotor phenotype that could be rescued by expression of human CELF4.28
Implication of CELF Proteins in Triplet Repeat Expansion Neurodegenerative Disorders
A number of inherited neurological disorders are caused by translated or untranslated repeat expansions in the disease gene. Current evidence points out to an, at least partial, implication of toxicity at the RNA level in most, if not all, of these disorders.29
DM1, caused by a CTG repeat expansion in the 3′-UTR of the DMPK gene, is primarily a muscle wasting disease, but, in addition, aged patients develop a neurodegenerative phenotype.30 Neurodegeneration in DM1 is associated with aggregation of the tau protein into neurofibrillar lesions reminiscent of the pathology of Alzheimer's disease.31,32 A similar tau pathology has been reported in a patient with myotonic dystrophy, type 2 (DM2), caused by an intronic CCTG tetramer repeat expansion in the ZFN9 gene.33 Alternative splicing of tau exon 10 (E10) generates tau isoforms with three (3R tau, E10−) or four (4R tau, E10+) microtubule-binding repeats in the C-terminus of the protein; alternative splicing of exons 2 (E2) and 3 (E3) produces tau isoforms with no, one or two inserts in the N-terminus. 3R tau and 4R tau are expressed at approximately equal levels in adult human brain. The pathogenic importance of tau splicing in dementia is demonstrated by a number of FTDP-17 MAPT mutations affecting cis-elements controlling E10 splicing and resulting in aberrant 4R/3Rtau isoform ratios.34 Levels of E2-containing tau transcripts are reduced in cortical neurons in DM131,32 and misregulation of E10 splicing has also been reported.35 Interestingly, tau E2 inclusion is repressed in vitro by CELF2.36 Also, inclusion of E10 is promoted at high efficiency in vitro by CELF3 and CELF4.26,37 Thus, an abnormal activity of CELF proteins, including CELF2, 3 and 4, on tau splicing may be a factor contributing to neuronal pathology of DM1. Repression of tau E2/3 inclusion can be obtained by RNAi downregulation of MBNL1 in HeLa cells, suggesting that altered MBNL1 activity may also contribute to changes in tau splicing occurring in DM1.38 Thus, like muscle and heart transcripts, abnormal tau splicing observed in DM1 might result from the combined effect of loss of MBNL1 activity and upregulation of CELF protein activity. Many transgenic mouse models of DM1 have been generated, and some tau abnormalities in the brain have been reported.39 It should be noted that, if aberrant splicing is central to tau pathology in DM1, it might be difficult to model in animals as the adult pattern of tau splicing differs between humans and mice.40
Fragile X syndrome is an X-linked form of inherited mental retardation, with mild to moderate cognitive and behavioural deficit but no neuronal loss. Fragile X syndrome is caused by expansion over 200 and hypermethylation of a CGG repeat domain in the 5′-UTR of the FMR1 gene, resulting in its silencing and the loss of its protein product, the FMRP protein, itself an RNA-binding protein.41,42 Carriers of CGG repeat expansions in the FMR1 gene, larger than 60, but below the disease threshold of 200 (so called pre-mutations) develop in old age a specific neurodegenerative syndrome referred to as fragile-X-associated tremor/ataxia syndrome (FXTAS). FXTAS is characterized by parkinsonism, brain atrophy and the presence of intranuclear inclusions in neurons and astrocytes.43,44 The levels of FMR1 mRNA are elevated by at least five times the normal levels in FXTAS individuals suggesting the involvement of an RNA gain-of-function mechanism.45,46 In support of an RNA-dependent toxic mechanism, transgenic mice47 or transgenic Drosophila48 expressing 90 CGG RNA repeats, within the premutation range, develop a neurodegenerative phenotype. Expanded CGG repeat RNA associated with FXTAS form intranuclear inclusions in transfected cells that recruit several RNA-binding proteins, including MBNL1 at late stages of their formation.49 MBNL1 also colocalizes with inclusions in FXTAS patients.50 Overexpression of hnRNP A2/B1 and CELF1 suppresses the degenerative eye phenotype induced by 90 CGG RNA repeats in Drosophila.51 HnRNP A2/B1 interacts directly with CGG RNA repeats but CELF1 exerts its effect through its interaction with hnRNP A2/B1.51 The mechanism of FXTAS would therefore bear of similarities with DM1 in that expanded triplet repeat RNA might alter the activity specific RNA-binding proteins, eventually affecting the processing of their targets.52
A number of inherited progressive neurodegenerative diseases are caused by CAG repeat expansions in the disease gene translated into a long polyQ tract in the protein product. An increase above approximately 35 in the number of CAG repeats defines the threshold between normal and disease phenotypes. PolyQ-expansion diseases include several spinocerebellar ataxias (e.g., SCA1, 2, 3, 6, 7, 17), Huntington's disease and spinal and bulbar muscular atrophy.53,54 A screen for modifiers of the neurodegenerative phenotype induced by polyQ-expanded ataxin-3, the SCA3 protein, in Drosophila identified an insertion in the promoter of the muscleblind (mbl) gene that enhanced photoreceptor neurodegeneration.55 Accordingly, overexpression of muscleblind as well as human MBNL1 enhanced toxicity. Furthermore, expression of a long untranslated CAG repeat induced neuronal degeneration; conversely, expression of the polyQ sequence through a mixed CAG/CAA sequence had a reduced toxicity.55 Another study suggested that RNA was unlikely to be involved to a major extent in polyQ expansion diseases.56 However, the effect of muscleblind on toxicity argues in favour of a role for RNA in the pathogenicity of CAG repeat disorders. Of note, MBNL1 interacts with CAG repeat RNA and co-localizes with CAG repeat RNA foci in cultured cells.22 Furthermore, a transcriptomic analysis revealed a three-fold downregulation of CELF2 in SCA3 transgenic mice as compared to non-transgenic controls.57 A trans-dominant effect of CAG repeat RNA might be elicited through abnormal processing of muscleblind targets. Considering that CELF1 and MBNL1 share a number of RNA targets, one might expect, as in FXTAS, that upregulation of CELF1 would reduce the SCA3 phenotype in flies, however, this remains to be tested. It is interesting to note that one member of the CELF family, CELF3, (also know as Trinucleotide Repeat Containing 4 (TNRC4)) contains a 15–18-repeat long polyQ sequence in its divergent domain12,58. The CAG repeat of the TNRC4 gene shows little variation in the normal human population and has not been associated with any pathological condition.59
Implication of CELF Proteins in Other Neurological Disorders
Generation of transgenic mouse lines expressing an unrelated gene led to the identification of a line, named the frequent-flyer (Ff) mouse, developing seizures in heterozygous mutants.27 Mapping of the insertion in Ff mice revealed that the transgene was inserted in intron 1 of the Celf4 gene.27 The contribution to Celf4 disruption to the Ff phenotype was confirmed through the generation of Celf4 knock-out mice which have a phenotype resembling that of the Ff mice.27 Absence of CELF4 results in downregulation of several genes involved in excitability27 but whether the splicing of specific transcript is affected is yet to be determined.
Spinal muscular atrophy (SMA) is an autosomal recessive disorder of childhood caused by the homozygous deletion of the survival motor neuron (SMN) gene and characterized by muscular atrophy and weakness resulting from the degeneration of lower motor neurons.60 The SMN gene encodes SMN, a 38 kDa protein with both cytoplasmic and nuclear localizations involved in various aspects of RNA processing.61 In the nucleus, SMN plays a role in the assembly of snRNP and regulates pre-mRNA splicing.62 In the cytoplasm, SMN has been shown to be necessary for the transport of specific mRNAs in the axon, such as actin mRNA.63,64 SMN interacts with a number of RNA-binding proteins; in particular CELF2 interacts with SMN in mouse brain and localizes with SMN in the nucleus of NSC-34 cells.65 Furthermore, CELF2 is upregulated in muscle cultures from SMA patients as well as in several mouse models of SMA.65 Anderson et al.65 suggested that the interaction of CELF2 with SMN together with its high expression in the neuromuscular system could partially explain the detrimental effect of SMN deficiency to motor neurons.
Neurofibromatosis type 1 (NF1) is characterized by the development of tumours of the nervous system: neurofibromas and schwannomas in the peripheral nervous system and gliomas and astrocytomas in the central nervous system.66 NF1 patients also display cognitive impairment and learning disabilities.67 NF1 is caused by loss-of-function mutations in the NF1 gene, encoding neurofibromin, a tumor suppressor with homologies with GTPase activating proteins (GAP) that represses the Ras kinase. Exon 23a in NF1 encodes a portion of the GAP domain and is differentially spliced between neurons and non-neuronal tissues. Mice with a targeted deletion in exon 23a display a cognitive impairment reminiscent of the phenotype observed in patients with NF1 mutations.68 All CELF proteins, with the exception of CELF6, promote exon 23a exclusion in cultured cells.69 Thus CELF proteins are involved in the regulation of a neurofibromin exon that might be important in regulating higher cognitive functions.
Conclusion
Although a number of neurological disorders involve altered RNA processing, most of them have not been directly linked to clear cis- and trans-acting effects, unlike those seen in FTDP-17 or ALS. A notion that has emerged recently is that proteins causing similar diseases may be part of networks of interacting proteins and genetic modifiers, as shown for SCAs, implying that diseases of the same class can share molecular mechanisms.70 Interestingly, a number of components of the SCA network are RNA-binding proteins, adding weight to the general implication of abnormal RNA processing in neurological, and particularly neurodegenerative, disorders.70 Most disease-causing proteins are yet to be assigned to functional networks, however, it could be speculated that CELF proteins/MBNL1 would be consistent components of such networks. Nonetheless, the implication of CELF proteins in many neurological diseases generalizes the pathogenic importance of impaired RNA processing in neurological diseases. In many cases, impaired RNA processing would not be causative, but could contribute to specific aspects of the phenotype of individual conditions.
Acknowledgements
Work in our laboratory is supported by the Wellcome Trust, the Biotechnology and Biological Sciences Research Council, GlaxoSmithKline, the Medical Research Council, the Alzheimer's Research Trust, the Psychiatry Research Trust and the NIHR Biomedical Research Centre for Mental Health at the South London & Maudsley National Health Service Foundation Trust.
Abbreviations
- ALS
amyotrophic lateral sclerosis
- CELF
CUG-BP and ETR-3-like factors
- DM1
myotonic dystrophy, type 1
- FTDP-17
frontotemporal dementia with parkinsonism linked to chromosome 17
- FXTAS
fragile-X-associated tremor/ataxia syndrome
- GAP
GTPase activating protein
- MBNL1
muscleblind-like 1
- NF1
neurofibromatosis type 1
- RRM
RNA recognition motif
- SCA
spinocerebellar ataxia
- SMA
spinal muscular atrophy
- SMN
survival motor neuron
- UTR
untranslated region
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/12345
References
- 1.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:1–5. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 3.Gallo J-M, Jin P, Thornton CA, Lin H, Robertson J, D'Souza I, et al. The role of RNA and RNA processing in neurodegeneration. J Neurosci. 2005;25:10372–10375. doi: 10.1523/JNEUROSCI.3453-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 5.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 8.O'Rourke JR, Swanson MS. Mechanisms of RNA-mediated disease. J Biol Chem. 2009;284:7419–7423. doi: 10.1074/jbc.R800025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem Soc Trans. 2009;37:1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, et al. Identification of a (CUG) n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timchenko LT, Timchenko NA, Caskey CT, Roberts R. Novel proteins with binding specificity for DNA CTG repeats and RNA CUG repeats: implications for myotonic dystrophy. Hum Mol Genet. 1996;5:115–121. doi: 10.1093/hmg/5.1.115. [DOI] [PubMed] [Google Scholar]
- 12.Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol Cell Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ladd AN, Nguyen NH, Malhotra K, Cooper TA. CELF6, a member of the CELF family of RNA-binding proteins, regulates muscle-specific splicing enhancer-dependent alternative splicing. J Biol Chem. 2004;279:17756–17764. doi: 10.1074/jbc.M310687200. [DOI] [PubMed] [Google Scholar]
- 14.Good PJ, Chen Q, Warner SJ, Herring DC. A family of human RNA-binding proteins related to the Drosophila Bruno translational regulator. J Biol Chem. 2000;275:28583–28592. doi: 10.1074/jbc.M003083200. [DOI] [PubMed] [Google Scholar]
- 15.Faustino NA, Cooper TA. Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol Cell Biol. 2005;25:879–887. doi: 10.1128/MCB.25.3.879-887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Culler SJ, Hoff KG, Voelker RB, Berglund JA, Smolke CD. Functional selection and systematic analysis of intronic splicing elements identifies active sequence motifs and associated splicing factors. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq248. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukhopadhyay D, Houchen CW, Kennedy S, Dieckgraefe BK, Anant S. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol Cell. 2003;11:113–126. doi: 10.1016/s1097-2765(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 18.Anant S, Henderson JO, Mukhopadhyay D, Navaratnam N, Kennedy S, Min J, et al. Novel role for RNA-binding protein CUGBP2 in mammalian RNA editing. CUGBP2 modulates C to U editing of apolipoprotein B mRNA by interacting with apobec-1 and ACF, the apobec-1 complementation factor. J Biol Chem. 2001;276:47338–47351. doi: 10.1074/jbc.M104911200. [DOI] [PubMed] [Google Scholar]
- 19.Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, et al. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 22.Ho TH, Savkur RS, Poulos MG, Mancini MA, Swanson MS, Cooper TA. Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J Cell Sci. 2005;118:2923–2933. doi: 10.1242/jcs.02404. [DOI] [PubMed] [Google Scholar]
- 23.Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, et al. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de la Grange P, Gratadou L, Delord M, Dutertre M, Auboeuf D. Splicing factor and exon profiling across human tissues. Nucleic Acids Res. 2010;38:2825–2838. doi: 10.1093/nar/gkq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dev A, Nayernia K, Meins M, Adham I, Lacone F, Engel W. Mice deficient for RNA-binding protein brunol1 show reduction of spermatogenesis but are fertile. Mol Reprod Dev. 2007;74:1456–1464. doi: 10.1002/mrd.20742. [DOI] [PubMed] [Google Scholar]
- 26.Chapple JP, Anthony K, Martin TR, Dev A, Cooper TA, Gallo J-M. Expression, localization and tau exon 10 splicing activity of the brain RNA-binding protein TNRC4. Hum Mol Genet. 2007;16:2760–2769. doi: 10.1093/hmg/ddm233. [DOI] [PubMed] [Google Scholar]
- 27.Yang Y, Mahaffey CL, Berube N, Maddatu TP, Cox GA, Frankel WN. Complex seizure disorder caused by Brunol4 deficiency in mice. PLoS Genet. 2007;3:124. doi: 10.1371/journal.pgen.0030124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loria PM, Duke A, Rand JB, Hobert O. Two neuronal, nuclear-localized RNA binding proteins involved in synaptic transmission. Curr Biol. 2003;13:1317–1323. doi: 10.1016/s0960-9822(03)00532-3. [DOI] [PubMed] [Google Scholar]
- 29.Li LB, Bonini NM. Roles of trinucleotide-repeat RNA in neurological disease and degeneration. TrendsNeurosci. 2010 doi: 10.1016/j.tins.2010.03.004. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Leon MB, Cisneros B. Myotonic dystrophy 1 in the nervous system: from the clinic to molecular mechanisms. J Neurosci Res. 2008;86:18–26. doi: 10.1002/jnr.21377. [DOI] [PubMed] [Google Scholar]
- 31.Vermersch P, Sergeant N, Ruchoux MM, Hofmann-Radvanyi H, Wattez A, Petit H, et al. Specific tau variants in the brains of patients with myotonic dystrophy. Neurology. 1996;47:711–717. doi: 10.1212/wnl.47.3.711. [DOI] [PubMed] [Google Scholar]
- 32.Sergeant N, Sablonniere B, Schraen-Maschke S, Ghestem A, Maurage CA, Wattez A, et al. Dysregulation of human brain microtubule-associated tau mRNA maturation in myotonic dystrophy type 1. Hum Mol Genet. 2001;10:2143–2155. doi: 10.1093/hmg/10.19.2143. [DOI] [PubMed] [Google Scholar]
- 33.Maurage CA, Udd B, Ruchoux MM, Vermersch P, Kalimo H, Krahe R, et al. Similar brain tau pathology in DM2/PROMM and DM1/Steinert disease. Neurology. 2005;65:1636–1638. doi: 10.1212/01.wnl.0000184585.93864.4e. [DOI] [PubMed] [Google Scholar]
- 34.Gallo J-M, Noble W, Rodriguez Martin T. RNA and protein-dependent mechanisms in tauopathies: consequences for therapeutic strategies. Cell Mol Life Sci. 2007;64:1701–1714. doi: 10.1007/s00018-007-6513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 36.Leroy O, Dhaenens CM, Schraen-Maschke S, Belarbi K, Delacourte A, Andreadis A, et al. ETR-3 represses Tau exons 2/3 inclusion, a splicing event abnormally enhanced in myotonic dystrophy type I. J Neurosci Res. 2006;84:852–859. doi: 10.1002/jnr.20980. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Gao QS, Wang Y, Lafyatis R, Stamm S, Andreadis A. Tau exon 10, whose missplicing causes frontotemporal dementia, is regulated by an intricate interplay of cis elements and trans factors. J Neurochem. 2004;88:1078–1090. doi: 10.1046/j.1471-4159.2003.02232.x. [DOI] [PubMed] [Google Scholar]
- 38.Dhaenens CM, Schraen-Maschke S, Tran H, Vingtdeux V, Ghanem D, Leroy O, et al. Overexpression of MBNL1 fetal isoforms and modified splicing of Tau in the DM1 brain: two individual consequences of CUG trinucleotide repeats. Exp Neurol. 2008;210:467–478. doi: 10.1016/j.expneurol.2007.11.020. [DOI] [PubMed] [Google Scholar]
- 39.Seznec H, Agbulut O, Sergeant N, Savouret C, Ghestem A, Tabti N, et al. Mice transgenic for the human myotonic dystrophy region with expanded CTG repeats display muscular and brain abnormalities. Hum Mol Genet. 2001;10:2717–2726. doi: 10.1093/hmg/10.23.2717. [DOI] [PubMed] [Google Scholar]
- 40.Takuma H, Arawaka S, Mori H. Isoforms changes of tau protein during development in various species. Brain Res Dev Brain Res. 2003;142:121–127. doi: 10.1016/s0165-3806(03)00056-7. [DOI] [PubMed] [Google Scholar]
- 41.Garber KB, Visootsak J, Warren ST. Fragile X syndrome. Eur J Hum Genet. 2008;16:666–672. doi: 10.1038/ejhg.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ranum LP, Day JW. Pathogenic RNA repeats: an expanding role in genetic disease. Trends Genet. 2004;20:506–512. doi: 10.1016/j.tig.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 43.Hagerman RJ, Leehey M, Heinrichs W, Tassone F, Wilson R, Hills J, et al. Intention tremor, parkinsonism and generalized brain atrophy in male carriers of fragile X. Neurology. 2001;57:127–130. doi: 10.1212/wnl.57.1.127. [DOI] [PubMed] [Google Scholar]
- 44.Hagerman PJ, Hagerman RJ. The fragile-X premutation: a maturing perspective. Am J Hum Genet. 2004;74:805–816. doi: 10.1086/386296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tassone F, Hagerman RJ, Taylor AK, Gane LW, Godfrey TE, Hagerman PJ. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am J Hum Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia-Arocena D, Hagerman PJ. Advances in understanding the molecular basis of FXTAS. Hum Mol Genet. 2010;19:R83–R89. doi: 10.1093/hmg/ddq166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hashem V, Galloway JN, Mori M, Willemsen R, Oostra BA, Paylor R, et al. Ectopic expression of CGG containing mRNA is neurotoxic in mammals. Hum Mol Genet. 2009;18:2443–2451. doi: 10.1093/hmg/ddp182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin P, Zarnescu DC, Zhang F, Pearson CE, Lucchesi JC, Moses K, et al. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron. 2003;39:739–747. doi: 10.1016/s0896-6273(03)00533-6. [DOI] [PubMed] [Google Scholar]
- 49.Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, et al. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010;29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iwahashi CK, Yasui DH, An HJ, Greco CM, Tassone F, Nannen K, et al. Protein composition of the intranuclear inclusions of FXTAS. Brain. 2005;129:256–271. doi: 10.1093/brain/awh650. [DOI] [PubMed] [Google Scholar]
- 51.Sofola OA, Jin P, Qin Y, Duan R, Liu H, de HM, et al. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swanson MS, Orr HT. Fragile X tremor/ataxia syndrome: blame the messenger! Neuron. 2007;55:535–537. doi: 10.1016/j.neuron.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 53.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 54.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 55.Li LB, Yu Z, Teng X, Bonini NM. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453:1107–1111. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McLeod CJ, O'Keefe LV, Richards RI. The pathogenic agent in Drosophila models of ‘polyglutamine’ diseases. Hum Mol Genet. 2005;14:1041–1048. doi: 10.1093/hmg/ddi096. [DOI] [PubMed] [Google Scholar]
- 57.Menzies FM, Huebener J, Renna M, Bonin M, Riess O, Rubinsztein DC. Autophagy induction reduces mutant ataxin-3 levels and toxicity in a mouse model of spinocerebellar ataxia type 3. Brain. 2010;133:93–104. doi: 10.1093/brain/awp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Margolis RL, Abraham MR, Gatchell SB, Li SH, Kidwai AS, Breschel TS, et al. cDNAs with long CAG trinucleotide repeats from human brain. Hum Genet. 1997;100:114–122. doi: 10.1007/s004390050476. [DOI] [PubMed] [Google Scholar]
- 59.Butland SL, Devon RS, Huang Y, Mead CL, Meynert AM, Neal SJ, et al. CAG-encoded polyglutamine length polymorphism in the human genome. BMC Genomics. 2007;8:126. doi: 10.1186/1471-2164-8-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Talbot K, Davies KE. Spinal muscular atrophy. Semin Neurol. 2001;21:189–197. doi: 10.1055/s-2001-15264. [DOI] [PubMed] [Google Scholar]
- 61.Monani UR. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron. 2005;48:885–895. doi: 10.1016/j.neuron.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 62.Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell. 1998;95:615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- 63.Rossoll W, Jablonka S, Andreassi C, Kroning AK, Karle K, Monani UR, et al. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of β-actin mRNA in growth cones of motoneurons. J Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang HL, Pan F, Hong D, Shenoy SM, Singer RH, Bassell GJ. Active transport of the survival motor neuron protein and the role of exon-7 in cytoplasmic localization. J Neurosci. 2003;23:6627–6637. doi: 10.1523/JNEUROSCI.23-16-06627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anderson KN, Baban D, Oliver PL, Potter A, Davies KE. Expression profiling in spinal muscular atrophy reveals an RNA binding protein deficit. Neuromuscul Disord. 2004;14:711–722. doi: 10.1016/j.nmd.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 66.Jett K, Friedman JM. Clinical and genetic aspects of neurofibromatosis 1. Genet Med. 2010;12:1–11. doi: 10.1097/GIM.0b013e3181bf15e3. [DOI] [PubMed] [Google Scholar]
- 67.Shilyansky C, Lee YS, Silva AJ. Molecular and cellular mechanisms of learning disabilities: A focus on NF1. Annu Rev Neurosci. 2010 doi: 10.1146/annurev-neuro-060909-153215. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Costa RM, Yang T, Huynh DP, Pulst SM, Viskochil DH, Silva AJ, et al. Learning deficits, but normal development and tumor predisposition, in mice lacking exon 23a of Nf1. Nat Genet. 2001;27:399–405. doi: 10.1038/86898. [DOI] [PubMed] [Google Scholar]
- 69.Barron VA, Zhu H, Hinman MN, Ladd AN, Lou H. The neurofibromatosis type I pre-mRNA is a novel target of CELF protein-mediated splicing regulation. Nucleic Acids Res. 2010;38:253–264. doi: 10.1093/nar/gkp766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 71.Singh G, Charlet B, Han J, Cooper TA. ETR-3 and CELF4 protein domains required for RNA binding and splicing activity in vivo. Nucleic Acids Res. 2004;32:1232–1241. doi: 10.1093/nar/gkh275. [DOI] [PMC free article] [PubMed] [Google Scholar]