Abstract
The role of aging in the pathogenesis of type 2 diabetes remains poorly understood. In the past adult β-cells were assumed to undergo frequent turnover. However, we find that β-cell turnover declines to very low levels in middle-aged mice. We therefore hypothesized that aged islets could exhibit a distinct gene expression program. We compared gene expression in islets from young mice to islets from aged mice under basal conditions. Aging was associated with differential expression of many genes in islets, including mRNAs encoding for chromatin remodeling components, RNA binding proteins and pancreatic endocrine transcription factors. We previously observed that cell cycle entry of β-cells is severely restricted by middle age, with minimal of β-cell proliferation in response to regenerative stimuli such as 50% partial pancreatectomy. To characterize the effect of age in adaptive β-cell proliferation, we measured gene expression in islets from young mice after pancreatectomy. As expected, partial pancreatectomy induced differential expression of many genes, including those encoding Reg (regenerating) proteins. Surprisingly, partial pancreatectomy also induced expression of Reg genes in islets from aged mice, which have greatly reduced capacity for adaptive β-cell proliferation. However, there was little overlap (besides the Reg genes) in between the partial pancreatectomy induced islet genes in young mice versus old mice. Thus, partial pancreatectomy does not induce the same gene expression program in young mice vs. old mice. Taken together, our results reveal that aged islets exhibit a unique gene expression signature that could contribute to the limited regenerative capacity of mature β-cells.
Key words: islets, regeneration, aging, gene expression, β-cells, reg genes
Introduction
β-cell regeneration remains a fundamental challenge for diabetes research. Consequently, major research initiatives towards β-cell regeneration have been launched over the past decades. A general consensus of β-cell mass expansion has emerged from this work. Several putative mechanisms have been invoked to explain adult β-cell mass expansion, including neogenesis from pancreatic ducts or hematopoietic tissues, replication of specialized β-cell progenitors and self-renewal by β-cells.1 However, recent studies by several groups (including ours) indicate that normal β-cell growth primarily occurs by division of cells with insulin promoter activity, not by replication of specialized progenitors.2,3 β-cell mass also appears to be dynamically regulated, at least in young rodents. β-cell mass expansion can accelerate in response to increased insulin requirements4 or during pregnancy.5 These observations have led to speculation that β-cell regeneration therapies for diabetes patients might someday become a reality. However, human β-cell regeneration is an ambitious long-term goal, and has yet to be convincingly demonstrated in vitro nor in vivo.6,7
Aging has emerged over the past few years as a key factor to limit β-cell regeneration. In the past rodent β-cells were reported to undergo frequent turnover (every 1–3 months) and assumed to have a relatively short lifespan.8 This observation implied that β-cell mass expansion could highly responsive to metabolic stimuli and that small changes in β-cell turnover might result in large changes in β-cell mass expansion. However, we discovered that mice have very little evidence of β-cell turnover at one year of age (approximately 1:1400 BrdU positive β-cells per 24 hours).9 Given that β-cell mass continues to expand into the second year, our observation implies that β-cells could be very long lived. Indeed, Clark and colleagues have recently reported that human β-cells are extremely long lived, as estimated by lipofuscin accumulation in cadaveric β-cells.10 Similarly, Perl and colleagues recently reported that β-cell turnover is limited to the first three decades of life as determined by in vivo thymidine analog incorporation and radiocarbon dating.11 Minimal β-cell turnover also implies that cell cycle entry of β-cells might be restricted with age. Consequently, we recently analyzed the effect of aging upon β-cell regeneration.12 We found that β-cell regeneration is severely and abruptly restricted by middle age in our cohort of mice. 50% partial pancreatectomy potently stimulated β-cell proliferation in young mice (2 or 8 months of age). But, partial pancreatectomy had little effect upon β-cell proliferation at 12 months, and no effect by 14 months or 19 months of age. Moreover, β-cell proliferation was stimulated by low dose streptozotocin (a β-cell toxin) in young mice but not in aged mice. Similarly, β-cell proliferation was stimulated by exendin-4 in young mice but not in aged mice. Taken together, these results reveal that adaptive β-cell proliferation is severely restricted with advanced age. Independent experiments by Bhushan and colleagues also showed that aged mice had little proliferative response to high fat diet, low dose streptozotocin or exendin-4.13 Thus, aging seems to be a previously unrealized factor that limits adaptive β-cell proliferation.
Much remains to be learned regarding the role of aging in β-cell proliferation. We recently studied the effect of aging upon islet gene expression using a candidate gene approach to measure mRNAs of cell cycle components.12 Several negative regulators of cell cycle progression were increased in aged islets compared to young islets, including p21, p16Ink4a, p15Ink4b and p19Ink4d. These results suggest that genomic studies could identify unique ageregulated genes or signals in islets. Moreover, truly important genes would be predicted to follow a pattern that correlates with the ability for β-cell regeneration. Consequently, we set out to interrogate the role of aging in β-cell proliferation using genomic expression studies in mice of various ages. Here we show that aging imparts a distinct gene expression program in aging mouse islets, with many genes that are differentially expressed in young vs. old islets. Partial pancreatectomy induces distinct gene expression patterns in both young and old islets. Surprisingly, Reg family genes were induced by partial pancreatectomy in mice of all ages, despite the lack of β-cell regeneration in aged mice. Still, few other genes were induced by partial pancreatectomy in both young and old mice.
Results
Aged islets exhibit a distinct gene expression pattern.
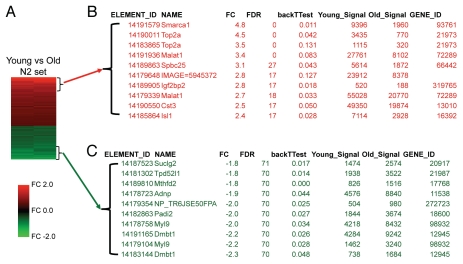
Based on our previous observations that β-cell proliferation declines in aged mice9,12,14, we compared basal gene expression in islets from young mice versus islets from aged mice. Islets were harvested under basal conditions from young mice (1.5 months) and aged mice (14 or 19 months) on a mixed genetic background (F1 hybrid B6129SF1/J). Purified total RNA was purity matched, as defined by endocrine/exocrine ratios. Samples were processed into cDNA and hybridized on spotted arrays (Mouse PancChip 6). Pair-wise gene expression analysis was performed on islets from young mice compared to islets from aged mice. Of the original 30 samples, eight pairs were selected for expression analysis. A total of eight pairs of hybridizations were performed, but four of the data sets were rejected for purity issues. Four samples remained, which were then more closely examined to match for purity. But, two of the remaining four samples exhibited modest exocrine contamination. The two sets that were the most highly matched for purity were designated as the N2 data set, and represented the main data set analyzed (Fig. 1 and Sup. Table 1). The larger gene list (the N4 data set) contained all four pairs, with the caveat that two of the samples included modest contamination by exocrine genes (Sup. Fig. 1 and Sup. Table 2).
Figure 1.
Aging is associated with distinct gene expression patterns in islets. Genomic microarray mRNA expression analysis of islets in the basal state, comparing islets from young mice (1.5 months of age) versus those from old mice (14 or 19 months of age). The two pair-wise arrays comprising in this data set (N2) represented the expression sets that exhibited the least exocrine contamination. (A) Heat map of the 91 elements of the PancChip microarray that were differentially expressed 1.5-fold change (FC) or more. Red: genes with higher expression in islets from young versus old mice. Green: genes with lower expression in islets from young versus old mice. (B and C) The most differentially expressed genes in islets from young versus old mice. (B) Genes that were expressed at higher levels in young mice. (C) Genes that were expressed at lower levels in young mice.
Aging was associated with distinct gene expression patterns in islets; many genes were expressed at higher levels in young islets compared to old islets. In all, 54 unique elements were more than 1.5-fold changed in young islets compared to old islets (Fig. 1 and Sup. Table 1). The most prominent was Smarca1 (expressed 4.8-fold higher in young islets). Smarca1 (a.k.a. SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1; Snf2l) encodes a component of chromatin remodeling complex, originally identified as the trithorax group (trxG) activator, which apposes the polycomb (PcG)-mediated silencing of gene expression.15 Decreased Smarca1 expression could allow transcription of target genes in aged islets. SNF complexes have been widely implicated in the pathogenesis of aging. Smarca6 deficient mice exhibit growth retardation and a premature aging phenotype.16 Smarca1 expression has been reported to decrease with age in developing heart, kidney and lung.17 Given that chromatin remodeling complexes potently regulate endocrine pancreas development,18 it seems likely that Smarca1 might influence β-cell growth or regeneration. Top2a expression was also present in greater amounts in young islets (3.5–4.5-fold). Top2a [a.k.a. topoisomerase (DNA) II alpha; Top2] is induced in islets from GK rats compared to islets from Wistar rats.19 Expression of spbc25 was also increased in young islets (3.1-fold). Spbc25 (a.k.a. spindle pole body component 25 homolog) is part of the NDC80 kinetochore complex that mediates segregation of chromosomes during mitosis.20 Igf2bp2 was also reduced in aged islets (2.8-fold, p < 0.05). Igf2bp2 (a.k.a. insulin-like growth factor 2 mRNA binding protein 2) regulates Igf2 expression in late embryonic development. Igf2bp2 has been identified in genome wide association studies to confer risk to type 2 diabetes mellitus.21 Several other RNA binding or ribosomal proteins were also expressed in greater amounts in young islets compared to old islets, including Eif4e (1.9-fold), Rpl4 (1.8-fold) and Rbm39 (1.5-fold). Two pancreatic endocrine transcription factors were also present in higher levels in islets from young mice: Isl1 (2.4-fold) and Pax4 (1.8-fold).
Transcripts for many genes (37 elements) were expressed at reduced levels 1.5-fold or greater in young islets compared to old islets (Table 1). Of the age-reduced genes, the most prominent was Dmbt1 (2.2–2.3-fold reduced with age). Dmbt1 (a.k.a. deleted in malignant brain tumors, CRP, Crpd, hensin) encodes a large scavenger receptor cysteine rich (SRCR) B protein that is highly expressed in liver and intestine.22,23 Myl9 is another gene that also differentially expressed (2.0–2.2-fold reduced with age). Myl9 (a.k.a. myosin, light polypeptide 9, regulatory; LC20; MLC2; MRLC1; MYRL2) is a 20 kDa myosin light chain that is known to be expressed at low levels in islets from young mice compared to other tissues.24 Interestingly, Myl9 expression in whole blood correlates with the presence of an established high-risk coronary artery disease risk allele in the 9p21.3 region.25 Padi2 is another gene that was differentially expressed (2.0-fold reduced with age). Padi2 (a.k.a. RP1-37C10.6, KIAA0994, PAD-H19, PAD2, PDI2) is a peptidylarginine deiminase. Padi2 has been observed to be more highly expressed in ngn3 expressing progenitors, and to progressively decline with islet development.26 Adnp is a homeodomain-class gene that was differentially expressed (1.9-fold reduced with age).
Partial pancreatectomy induces islet cell cycle component gene expression in young mice.
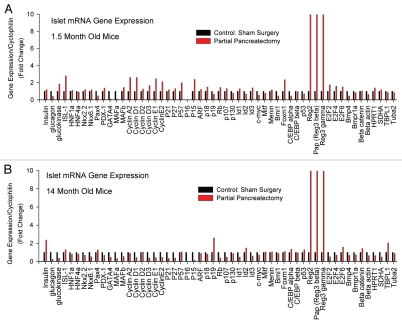
To define the effect of age upon β-cell regeneration we first measured expression of candidate genes by quantitative PCR (qPCR). We compared mRNA expression in islets after 50% partial pancreatectomy versus sham-operated controls, performing the entire experiment in young mice (1.5 months) and also in old mice (14 months). We measured expression across a broad range of genes, including islet hormones and enzymes (insulin, glucagon, etc.,), islet transcription factors (Pdx1, Isl1, HNF1a, etc.,), cell cycle components (cyclin A2, Rb, etc.,), negative regulators of cell cycle (p21 CIP1, p16 Ink4a, etc.,), transcriptional regulators of cell cycle (Bmi1, Men1, etc.,), and control genes (cyclophilin, β actin, etc.). In all, mRNAs of 55 genes were measured (Fig. 2). Partial pancreatectomy induced islet gene expression of several cell cycle components in young mice but not in old mice. For example, partial pancreatectomy induced cyclin A2 expression in islets from young mice. Cyclin A2 partners with Cdk2 to promote progression through the S and G2 phases of cell cycle. But, cyclin A2 was not induced in islets from aged mice. mRNA for cyclin D1, which participates in G1 cell cycle progression in β-cells,27 was also induced by partial pancreatectomy in islets from young mice. But, cyclin D1 was not induced by partial pancreatectomy in islets from aged mice. Similarly, mRNA for proproliferative transcription factor Foxm1 was also induced two-fold in islets from young mice but not in islets from aged mice. Foxm1 is an important regulator of β-cell proliferation.28 Interestingly, partial pancreatectomy induced insulin gene expression in old mice but not young mice. This result suggests that β-cells in aged mice may try to compensate for increased metabolic demand, but we have not yet tested this notion. Still, these candidate gene studies reveal that aging islets exhibit distinct gene expression patterns of cell cycle components. These gene expression changes may underlie the reduced regenerative capacity of aged β-cells.
Figure 2.
Partial pancreatectomy induces cell cycle component gene expression in islets from young mice but not in old mice. Candidate qPC R mRNA expression studies of islets after 50% partial pancreatectomy versus sham-operated controls. (A) Studies performed in young mice (1.5 months). (B) Studies performed in old mice (14 months). Results expressed as fold change from control.
Partial pancreatectomy induces reg gene expression in islets harvested from both young and old mice.
To test for the presence of regenerative signals we measured expression of mRNAs encoding the Reg (Regenerating) family proteins in islets from young mice and old mice after 50% partial pancreatectomy. We chose genes from the Reg family proteins because they have been repeatedly associated with β-cell regeneration.29 As expected, Reg2 (a.k.a islet of langerhans regenerating protein 2, pancreatic stone protein 2) was potently induced in islets from young mice (∼211-fold, Fig. 2). Surprisingly, Reg2 was also induced in islets from aged mice (∼34-fold, Fig. 2). Similarly, islet expression of Pap (a.k.a. pancreatitis-associated protein; HIP; INGAP; REG-III; Reg3b) was induced by partial pancreatectomy in both young and old mice (∼48-fold and ∼65-fold, Fig. 2). Pap is a Reg family protein that was originally cloned as a pancreatic islet neogenesis associated protein, and has been widely suggested as a potential therapeutic target to promote β-cell regeneration in diabetes.30,31 Reg3g expression in islets was also induced by partial pancreatectomy in both young and old mice (∼11-fold and ∼33-fold, Fig. 2). Thus, islet expression of Reg genes can be induced in both young and old mice. This result is surprising given the impaired β-cell regeneration after partial pancreatectomy in old mice, and implies that Reg gene expression may be dissociated from β-cell regeneration in aged mice.12
Partial pancreatectomy induces a distinct gene expression program in islets from young mice.
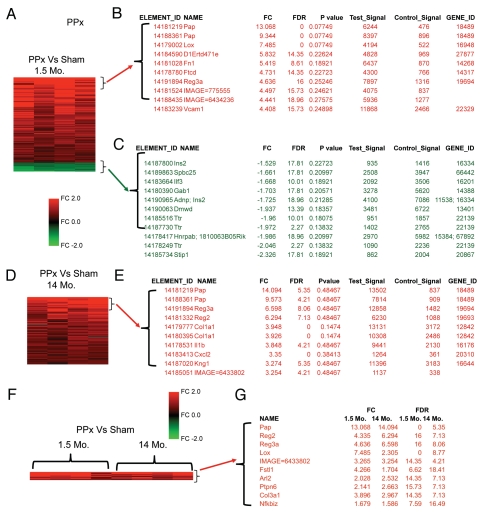
To further define the effect of age upon β-cell regeneration we performed genome-wide mRNA expression studies, comparing islets from young mice (1.5 months of age) after 50% partial pancreatectomy versus sham-operated controls (n = 4). As expected, partial pancreatectomy was associated with differential expression of many genes (104 elements were increased more than 1.5-fold) (Fig. 3A–C and Sup. Table 3). The most prominently induced genes were from the Reg family of proteins. At the top of the list of induced genes was Pap (9.3–13.0-fold change compared to sham). Reg3a was also stimulated by partial pancreatectomy (4.6-fold induced). Similarly, Reg2 was also induced in islets after partial pancreatectomy (4.3-fold induced). These results confirm our candidate gene qPCR gene expression studies and validate the use of genomic microarrays to interrogate islet gene expression.
Figure 3.
Partial pancreatectomy induces distinct gene expression patterns in islets from young mice versus old mice, except for Reg family genes. Genomic microarray mRNA expression analysis of islets after partial pancreatectomy (PPx) versus sham operated controls (Sham). (A–C) Partial pancreatectomy studies in young mice (1.5 months of age). (A) Heat map of the 115 elements of the PancChip microarray that were differentially expressed 1.5-fold or more. Red: genes with higher expression in islets after PPx. Green: genes with lower expression in islets after PPx. (B and C) The most differentially expressed genes in islets after PPx. (B) Genes that were expressed at higher levels after PPx. (C) Genes that were expressed at lower levels after PPx. (D and E) Partial pancreatectomy studies in old mice (14 months of age). (D) Heat map of the 83 elements of the PancChip microarray that were differentially expressed 1.5-fold or more. (E) Genes that were expressed at higher levels after PPx. (F and G) Overlap analysis of PPx induced genes in both young and old mice. (F) Heat map of the 10 elements of the PancChip microarray that were commonly induced 1.5-fold or more. (G) Genes that were commonly induced by PPx in both young and old mice.
Several TGFβ related genes were differentially expressed in islets from young mice after partial pancreatectomy, including Fstl1 (induced 4.2-fold). Fstl1 (a.k.a. follistatin-like 1) is an activin and myostatin antagonist. Fstl1 is closely related to Fstl3, which has established roles in glucose homeostasis in mice.32 Notably, follistatin and activins have been recently discovered to have roles in β-cell maturity, with potent effects to regulate insulin gene expression and insulin secretion.33 Tgfbr2 (a.k.a. transforming growth factor, beta receptor II) was also differentially expressed after partial pancreatectomy (3.8-fold induced). Lox was potently upregulated by partial pancreatectomy (7.4-fold induced). Lox (a.k.a. lysyl oxidase, TSC-160) is a copper containing amine oxidase that participates in collagen and elastin cross linking within the extracelluar matrix.34 Lox is cleaved and thus activated by BMP-1 and other related metalloproteinases.
Components of the IGF signaling cascade were differentially expressed in islets from young mice after partial pancreatectomy, including H19 (4.1-fold induced). H19 is an imprinted gene in the genetic region associated with Beckwith Weidemann syndrome, which is associated with hyperinsulinemia and hypoglycemia in affected children.35 Igfbp4 was also differentially expressed in islets from young mice after partial pancreatectomy (3.2-fold increased).
Tissue specific transcription factors were induced in islets from young mice after partial pancreatectomy, including TGIF (3.7-fold increased). TGIF (a.k.a. transforming growth interacting factor; Hpe4) is a three-amino acid loop extension (TALE) class of homeodomain proteins that act as transcriptional repressors in retinoid and TGFβ signals.36,37 Foxa1 is another transcription factor that was induced by partial pancreatectomy (2.4-fold). Foxa1 (a.k.a. Hnf3a, Tcf3a) is a winged-helix transcription factor that regulates insulin secretion in the mature β cell.38 TCF7L2 was also differentially expressed by partial pancreatatectomy (1.9-fold increased). TCF7L2 (a.k.a. Tcf4) is a high mobility group (HMG) box-containing transcription factor that interacts with β-catenin. Genetic changes in the TFC7L2 the genetic region are associated with type 2 diabetes.39
Several chromatin-regulating proteins were induced after partial pancreatectomy, including UHRF1 (2.3-fold). UHRF1 (a.k.a. NP95, ICBP90) is a PHD and ring finger containing protein that maintains DNA methylation by interacting with DNMT1.40 DNMT1 appears to be essential for pancreas development and β-cell regeneration in zebrafish.41
Islet mRNAs for a few genes were reduced by partial pancreatectomy in young mice (11 elements with 1.5-fold change). Reduced genes include Stip1 (2.3-fold decreased). Stip1 (a.k.a. stress-induced phosphoprotein 1; Hsp70–Hsp90 organizing protein; Hop; STI1; p60) is a cochaperone protein that has been implicated with protein folding.42 Stip1 has been indirectly implicated in β-cells. Pdx1 is known to occupy a site near Stip1.43 Stip1 may also be downstream of Foxo1 in β-cells.44 Ttr was also sharply downregulated after partial pancreatectomy in young mice (2.0-fold decreased). Hnrpab was also downregulated (2.0-fold decreased). Hnrpab (a.k.a. heterogeneous nuclear ribonucleoprotein A/B; Abbp1) is a heterogeneous nuclear ribonucleoprotein that is expressed in the developing endocrine pancreas.45 Other notable genes downregulated by partial pancreatectomy include insulin 2 (1.7-fold reduced).
Partial pancreatectomy induces limited changes in gene expression in islets from aged mice.
Based on our previous observation that adaptive β-cell proliferation is severely reduced in aged mice,12 we hypothesized that aging islets might exhibit reduced differential gene expression in response to partial pancreatectomy. We compared gene expression in islets from old mice (14 months of age) after 50% partial pancreatectomy versus sham-operated controls (n = 4 of each). Partial pancreatectomy in aged mice induced broad changes in gene expression (83 elements changed more than 1.5-fold) (Fig. 3D and E; Sup. Table 4). As expected from our qPCR based candidate gene studies (see above), partial pancreatectomy induced expression of Reg family genes in the aged mice. Pap was highly induced in islets from old mice (14.1-fold). Similarly, Reg3a was also induced in aged mice by partial pancreatectomy (6.6-fold). Reg 2 was also induced in islets after partial pancreatectomy (6.3-fold). These results confirm our candidate based qPCR gene expression studies. Thus, Reg family genes are potently induced in aged islets, despite their profoundly reduced capacity for adaptive β-cell proliferation. Partial pancreatectomy in aged mice induced islet mRNA expression of several other notable genes, including Col1a1 (3.9-fold), Il1b (3.8-fold) and Cxcl2 (3.2-fold).
Partial pancreatectomy induced islet gene expression changes in young mice were mostly unique. Of the 104 elements increased more than 1.5-fold in young mice, only 11 were also increased in old mice (Fig. 3F and G). Genes that were commonly induced by partial pancreatectomy in both young and old mice include Lox, Fstl1, Arl2, Ptpn6, Col3a1 and Nfkbiz (along with the aforementioned Reg genes).
We then tested for overlap across ages in the partial pancreatectomy induced genes from young mice. But, the vast majority of other genes induced by partial pancreatectomy were not also induced by partial pancreatectomy in old mice (Sup. Fig. 2A–C). We further for overlap in between the partial pancreatectomy induced gene set and those genes differentially expressed with age at the basal state. There was no correlation between the partial pancreatectomy induced genes at 1.5 months and the direction of change in the same genetic elements associated with young islets (Sup. Fig. 2D and E). Interestingly, some of the genes that were reduced by partial pancreatectomy in young mice were expressed at higher levels in young mice (Sup. Fig. 2F). These differentially expressed genes include Ins2, which encodes insulin. Taken together, these results indicate that aging a distinct gene expression program in islets.
Discussion
Aging is associated with differential mRNA islet expression of several classes of genes, including chromatin remodeling factors, RNA binding proteins and transcription factors. We used partial pancreatectomy to model regeneration-associated gene expression in islets. Partial pancreatectomy induced islet expression of cell cycle component mRNAs in young mice, including cyclin A2 and cyclin D1. But, partial pancreatectomy had minimal (if any) effect upon islet expression of cell cycle component mRNAs in aged mice. As expected, partial pancreatectomy induced islet expression of Reg genes in young mice. Surprisingly, partial pancreatectomy also induced islet expression of Reg genes in aged mice. Genome-wide mRNA expression studies revealed that partial pancreatectomy induced a broad range of gene expression changes in islets from both young and old mice. However, very few genes (besides those encoding Reg family proteins) were commonly induced in islets from young and old mice after partial pancreatectomy. Thus, aging imparts a distinct gene expression program in mouse islets.
Our study represents the first genomic microarray study of aging and regeneration in islets. The main strength of our study lies in the novel approach to interrogate the intersection between aging and islet regeneration. We identify several new genes that are differentially expressed with age and interrogate the role of aging in regeneration induced islet gene expression. The main weakness of our study is that we used whole islets instead of purifed β-cells to maximize yield (islet yields after partial pancreatectomy are severely reduced, necessitating the whole islet approach). Although we were careful to minimize contamination by non-islet components, we cannot exclude formally the possibility that contamination by nonendocrine cells within our islet preps might have altered gene expression patterns. In the future we anticipate that our study will inspire others to perform more detailed studies of purified cells to quantify the effect of aging in specific islet components (α-cells, β-cells, δ-cells, ductal cells, etc). Similarly, it might someday even be possible to perform gene expression studies of β-cells that are preparing to replicate, which could identify β-cell specific gene expression patterns that are associated with mitogenic signals.
Our results reveal that aging imparts distinct gene expression patterns upon islets. As expected, partial pancreatectomy failed to induce cell cycle component mRNA expression in islets of aged mice. These results are consistent with the lack of partial pancreatectomy induced β-cell regeneration previously observed in aged mice.12 However, partial pancreatectomy robustly induced Reg family gene expression in islets of aged mice. Thus, our data uncouple Reg gene expression from β-cell regeneration in aged mice. This result implies that Reg gene expression is insufficient to promote β-cell regeneration in aged mammals. Further, our work challenges the hypothesis that Reg family proteins such as INGAP or HIP might have therapeutic benefit in adult human islets. We hope our studies will inspire investigators to test Reg family protein based therapies in older rodents, which have less regenerative capacity than young rodents and thus may serve as more accurate models of human β-cell regeneration capacity.
The major implication of our study is that aging imparts a distinct gene expression program upon islets. Type 2 diabetes is largely a disease of the elderly. Thus, the role of aging in islet physiology and regeneration remains a central question in diabetes research. Putative β-cell regeneration therapies should be tested in both young and old animal models with accompanying genomic microarray gene expression studies to fully understand the potential role of such therapies in mature human β-cells.
Materials and Methods
Mice.
All experiments with mice were performed in the animal facility at The Children's Hospital of Philadelphia according to the guidelines of the IACUC. Male F1 hybrid B6129SF1/J mice (stock 101043) were obtained at 1 and 8 months of age from the Jackson Laboratory (Bar Harbor, Maine). The Jackson B6129SF1/J hybrid is the product of an intercross between C57BL/6J (000664) female mice and 129S1/SvImJ (002448) male mice from the Jackson lab's commercial colonies.
Partial pancreatectomy.
Partial pancreatectomy was performed as previously.3,12 The splenic portion of the pancreas was surgically removed, resulting in a ∼50% pancreatectomy. Sham operation was performed by opening the abdomen but leaving the pancreas intact.
Islet Isolation and mRNA preparation.
Partial pancreatectomy or sham operation was performed on mice of various ages (1.5 months, 8 months, 14 months and 19 months). We harvested islets five days later. Islets were isolated from individual mice and processed into cDNA, as previously.12 A total of 78 RNA samples were processed into cDNA.
Sample selection and cDNA processing.
Islet purity matching was then performed as previously described by Kaestner and colleagues.46 Briefly, qPCR was used to determine the relative levels of endocrine (insulin and pro-hormone convertase) and exocrine (amylase and chymotrypsin) gene expression levels in each sample. Samples were then matched for purity, pairing one test and one control condition of interest. Ten to twenty-five nanograms of total RNA was amplified using the Ovation™ RNA Amplification System V2 (Nugen Inc., CA).
Arrays.
Samples were hybridized to Mouse PancChip 6, which contains 13,059 mouse cDNAs chosen for their expression in various stages of pancreatic development, many of which are not found on commercially available arrays. Microarray slides were hybridized overnight, then washed and scanned with Agilent G2565BA microarray scanner.
For basal gene expression analysis to study gene expression in young mice versus old mice, a total of 30 samples from 12 biological replicates were used. After purity matching the final four pairs of islet samples from young mice versus old mice were selected (designated the N4 group). Of those four pairs of samples, two pairs exhibited some degree of exocrine contamination. The two most highly pure samples were used for further analysis (designated the N2 group).
Hybridizations for adaptive β-cell proliferation analysis worked very well in the samples from 1.5 months and 14 months. Other hybridizations were rejected due to purity issues, with only two samples remaining from 8 months and one sample from 19 months of age. Consequently, we only analyzed the results of samples obtained at 1.5 months and 14 months.
Analysis of arrays.
Median foreground intensities were obtained for each spot and imported into the mathematical software package “R” (www.R-project.org/). Statistical analysis was performed in “R” using both the LIMMA and SAM packages. SAM uses a False Discovery Rate (FDR) and q-value to predict false positives.47 A one class unpaired analysis with an FDR of 10% was used as a cutoff. In this method, the lower the q-value (which describes the false discovery rate), the more robust the result. In this report we only reported results with FDR q-value of 20% or less to maximize reporting of genes that were reliably differentially expressed. Sub-analyses of the two purest basal gene expression sets were then performed, which limited the utility of FDR reporting for this data set (designated N2). As a result, the N2 samples were reported a back calculated T test as permutation-based method to estimate false discovery rates.
Candidate gene based RT-PCR.
Partial pancreatectomy or sham operation was performed on mice at 1.5 months or 14 months of age. Five days later, islets were isolated from pools of at least four mice and processed into cDNA. Real time quantitative dual fluorescent labeled FRET PCR was performed as previously.12 Relative gene product amounts were reported for each gene compared with cyclophilin and confirmed in separate studies with a large panel of control genes. See Supplemental Table 5 for primer sequences. Results are reported from a single experiment with technical repeats averaged and reported as a mean. Representative RT-PCR data for Reg genes was confirmed in an independent experiment with islet mRNAs derived from equivalent groups of mice (data not shown).
Acknowledgements
We thank Peter White and Alan J. Fox for their expert assistance with carrying out the microarray work. We also thank Klaus Kaestner for helpful advice. We also thank Jonathan Schug for help with expression data archiving.
Footnotes
Previously published online: www.landesbioscience.com/journals/islets/article/13376
Accession Code
ArrayExpress E-MTAB-433
Author Contributions
Conceived and designed the experiments: M.M.R. and J.A.K. Performed the experiments: M.M.R. Analyzed the data: M.M.R and J.A.K. Wrote the paper: J.A.K.
Financial Support
Supported by the JDRF, the NIH (R01-DK081469), the Commonwealth of Pennsylvania (Center for Excellence in Regenerative Medicine grant 4100043362), the March of Dimes (Basil O'Connor Starter Scholar Research Award), and a University of Pennsylvania DERC pilot and feasibility grant (DK19525). Microarrays were carried out by the Functional Genomics Core, supported by the NIH (P30-DK050306).
Precis
mRNA islet expression studies reveal differential expression of many genes with age in the basal state and during regeneration.
Supplementary Material
References
- 1.Ku HT. Minireview: pancreatic progenitor cells—recent studies. Endocrinology. 2008;149:4312–4316. doi: 10.1210/en.2008-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by selfduplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- 3.Teta M, Rankin MM, Long SY, Stein GM, Kushner JA. Growth and regeneration of adult Beta cells does not involve specialized progenitors. Dev Cell. 2007;12:817–826. doi: 10.1016/j.devcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 4.Bock T, Pakkenberg B, Buschard K. Increased islet volume but unchanged islet number in ob/ob mice. Diabetes. 2003;52:1716–1722. doi: 10.2337/diabetes.52.7.1716. [DOI] [PubMed] [Google Scholar]
- 5.Parsons JA, Brelje TC, Sorenson RL. Adaptation of islets of Langerhans to pregnancy: increased islet cell proliferation and insulin secretion correlates with the onset of placental lactogen secretion. Endocrinol. 1992;130:1459–1466. doi: 10.1210/endo.130.3.1537300. [DOI] [PubMed] [Google Scholar]
- 6.Scharfmann R. Expanding human beta cells. Diabetologia. 2008;51:692–693. doi: 10.1007/s00125-007-0909-4. [DOI] [PubMed] [Google Scholar]
- 7.Menge BA, Tannapfel A, Belyaev O, Drescher R, Muller C, Uhl W, et al. Partial pancreatectomy in adult humans does not provoke beta-cell regeneration. Diabetes. 2008;57:142–149. doi: 10.2337/db07-1294. [DOI] [PubMed] [Google Scholar]
- 8.Finegood DT, Scaglia L, Bonner Weir S. Dynamics of beta-cell mass in the growing rat pancreas. Estimation with a simple mathematical model. Diabetes. 1995;44:249–256. doi: 10.2337/diab.44.3.249. [DOI] [PubMed] [Google Scholar]
- 9.Teta M, Long SY, Wartschow LM, Rankin MM, Kushner JA. Very slow turnover of beta-cells in aged adult mice. Diabetes. 2005;54:2557–2567. doi: 10.2337/diabetes.54.9.2557. [DOI] [PubMed] [Google Scholar]
- 10.Cnop M, Hughes SJ, Igoillo-Esteve M, Hoppa MB, Sayyed F, van de Laar L, et al. The long lifespan and low turnover of human islet beta cells estimated by mathematical modelling of lipofuscin accumulation. Diabetologia. 53:321–330. doi: 10.1007/s00125-009-1562-x. [DOI] [PubMed] [Google Scholar]
- 11.Perl SKJ, Buchholz BA, Meeker AK, Stein GM, Hsieh M, Kirby M, et al. Significant human beta-cell turnover is limited to the first three decades of life as determined by in-vivo thymidine analog incorporation and radiocarbon dating. J Clin Endocrinol Metab. 2010;95:E234–E239. doi: 10.1210/jc.2010-0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rankin MM, Kushner JA. Adaptive beta-cell proliferation is severely restricted with advanced age. Diabetes. 2009;58:1365–1372. doi: 10.2337/db08-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tschen SI, Dhawan S, Gurlo T, Bhushan A. Age-dependent decline in beta-cell proliferation restricts the capacity of beta-cell regeneration in mice. Diabetes. 2009;58:1312–1320. doi: 10.2337/db08-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He LM, Sartori DJ, Teta M, Opare-Addo LM, Rankin MM, Long SY, et al. Cyclin D2 protein stability is regulated in pancreatic beta-cells. Mol Endocrinol. 2009;23:1865–1875. doi: 10.1210/me.2009-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Sun LQ, Lee DW, Zhang Q, Xiao W, Raabe EH, Meeker A, et al. Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes Dev. 2004;18:1035–1046. doi: 10.1101/gad.1176104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finkielstain GP, Forcinito P, Lui JC, Barnes KM, Marino R, Makaroun S, et al. An extensive genetic program occurring during postnatal growth in multiple tissues. Endocrinology. 2009;150:1791–1800. doi: 10.1210/en.2008-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haumaitre C, Lenoir O, Scharfmann R. Directing cell differentiation with small-molecule histone deacetylase inhibitors: the example of promoting pancreatic endocrine cells. Cell Cycle. 2009;8:536–544. doi: 10.4161/cc.8.4.7610. [DOI] [PubMed] [Google Scholar]
- 19.Homo-Delarche F, Calderari S, Irminger JC, Gangnerau MN, Coulaud J, Rickenbach K, et al. Islet inflammation and fibrosis in a spontaneous model of type 2 diabetes, the GK rat. Diabetes. 2006;55:1625–1633. doi: 10.2337/db05-1526. [DOI] [PubMed] [Google Scholar]
- 20.Bharadwaj R, Qi W, Yu H. Identification of two novel components of the human NDC80 kinetochore complex. J Biol Chem. 2004;279:13076–13085. doi: 10.1074/jbc.M310224200. [DOI] [PubMed] [Google Scholar]
- 21.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mollenhauer J, Wiemann S, Scheurlen W, Korn B, Hayashi Y, Wilgenbus KK, et al. DMBT1, a new member of the SRCR superfamily, on chromosome 10q25.3-26.1 is deleted in malignant brain tumours. Nat Genet. 1997;17:32–39. doi: 10.1038/ng0997-32. [DOI] [PubMed] [Google Scholar]
- 23.Bisgaard HC, Holmskov U, Santoni-Rugiu E, Nagy P, Nielsen O, Ott P, et al. Heterogeneity of ductular reactions in adult rat and human liver revealed by novel expression of deleted in malignant brain tumor 1. Am J Pathol. 2002;161:1187–1198. doi: 10.1016/S0002-9440(10)64395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pullen TJ, Kahn AM, Baron G, Butcher SA, Sun S, Rutter GA. Identification of genes selectively disallowed in the pancreatic islet. Islets. 2010;2:1–8. doi: 10.4161/isl.2.2.11025. [DOI] [PubMed] [Google Scholar]
- 25.Jarinova O, Stewart AF, Roberts R, Wells G, Lau P, Naing T, et al. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol. 2009;29:1671–1677. doi: 10.1161/ATVBAHA.109.189522. [DOI] [PubMed] [Google Scholar]
- 26.White P, May CL, Lamounier RN, Brestelli JE, Kaestner KH. Defining pancreatic endocrine precursors and their descendants. Diabetes. 2008;57:654–668. doi: 10.2337/db07-1362. [DOI] [PubMed] [Google Scholar]
- 27.Kushner JA, Ciemerych MA, Sicinska E, Wartschow LM, Teta M, Long SY, et al. Cyclins D2 and D1 Are Essential for Postnatal Pancreatic {beta}-Cell Growth. Mol Cell Biol. 2005;25:3752–3762. doi: 10.1128/MCB.25.9.3752-3762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Ackermann AM, Gusarova GA, Lowe D, Feng X, Kopsombut UG, et al. The FoxM1 transcription factor is required to maintain pancreatic beta-cell mass. Mol Endocrinol. 2006;20:1853–1866. doi: 10.1210/me.2006-0056. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto H, Takasawa S. Recent advances in the Okamoto model: the CD38-cyclic ADP-ribose signal system and the regenerating gene protein (Reg)-Reg receptor system in betacells. Diabetes. 2002;51:462–473. doi: 10.2337/diabetes.51.2007.s462. [DOI] [PubMed] [Google Scholar]
- 30.Rafaeloff R, Pittenger GL, Barlow SW, Qin XF, Yan B, Rosenberg L, et al. Cloning and sequencing of the pancreatic islet neogenesis associated protein (INGAP) gene and its expression in islet neogenesis in hamsters. J Clin Invest. 1997;99:2100–2109. doi: 10.1172/JCI119383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madrid V, Del Zotto H, Maiztegui B, Raschia MA, Alzugaray ME, Boschero AC, et al. Islet neogenesis-associated protein pentadecapeptide (INGAP-PP): mechanisms involved in its effect upon beta-cell mass and function. Regul Pept. 2009;157:25–31. doi: 10.1016/j.regpep.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 32.Mukherjee A, Sidis Y, Mahan A, Raher MJ, Xia Y, Rosen ED, et al. FSTL3 deletion reveals roles for TGFbeta family ligands in glucose and fat homeostasis in adults. Proc Natl Acad Sci USA. 2007;104:1348–1353. doi: 10.1073/pnas.0607966104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szabat M, Johnson JD, Piret JM. Reciprocal modulation of adult beta cell maturity by activin A and follistatin. Diabetologia. 53:1680–1689. doi: 10.1007/s00125-010-1758-0. [DOI] [PubMed] [Google Scholar]
- 34.Ge G, Greenspan DS. Developmental roles of the BMP1/TLD metalloproteinases. Birth Defects Res C Embryo Today. 2006;78:47–68. doi: 10.1002/bdrc.20060. [DOI] [PubMed] [Google Scholar]
- 35.Catchpoole D, Lam WW, Valler D, Temple IK, Joyce JA, Reik W, et al. Epigenetic modification and uniparental inheritance of H19 in Beckwith-Wiedemann syndrome. J Med Genet. 1997;34:353–359. doi: 10.1136/jmg.34.5.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bertolino E, Reimund B, Wildt-Perinic D, Clerc RG. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J Biol Chem. 1995;270:31178–31188. doi: 10.1074/jbc.270.52.31178. [DOI] [PubMed] [Google Scholar]
- 37.Wotton D, Lo RS, Lee S, Massague J. A Smad transcriptional corepressor. Cell. 1999;97:29–39. doi: 10.1016/s0092-8674(00)80712-6. [DOI] [PubMed] [Google Scholar]
- 38.Vatamaniuk MZ, Gupta RK, Lantz KA, Doliba NM, Matschinsky FM, Kaestner KH. Foxa1-deficient mice exhibit impaired insulin secretion due to uncoupled oxidative phosphorylation. Diabetes. 2006;55:2730–2736. doi: 10.2337/db05-0470. [DOI] [PubMed] [Google Scholar]
- 39.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 40.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 41.Anderson RM, Bosch JA, Goll MG, Hesselson D, Dong PD, Shin D, et al. Loss of Dnmt1 catalytic activity reveals multiple roles for DNA methylation during pancreas development and regeneration. Dev Biol. 2009;334:213–223. doi: 10.1016/j.ydbio.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi F, Doudevski I, Regan L. HOP is a monomer: investigation of the oligomeric state of the co-chaperone HOP. Protein Sci. 19:19–25. doi: 10.1002/pro.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sachdeva MM, Claiborn KC, Khoo C, Yang J, Groff DN, Mirmira RG, et al. Pdx1 (MODY4) regulates pancreatic beta cell susceptibility to ER stress. Proc Natl Acad Sci USA. 2009;106:19090–19095. doi: 10.1073/pnas.0904849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buteau J, Shlien A, Foisy S, Accili D. Metabolic diapause in pancreatic beta-cells expressing a gain-offunction mutant of the forkhead protein Foxo1. J Biol Chem. 2007;282:287–293. doi: 10.1074/jbc.M606118200. [DOI] [PubMed] [Google Scholar]
- 45.Juhl K, Sarkar SA, Wong R, Jensen J, Hutton JC. Mouse pancreatic endocrine cell transcriptome defined in the embryonic Ngn3-null mouse. Diabetes. 2008;57:2755–2761. doi: 10.2337/db07-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White P, Kaestner KH. Gene expression analysis in diabetes research. Methods Mol Biol. 2009;560:239–261. doi: 10.1007/978-1-59745-448-3_16. [DOI] [PubMed] [Google Scholar]
- 47.Storey JD. A direct approach to false discovery rates. J R Statist Soc B. 2002;64:479–498. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.