Abstract
Recent results from deep-sequencing and tiling array studies indicated the existence of a large number of short, metabolically stable, non-coding RNAs. Some of these short RNAs derive from known RNA classes like snoRNA or tRNAs. There are intriguing similarities between short non-coding nuclear RNAs and oligonucleotides used to change alternative splicing events, which usually target a disease-relevant RNA. We review the current knowledge of this emerging class of RNAs and discuss evidence that some of these short RNAs could function in alternative splice site selection.
Key words: small nucleolar RNA, oligonucleotide, miRNA, processed snoRNAs, Prader-Willi Syndrome
Introduction
The involvement of short RNAs in splice site selection is evident from the well-established role that U1 and U2 snRNPs play in this process.1 The discovery of miRNAs further demonstrated that short non-coding RNAs play a crucial role in RNA metabolism.2 Common to these short RNAs is that their interaction with their RNA targets is governed by short and often interrupted sequence complementarities. It is therefore difficult to determine their targets bioinformatically. Recent results indicate that snoRNAs and tRNAs are further processed into shorter RNAs. Some of these shorter RNAs could function in pre-mRNA processing, most likely similar to oligonucleotides that are used to manipulate exon usage.
Deep Sequencing and Array Analysis Revealed Abundant Expression of Small Non-coding RNAs
snoRNAs.
Small nucleolar RNAs (snoRNA) are short, non-coding RNAs that accumulate in the nucleolus. Based on characteristic sequence elements, they can be subdivided into two groups: H/ACA box snoRNAs that contain the H (ANANNA) and ACA (ACANNN) boxes and the C/D box snoRNAs that contain the C (RUGAUGA) and D (CUGA) boxes. The known function of snoRNAs is to guide RNA modifying enzyme complexes to RNA targets. H/ACA box snoRNAs guide pseudouridinylation activity and C/D box snoRNAs guide 2′-O-methylation activity. In higher eukaryotes, snoRNAs reside in introns from which they are released during pre-mRNA processing of the hosting genes through nuclease action.3
The guiding activity of the snoRNAs is achieved by the formation of a specific RNA:RNA duplex between the snoRNA and its target. Most C/D-box and H/ACA snoRNAs contain two regions to interact with other RNAs, which are called the antisense boxes. Each antisense box exhibits sequence complementarity to its target and forms a short, transient double strand with it. The antisense elements of H/ACA box snoRNAs interact with target RNA-regions that flank an unpaired uracil base that is modified to a pseudouracil. C/D box snoRNAs cause 2′-O-methylation of a nucleotide on the target RNA that is located 5 nt upstream of the D box (reviewed in ref. 4).
Several snoRNAs show complementarity towards prerRNA, but the rRNA is not 2′-O-methylated at the predicted positions.5 Numerous C/D Box snoRNAs were discovered that show no sequence complementarity to other RNAs, suggesting that C/D box snoRNAs might have functions other than 2′-O-methylation.6
psnoRNAs/sdRNAs.
Deep sequencing of RNAs from human, mouse, chicken, Drosophila, Arabidopsis and Schizosaccharomyces pombe revealed the existence of shorter RNAs that were derived from known H/ACA snoRNAs.7,8 The shorter RNAs are predominantly 20–24 nts in length. These studies were supported by bioinformatic analyses that showed that numerous H/ACA snoRNAs act as precursors for experimentally confirmed miRNAs.9 The abundant expression of shorter snoRNAs raised the question of whether they are just degradation products or have a function. A functional role was suggested by the association of H/ACA snoRNA fragments with argonaute proteins.10 Argonaute proteins are part of the cytosolic RISC complex that processes miRNAs and the presence of snoRNA fragments in this complex suggested that the fragments assume miRNA-like functions.
C/D box snoRNAs also give rise to shorter RNA forms, including RNAs that are longer than 27 nt. Analysis of deep sequencing data revealed that C/D box derived smaller RNAs fall into two length classes: those of a length of 17–19 nt and those larger than 27 nt.7 Two further reports supported the existence of C/D box derived shorter RNAs: The sequencing of small RNAs libraries made from the ancient eukaryote Giardia lamblia revealed four RNA fragments that were derived from C/D box snoRNAs.11 The direct analysis of the brain-specific C/D box snoRNA HBII-52 (SNORD115) by RNAse protection showed that it is mostly processed into at least five shorter RNAs,12 ranging from 37 to 73 nt. Since RNase protection is quantitative, the data argue that the shorter RNAs are actually the major products from the snoRNA expressing unit. Mutational studies of one snoRNA, MBII-52/SNORD115 showed that the shorter RNAs are only formed when a full length snoRNA is formed, indicating that the shorter RNAs derive from processing of the full length snoRNA.12
Together these data indicate that snoRNA expressing units give rise to smaller RNAs that were termed psnoRNAs (for processed snoRNAs) or sdRNAs (for sno-derived RNAs). It is possible that there is a size difference between shorter RNAs derived from H/ACA or C/D snoRNAs, which might reflect different processing enzymes.
SnoRNAs are not the only source of short non-coding RNAs. The analysis of deep-sequencing data also revealed the presence of shorter tRNA-derived RNAs that are on average 19 nt long.8 Furthermore, short RNAs can be larger than the 21 nt long fragments generated by dicer, as illustrated by Piwi associated short RNAs that have lengths between 27 to 31 nt.13,14 Finally, the analysis of tiling arrays revealed abundant expression of non-coding RNAs, which showed its highest complexity in the nucleus. Most of the non-coding longer RNAs were processed into shorter RNAs of less than 200 nt.15–17
In summary, these data indicate the existence of numerous short RNAs. Most of them are derived from longer precursors and are expressed in the nucleus.
Role of Short RNAs in Alternative Splice Site Selection
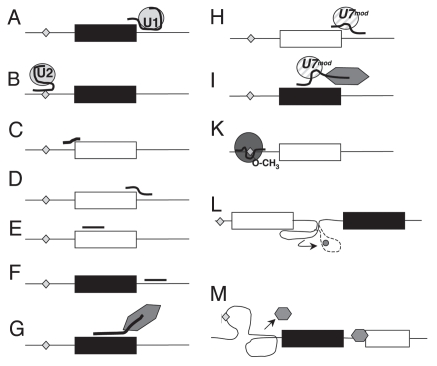
The abundance of nuclear RNAs raises the question of their function. It has been known for a long time that snRNAs form the backbone of the splicing machinery.18 The interaction between pre-mRNA substrate and the spliceosome depends at least in part on small RNAs, as the recognition of the 5′ splice site and the branchpoint depends on the interaction between the pre-mRNA and the U1 and U2 snRNAs, respectively (Fig. 1A and B). In both recognition events, there is only a partial base complementarity between the snRNA and its pre-mRNA target.
Figure 1.
Examples of regulation by short RNAs. Exons are shown as boxes, introns as lines. The branchpoint is shown as a diamond. Events that generally promote exon inclusion are indicated by black exons, events that lead to exon skipping are indicated by white exons. However, there are numerous exceptions to these rules. (A and B) U1 and U2 snRNP binding promotes exon inclusion. (C and D) Oligonucleotides that block 3′ or 5′ splice sites generally promote exon skipping. (E) Oligonucleotides that target exonic (shown) or intronic enhancers promote exon skipping. (F) Oligonucleotides that target exonic or intronic (shown) silencers promote exon skipping. (G) Bifunctional oligonucleotides can be used to target splicing enhancing proteins to exons, which generally promotes their inclusion. They bind both to RNA and to regulatory proteins (hexagon). (H) Modified U7 constructs can be used to target the 5′ splice site (shown) or splicing enhancers, which typically blocks exon usage. (I) Chimeric U7 constructs that bind both the RNA and to splicing regulators can target splicing enhancing proteins to exons, which generally promotes their inclusion. (K) C/D box snoRNAs and their associated RNPs (circle) can be modified to target the branchpoint where they cause 2′-O-methylation, which results in exon skipping. (L) Riboswitches change conformation upon ligand binding (indicated by the dotted structure and the small circle), which causes release of a previously blocked splice site and subsequent exon inclusion. (M) RNA can form secondary structures in cis, which can promote exon inclusion by removing repressing proteins (small hexagon).
Experimental use of synthetic short RNAs.
Since the interaction between a short RNA and its target can be easily predicted by its sequence, short DNAs or RNAs have been used for a long time experimentally to influence alternative splice site selection. Oligonucleotides have been used to modify splice site selection by blocking U1 binding to a 5′ splice site, by blocking the 3′ splice site and masking splicing enhancers necessary for exon inclusion (Fig. 1C–E). These approaches typically lead to exon skipping and are now in clinical trials, for example against Duchenne muscular dystrophy. Other splicing events targeted included APOB, tau and TNFRSF1B.19
To promote exon inclusion, oligonucleotides can be targeted to silencer regions of the pre-mRNA (Fig. 1F)20 or modified to contain binding sites for splicing regulatory proteins that promote exon inclusion (Fig. 1G). In the latter approach, a bifunctional oligonucleotide can be used. One part of the bifunctional oligonucleotide targets the pre-mRNA, the other part binds a regulatory protein that promotes exon inclusion.21
The use of bifunctional oligonucleotides shows that a short RNA that is introduced into a cell can assemble RNA binding proteins, which is reminiscent of natural ribonuclear protein complexes. A further development of the oligonucleotide approach is the use of modified U7 snRNPs.22–24 The U7 snRNP is necessary for cleaving the 3′ end of histone pre-mRNA. The U7 RNP is targeted to histone RNA via an 18 nt long stretch in the U7 RNA that is complementary to the histone pre-mRNA. The binding of U7 marks the histone pre-mRNA 3′ end for cleavage. The U7 snRNA contains an Sm protein binding site that is distinct from the Sm site found in the spliceosomal U1, U2, U4-U6 snRNA. If the U7 Sm site is mutated into a spliceosomal Sm site, the resulting snRNP loses its ability to cleave histone RNA, but can still interact with pre-mRNA. If the RNA binding site of U7 snRNA is changed to be complementary to splice sites or enhancer sequences, expression of this new U7-RNA causes skipping of targeted exons (Fig. 1H). The U7 snRNA can be further modified to contain a binding site for a splicing activator, like an SR-protein, which resembles the bifunctional oligonucleotide approach. These modified U7 snRNAs promote exon inclusion (Fig. 1I). The modified U7 RNAs have three functional parts: a targeting site for the pre-mRNA, an Sm binding site that promotes formation of an RNP and a protein-interaction site. These domains allow the RNA to form a scaffold for protein binding and to target these proteins to a pre-mRNA.
Finally, C/D box snoRNAs have been engineered to alter splice site selection by targeting the branchpoint. In the splicing reaction, an RNA-lariat is formed during intron removal. The ‘knot’ of this lariat is the branchpoint adenosine that forms phosphobonds at its 5′, 3′ and 2′ ribose hydroxyl groups. Blocking the 2′-hydroxyl group by methylation prevents lariat formation, as no phosphodiester bond can be made. C/D box snoRNAs target the methylation of the 2′-OH groups of specific ribose-residues. The ribose to be modified is attached to a base that is complementary to a snoRNA nucleotide that is located 5 bases upstream of its D box. It is therefore possible to engineer snoRNAs that target a particular branch point. For targeting, the antisense box of an existing snoRNA is changed to bind to the RNA of interest. A 2′-O-methylation of the branchpoint adenosine prevents the lariat formation and usage of the downstream exon. Experiments both in yeast25 and in mammalian systems26 showed that such engineered snoRNAs can change splice site selection (Fig. 1K).
Natural RNAs that change splice site selection.
Regulation of alternative splicing in cis by secondary RNA structures. Single stranded RNAs forms extensive secondary and tertiary structures in vitro. A secondary structure is caused by the folding of a short RNA segment on the same RNA molecule, i.e., an interaction between RNA parts in cis. In this sense, secondary structures resemble the interaction between a short RNA and a pre-mRNA molecule in trans.
It is experimentally very difficult to determine the secondary structures formed by pre-mRNAs in vivo, as they are transient and are removed when the processing proceeds. Studies in yeast suggested structures between the 5′ splice site and branch-points in vivo that help to commit an exon to the splicing reaction.27 In addition, studies of in vitro systems and of mutations causing human diseases provided evidence for a strong role of RNA secondary structures in alternative splice site selection (reviewed in ref. 28). Secondary structures that include the splice sites or the branchpoint can change exon usage, as found for the tau, HPRT and hnRNPA1 genes.29–31
The recent discovery of alternative splicing regulation by a ribo-switch in Neurospora crassa provides further evidence for the regulation of a 5′ splice site by a secondary structure in vivo. Three Neurospora crassa genes contain a thiamine aptamer in introns located at the 5′ end of genes involved in thiamine metabolism. Two of these introns have alternative 5′ splice sites. The thiamine aptamer sterically blocks one of the alternative 5′ splice sites in the absence of thiamine pyrophosphate. Upon binding of thiamine phosphate, the aptamer changes its conformation which makes the alternative 5′ splice site accessible to the splicing machinery. Thus, a metabolite can change alternative splicing by changing the secondary structure of a pre-mRNA32 (Fig. 1K). In contrast to other examples of splicing regulation by secondary structures, this system does not require direct base pairing between the splice site and the regulatory RNA element and is inducible by a metabolite.
The regulation of alternative splicing by secondary structure is not limited to the splice sites. The drosophila Dscam (Downsyndrome cell adhesion molecule) can potentially encode 38,016 proteins due to mutually exclusive exons located at four sites in the protein. The selection of one of 48 alternative exons at the second site that provides the 6th exon in the mRNA is controlled by the formation of a double stranded RNA structure between a region close to exon 5 (the docking site) and a region close to exon 6 (the selector site). The usage of the 48 exons at the second site is blocked by a protein. Upon interaction between the docking and the selector site, this blockage is interrupted by an unclear mechanism and the selected exon is included in the mRNA (Fig. 1L). Selector and docking sequences exhibit complementarities between 21 and 30 nt. None of the complementarities is perfect and there are mismatches and short bulges on both RNA elements.33
These examples demonstrate that interactions between different parts of a pre-mRNA molecule can change splice site selection. The interacting regions are always small (<50 nt), have numerous mismatches and can be interpreted as a regulation of an RNA in cis.
U1 snRNP binding at the 5′ splice site as an example for regulation in trans. Regulation at the 5′ splice site. By far the best understood small RNA system in the control of alternative splicing is the interaction between U1 snRNA and the 5′ splice site. The 5′ end of the U1 snRNA exhibits 9 nt of full complementarity to the 5′ splice site34 (Fig. 1A). However, in humans, only 5% of constitutive 5′ splice sites show the full 9-nucleotide complementarity. The majority of 5′ splice sites, representing about 41% of all constitutive splice sites have two mismatches towards the U1 snRNA.35 5% of all constitutive splice sites have 4 mismatches in the 9-nucleotide region.35 The binding of the U1 snRNP is therefore stabilized by contacts to other proteins assembled on the pre-mRNA to stabilize its binding to the 5′ splice site. The importance of the correct base complementarity at the 5′ splice site is underlined by human mutations that frequently lead to diseases caused by aberrant exon usage. Currently 431 mutations in human 5′ splice sites that are associated with more than 160 human diseases have been described.36 In most cases analyzed, mutating the 5′ splice site back into the consensus restored proper exon usage.
The U1 example shows that splice site selection can be influenced by short stretches of complementarity between a nuclear RNA and the pre-mRNA that tolerates an amazing degree of mismatches. The interaction between the short RNA and its target can be stabilized by interaction between associated protein factors.
Regulation of alternative splicing by processed snoRNAs (psnoRNAs). The Prader-Willi syndrome is a congenital disease that illustrates the importance of non-coding RNAs. The syndrome has an incidence of about 1 in 8,000 to 20,000 live births. A characteristic of the disease in older children is the inability to gain satiety after a meal and people with PWS are subsequently hyperphagic. The hyperphagia causes weight gain and makes PWS the most common genetic cause of marked obesity in humans.
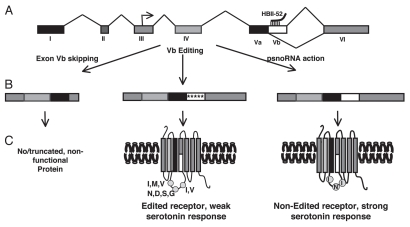
PWS is caused by the loss of gene expression from a maternally imprinted region on chromosome 15q11-q13 (reviewed in ref. 37). Recent human genetic evidence and several mouse models strongly suggest that the disease is caused by the loss of RNA expression from a cluster of snoRNAs located in the 3′ UTR of the SNURF-SNRPN locus.38–41 One of the snoRNAs in this region is HBII-52 (SNORD115), that has 48 expression units in human and over 130 units in mouse. Each expression unit consists of two exons flanking a hosting intron that contains the snoRNA. The snoRNAs are evolutionary highly conserved, whereas the hosting non-coding exons are poorly conserved. HBII-52 is expressed only in brain. A striking feature of HBII-52 is that its antisense box exhibits sequence complementarity to the serotonin receptor 5-HT2C pre-mRNA. The perfect 18 nt-long sequence complementarity is located in the alternative exon Vb of the receptor (Fig. 2A). Failing to include this exon into the pre-mRNA causes the generation of a non-functional receptor. Transfection experiments showed that cotransfecting the HBII-52 expression unit with a 5-HT2C reporter gene promoted exon Vb inclusion. Mutagenesis studies showed that exon Vb contains splicing silencers that normally prevent the inclusion of the exon. In addition, the exon forms an extended secondary structure that possibly masks some of the alternative splice site. Expression of the snoRNA blocks the action of the silencers and/or changes the secondary structure of the pre-mRNA, which promotes exon inclusion. The silencers located on the pre-mRNA can also be modified by RNA editing that changes adenosine to inosine residues.42 As a result, there are two ways of generating a full-length serotonin 5-HT2C receptor: blocking the silencers through expression of the snoRNA and weakening the silencers by editing some of its bases (Fig. 2B). However, the editing events change the amino acid composition of the receptor at three sites. These sites are located in an area critical for protein function, namely in a loop that couples to the G protein. The editing of the receptor pre-mRNA decreases the coupling of the G protein to the receptor and thus reduces its efficacy. The mRNA containing the non-edited version of the 5-HT2C receptor encodes a receptor that couples optimal to its effector G protein and shows the highest response to serotonin stimulation (Fig. 2C). Analysis of limited brain samples from PWS patients showed a reduction of the non-edited isoform,42 which has also been observed in mouse models lacking HBII-52 expression.43 A molecular link between a defect in the 5-HT2C production and PWS is an attractive hypothesis, as the 5-HT2C receptor plays a crucial role in hunger control and satiety, which is the major problem in PWS. Since HBII-52 promotes the generation of the most active receptor, it acts like a ‘genetic agonist’ of the serotonin receptor. The administration of selective 5-HT2CR agonists, such as d-fenfluramine has a strong appetite-suppressing effect,44 which shows the physiological relevance of this system. Underlining the importance of the 5-HT2C receptor for hunger control, the mouse knock-out of 5-HT2CR is hyperphagic and develops obesity. Expression of the 5-HT2CR in the arcuate nucleus, a major hunger control center, reverses the hyperphagic phenotype.45 Conversely, when a mutant of the receptor that represents the fully edited 5-HT2CR is expressed in knock-out mice, the resulting mice remain hyperphagic.46,47 Collectively, the data strongly support a model where the loss of HBII-52 causes a loss of the mRNA isoform that encodes the most active form of the receptor, which is necessary for proper hunger control.
Figure 2.
Regulation of serotonin receptor 5-HT2C by HBII -52 derived psnoRNAs. (A) The genomic structure of the 5-HT2C receptor. The arrow in exon III indicated the translational start point. HBII -52 derived psnoRNAs interact with an 18 nucleotide complementarity region in exon Vb. (B) Protein coding parts of the mRNAs derived from different pre-mRNA processing events. Exon Vb skipping results in a shortened mRNA that endodes a truncated protein but is most likely subject to nonsense-mediated mRNA decay. Exon Vb can be edited at five positions (indicated as arrows). The editing event promotes inclusion of the exon, but changes the amino acid sequence at three points. The psnoRNAs cause inclusion of exon Vb without editing, which generates a receptor with the highest agonist efficacy. (C) Structure of the encoded proteins. Editing of exon Vb leads to a change a potentially three amino acids, which are located in the second intracellular loop that couples to the effector G protein. The editing events weaken the receptor-G protein interaction and lead to a weak serotonin response. The non-edited receptor features the amino acids I, N and I at the positions that could be edited and shows the strongest coupling to the G protein and response to serotonin.
However, the exact molecular mechanism by which a snoRNA-expressing unit changes alternative splicing was hard to imagine, especially since HBII-52 is mainly localized in the nucleolus. The RNAs generated from the HBII-52 expression unit were therefore analyzed by RNase protection analysis, that directly quantifies the expressed RNAs. The data indicated that the HBII-52 expression unit generates several RNAs. Mutation studies showed that these shorter RNAs are only made when their precursor snoRNA contains intact C and D boxes. This indicates that they are most likely generated by further processing of the snoRNA and were therefore termed psnoRNAs (for processed snoRNAs).12 The main product of the 48 HBII-52 expressing units that are missing in PWS is therefore not a C/D box snoRNA, but a psnoRNA that lacks several nucleotides at the ends. This shorter version lacks the stem of the snoRNA that is crucial for the assembly of a functional snoRNPs, but still contains the antisense box needed for targeting to pre-mRNA. In addition to this form, three other shorter RNAs (60–37 nt) can be detected. The psnoRNAs were present in the nucleoplasma, where they could interact with pre-mRNA. The analysis of the protein composition showed that the RNAs associate with hnRNPs commonly implicated in splice site regulation, but not with the known structural C/D box snoRNA proteins or the 2′O-methylase.12,48 This strongly suggests that HBII-52 has a function different from traditional C/D box snoRNAs which function in 2′-O-methylation of RNA.
As the major HBII-52 psnoRNA form still contains the antisense box that targets the serotonin receptor exon Vb sequence, it is possible that this RNA form brings processing factors to this exon, similar to a bifunctional oligonucleotide (Fig. 1G). Studies of miRNAs, dscam selector RNA or U1 snRNAs showed that RNA:RNA interactions can tolerate multiple mismatches towards their targets. This indicates that HBII-52 could also regulate other splicing events.
A bioinformatic analysis predicted about 220 alternative exons that have evolutionary conserved sites which exhibit limited complementarity to the antisense box. Five of these exons were regulated by HBII-52 expression.12 In each of the identified exons there were three mismatches between the 18 nt antisense element and the target RNA, which is reminiscent of U1, where the majority of 5′ splice sites has a mismatch in 2 of the 9 possible bases.
Together, the data provide evidence that snoRNAs can be processed into shorter RNAs that regulate pre-mRNA splicing.
Generation of Short Non-Coding RNAs
The molecular mechanisms that are used to generate new classes of small RNAs are only beginning to emerge. Analysis of deep sequencing data and in vitro experiments showed that dicer is involved in the generation of small RNAs both from tRNAs8 and from H/ACA snoRNAs.7,10,11 In addition, these small RNAs were found to be associated with argonaute proteins.10,11 Argonaute proteins and dicer have been so far studied in the cytosol, but all of the investigated smaller RNAs derived from H/ACA snoRNAs localize mainly in the nucleus where they accumulate in the nucleoli.9,11 However, argonaute proteins can be detected in the nucleus of cells49,50 and it was recently shown that their nuclear localization depends on importin-8.51 It is unclear whether a regulated presence of argonautes represents an import of small RNAs back into the nucleus after processing in the cytosol, which would be similar to the U1, 2, 4–6 snRNA or whether there is a parallel nuclear processing system that uses argonautes variant.
Conclusion and Outlook
Given the high prevalence of short, non-coding nuclear RNAs, their emerging involvement in alternative splice site selection looks like the tip of the iceberg. The usage of oligonucleotides clearly showed that alternative splicing can be regulated by small RNAs, but also showed that there are unpredictable effects, like targeting seemingly unrelated mRNAs or silencing of genes. Proteins involved in splicing regulation have functions in other aspects of RNA metabolism. For example SF2/ASF works in translation and genome stability.52,53 Given the off-side effect of RNA oligonucleotides and the multiple role of splicing factors, it is therefore likely that short nuclear RNAs have multiple functions as well. One of these functions is a change in splice site selection. A better understanding of their mechanism of action will likely help in improving the design of oligonucleotides for therapeutic approaches.
There could be several biological advantages for using RNA molecules in alternative splicing regulation. The vast majority of short RNAs derive from non protein-coding RNAs and are not under the same selective pressure as protein-coding RNAs. This probably allows the cell to ‘recruit’ selected shorter RNAs for novel regulatory functions, which could enhance evolution. Relaxed evolutionary requirements could also indicate that bioinformatic predictions for RNA targets that rely heavily on evolutionary sequence conservation could be misleading.
It has long been an enigma how alternative splicing is regulated in a tissue-specific way by mostly ubiquitously expressed proteins. The example of HBII-52 derived psnoRNAs shows that tissue-specific short RNAs can regulate splice site selection, which contributes to tissue-specificity of gene regulation and is a new regulatory mechanism in alternative splicing.
Acknowledgements
This work was supported by EURASNET and the NIH (GM083187 to S.S.).
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/12746
References
- 1.Luhrmann R, Kastner B, Bach M. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim Biophys Acta. 1990;1087:265–292. doi: 10.1016/0167-4781(90)90001-i. [DOI] [PubMed] [Google Scholar]
- 2.Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Genet. 2005;14:121–132. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- 3.Brown JW, Marshall DF, Echeverria M. Intronic noncoding RNAs and splicing. Trends Plant Sci. 2008;13:335–342. doi: 10.1016/j.tplants.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 5.Steitz JA, Tycowski KT. Small RNA chaperones for ribosome biogenesis. Science. 1995;270:1626–1627. doi: 10.1126/science.270.5242.1626. [DOI] [PubMed] [Google Scholar]
- 6.Cavaille J, et al. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taft RJ, et al. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole C, et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput Biol. 2009;5:1000507. doi: 10.1371/journal.pcbi.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ender C, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 11.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kishore S, et al. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum Mol Genet. 2010;19:1153–1164. doi: 10.1093/hmg/ddp585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malone CD, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 15.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 16.Willingham AT, Gingeras TR. TUF love for “junk” DNA. Cell. 2006;125:1215–1220. doi: 10.1016/j.cell.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 18.Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991;253:157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- 19.Aartsma-Rus A, van Ommen GJ. Antisense-mediated exon skipping: a versatile tool with therapeutic and research applications. RNA. 2007;13:1609–1624. doi: 10.1261/rna.653607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh NN, Shishimorova M, Cao LC, Gangwani L, Singh RN. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol. 2009;6:341–350. doi: 10.4161/rna.6.3.8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skordis LA, Dunckley MG, Yue B, Eperon IC, Muntoni F. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc Natl Acad Sci USA. 2003;100:4114–4119. doi: 10.1073/pnas.0633863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer K, et al. Rescue of a severe mouse model for spinal muscular atrophy by U7 snRNA-mediated splicing modulation. Hum Mol Genet. 2009;18:46–55. doi: 10.1093/hmg/ddn382. [DOI] [PubMed] [Google Scholar]
- 23.Asparuhova MB, et al. Inhibition of HIV-1 multiplication by a modified U7 snRNA inducing Tat and Rev exon skipping. J Gene Med. 2007;9:23–34. doi: 10.1002/jgm.1027. [DOI] [PubMed] [Google Scholar]
- 24.Liu S, et al. Inhibition of HIV-1 multiplication by antisense U7 snRNAs and siRNAs targeting cyclophilin A. Nucleic Acids Res. 2004;32:752–759. doi: 10.1093/nar/gkh715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao X, Yu YT. Targeted pre-mRNA modification for gene silencing and regulation. Nat Methods. 2008;5:95–100. doi: 10.1038/nmeth1142. [DOI] [PubMed] [Google Scholar]
- 26.Semenov DV, Vratskih OV, Kuligina EV, Richter VA. Splicing by exon exclusion impaired by artificial box C/D RNA targeted to branch-point adenosine. Ann N Y Acad Sci. 2008;37:19–24. doi: 10.1196/annals.1448.037. [DOI] [PubMed] [Google Scholar]
- 27.Charpentier B, Rosbash M. Intramolecular structure in yeast introns aids the early steps of in vitro spliceosome assembly. RNA. 1996;2:509–522. [PMC free article] [PubMed] [Google Scholar]
- 28.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grover A, et al. 5′ splice site mutation in tau associated with the inherited dementia FTDP-17 affect a stemloop structure that regulates alternative splicing of exon 10. J Biol Chem. 1999;274:15134–15143. doi: 10.1074/jbc.274.21.15134. [DOI] [PubMed] [Google Scholar]
- 30.Tu M, Tong W, Perkins R, Valentine CR. Predicted changes in pre-mRNA secondary structure vary in their association with exon skipping for mutations in exons 2, 4 and 8 of the Hprt gene and exon 51 of the fibrillin gene. Mutat Res. 2000;432:15–32. doi: 10.1016/s1383-5726(99)00011-4. [DOI] [PubMed] [Google Scholar]
- 31.Blanchette M, Chabot B. A highly stable duplex structure sequesters the 5′ splice site region of hnRNP A1 alternative exon 7B. RNA. 1997;3:405–419. [PMC free article] [PubMed] [Google Scholar]
- 32.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 33.Graveley BR. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123:65–73. doi: 10.1016/j.cell.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhuang Y, Weiner AM. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- 35.Carmel I, Tal S, Vig I, Ast G. Comparative analysis detects dependencies among the 5′ splice-site positions. RNA. 2004;10:828–840. doi: 10.1261/rna.5196404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buratti E, et al. Aberrant 5′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2007;35:4250–4263. doi: 10.1093/nar/gkm402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butler MG, Hanchett JM, Thompson TE. Clinical Findings and Natural History of Prader-Willi Syndrome. In: Butler MG, Lee PDK, Whitman BY, editors. Managment of Prader-Willi Syndrome. Springer; 2006. pp. 3–48. [Google Scholar]
- 38.Sahoo T, et al. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet. 2008;40:719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skryabin BV, et al. Deletion of the MBII-85 snoRNA gene cluster in mice results in postnatal growth retardation. PLoS Genet. 2007;3:235. doi: 10.1371/journal.pgen.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding F, et al. SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice. PLoS ONE. 2008;3:1709. doi: 10.1371/journal.pone.0001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding F, et al. Lack of Pwcr1/MBII-85 snoRNA is critical for neonatal lethality in Prader-Willi syndrome mouse models. Mamm Genome. 2005;16:424–431. doi: 10.1007/s00335-005-2460-2. [DOI] [PubMed] [Google Scholar]
- 42.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 43.Doe CM, et al. Loss of the imprinted snoRNA mbii-52 leads to increased 5 htr2c pre-RNA editing and altered 5HT2CR-mediated behaviour. Hum Mol Genet. 2009;18:2140–2148. doi: 10.1093/hmg/ddp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vickers SP, Dourish CT, Kennett GA. Evidence that hypophagia induced by d-fenfluramine and d-norfenfluramine in the rat is mediated by 5-HT2C receptors. Neuropharmacology. 2001;41:200–209. doi: 10.1016/s0028-3908(01)00063-6. [DOI] [PubMed] [Google Scholar]
- 45.Xu Y, et al. 5-HT2CRs expressed by pro-opiomelanocortin neurons regulate energy homeostasis. Neuron. 2008;60:582–589. doi: 10.1016/j.neuron.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawahara Y, et al. Dysregulated editing of serotonin 2C receptor mRNAs results in energy dissipation and loss of fat mass. J Neurosci. 2008;28:12834–12844. doi: 10.1523/JNEUROSCI.3896-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morabito MV, et al. Mice with altered serotonin 2C receptor RNA editing display characteristics of Prader-Willi Syndrome. Neurobiol Dis. 2010;39(2):169–180. doi: 10.1016/j.nbd.2010.04.004. Neurobiol Dis 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soeno Y, et al. Identification of novel ribonucleoprotein complexes from the brain-specific snoRNA MBII-52. RNA. 2010;161(7):1293–1300. doi: 10.1261/rna.2109710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 50.Janowski BA, et al. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 51.Weinmann L, et al. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell. 2009;136:496–507. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 52.Sanford JR, Gray NK, Beckmann K, Caceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]