Abstract
Several engineered selenocysteine (Sec)-containing glutaredoxins (Grxs) and their enzymatic properties have been reported, but natural selenoprotein Grxs have not been previously characterized. We expressed a bacterial selenoprotein Grx from Clostridium sp. (also known as Alkaliphilus oremlandii) OhILAs in Escherichia coli and characterized this selenoenzyme and its natural Cys homologs in Clostridium and E. coli. The selenoprotein Grx had a 200-fold higher activity than its Sec-to-Cys mutant form, suggesting that Sec is essential for catalysis by this thiol-disulfide oxidoreductase. Kinetic analysis also showed that the selenoprotein Grx had a 10-fold lower Km than Cys homologs. Interestingly, this selenoenzyme efficiently reduced a Clostridium selenoprotein methionine sulfoxide reductase A (MsrA), suggesting that it is the natural reductant for the protein that is not reducible by thioredoxin, a common reductant for Cys-containing MsrAs. We also found that the selenoprotein Grx could not efficiently reduce a Cys version of Clostridium MsrA, whereas natural Clostridium and E. coli Cys-containing Grxs, which efficiently reduce Cys-containing MsrAs, poorly acted on the selenoprotein MsrA. This specificity for MsrA reduction could explain why Sec is utilized in Clostridium Grx and more generally provides a novel example of the use of Sec in biological systems.
Keywords: Glutaredoxin, methionine sulfoxide reductase, selenocysteine, selenoprotein, MsrA
Introduction
Glutaredoxin (Grx) and thioredoxin (Trx) systems are two major redox systems that maintain cellular redox homeostasis (Lillig et al., 2008, Lillig & Holmgren, 2007, Holmgren et al., 2005). Grxs are small, heat-stable thiol-disulfide oxidoreductases that were first discovered as a glutathione (GSH)-dependent hydrogen donor for Escherichia coli ribonucleotide reductase (Holmgren, 1976). The Grx system is composed of NADPH, GSH, GSH reductase and Grx, wherein electrons are transferred from NADPH to Grx via GSH. Grxs can be classified into two groups, dithiol and monothiol Grxs, according to the number of Cys residues in the active site. Dithiol Grxs involve two Cys residues in catalysis and contain a characteristic CPYC motif (or its close derivatives) in the active site. One of these Cys acts as a catalytic residue and the other as a resolving Cys. These enzymes primarily catalyze the reduction of protein disulfides. Monothiol Grxs lack the C-terminal active site thiol and instead CGFS sequence is typically present in the active site. They are mainly responsible for the reduction of disulfides between GSH and proteins leading to so-called deglutathionylation in proteins.
Selenocysteine (Sec) is a rare amino acid found in catalytic sites of certain oxidoreductases (Stadtman, 1996, Bock et al., 1991, Cone et al., 1976, Gladyshev et al., 1996). This amino acid is inserted into proteins during translation wherein UGA (normally a stop codon) is recoded for the insertion of Sec (Hatfield & Gladyshev, 2002, Driscoll & Copeland, 2003). A stem-loop structure, called SECIS (Sec insertion sequence) element, must be present in selenoprotein mRNA for co-translational Sec insertion. SECIS elements are located in the 3′-untranslational regions in archaea and eukaryotes, whereas these elements are present immediately downstream of the Sec UGA codons in bacteria. In addition to SECIS element, several trans-acting factors, such as SECIS binding protein, tRNASec, and Sec-specific elongation factor, are required for Sec incorporation. A consensus bacterial SECIS structural model has been reported in which a conserved G nucleotide is present in the apical loop of SECIS (Zhang & Gladyshev, 2005). Studies on the E. coli SECIS elements showed that a GU sequence on the tip of the apical loop and a bulged U at the bottom of the stem are important for the function of SECIS (Fourmy et al., 2002, Liu et al., 1998, Sandman et al., 2003). However, the bulged U is missing in most other bacterial SECIS elements (Zhang & Gladyshev, 2005). The differences in SECIS elements between E. coli and other bacteria result in barriers and often failure in expression of heterologous selenoproteins in E. coli (Tormay & Bock, 1997).
We previously characterized the selenoproteome of an anaerobic gram-positive bacterium, Clostridium sp. (also known as Alkaliphilus oremlandii) OhILAs (Kim et al., 2009). This bacterium was found to be a selenoprotein-rich organism containing at least 13 Sec-containing proteins. Grx was one of selenoproteins identified in Clostridium sp. OhILAs. Selenoprotein forms of this protein were also found in some other bacteria (Zhang et al., 2006). However, characterization of natural selenoprotein Grxs has not been reported. Instead, a few chemically or biologically engineered Sec-containing Grxs have recently been described including E. coli Grx1, Grx3, and a Grx domain of mouse Trx/GSH reductase (Casi et al., 2008, Ge et al., 2009, Metanis et al., 2006).
In this study, we characterized the Clostridium selenoenzyme Grx and its Sec-to-Cys mutant form, a natural Clostridium Cys-containing Grx, and an E. coli Grx. We found that, as expected, the use of Sec offered a higher catalytic competency to this thiol oxidoreductase. Most interestingly, the Clostridium selenoprotein Grx efficiently reduced selenoprotein methionine sulfoxide reductase A (MsrA), whereas the Clostridium Cys-containing homolog or E. coli Grx, which could efficiently reduce Cys-containing MsrAs, poorly acted on the selenoprotein MsrA.
Results
Clostridium selenoprotein Grx and Cys-containing Grx sequences
Grx is a thiol-disulfide oxidoreductase (also called thioltransferase) that can reduce disulfides in proteins or small molecules and mixed disulfides between small molecules and proteins (Lillig et al., 2008). Clostridium sp. OhILAs was previously found to be rich in Sec-containing proteins; i.e., it contains at least 13 selenoproteins (8 known and 5 predicted) (Kim et al., 2009). The known selenoproteins identified include formate dehydrogenase alpha subunit (FdhA), selenophosphate synthetase (SelD), proline reductase, glycine reductases A and B, HesB-like protein, MsrA, and Grx. Selenoprotein Grxs have also been identified in certain other bacteria (Supplementary Figure S1), but are absent in eukaryotes (Zhang et al., 2006).
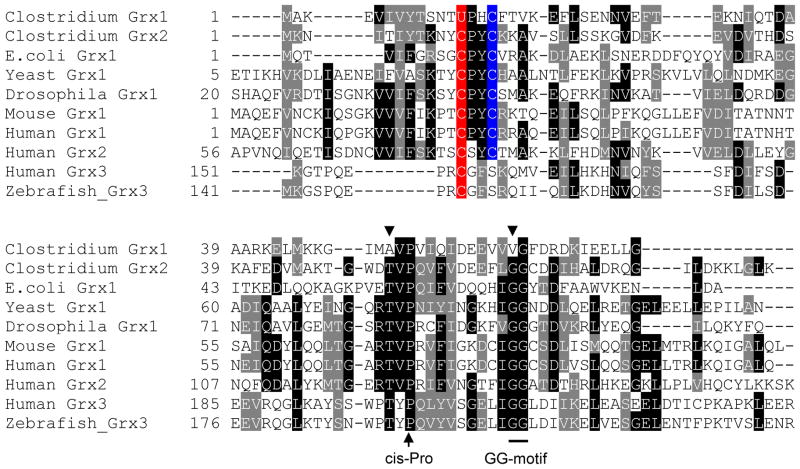
The Clostridium sp. OhILAs genome also contains a separate Cys-containing Grx. We designated the Clostridium selenoprotein Grx as cGrx1 and the Cys-containing Grx as cGrx2. cGrx1 has 76 amino acids and contains UPHC (U, Sec) catalytic motif in the N-terminal portion (Figure 1). Translation of cGrx2 appears to be initiated at the UUG codon. This protein has 85 amino acids including a typical catalytic CPYC motif present in the N-terminal region (Figure 1). Thus, both selenoprotein cGrx1 and Cys-containing cGrx2 belong to dithiol Grxs. Multiple sequence alignment revealed differences between selenoprotein cGrx1 and known Cys-containing Grxs in the GSH binding region (Figure 1): (i) The cis-Pro region. The consensus sequence is TVP, which is present in all Cys-containing dithiol Grxs. However, in cGrx1, this sequence changed to AVP; (ii) The GG-motif. As shown in Figure 1, cGrx1 contained a VG sequence instead of the conserved GG sequences in Cys-containing Grxs.
Figure 1. Multiple sequence alignment of Sec- and Cys-containing Grxs.
Catalytic Sec (U) and Cys residues are highlighted in red. Resolving Cys residues in the Sec- and Cys-containing dithiol Grxs are shown in blue. Two variant amino acid sequences in the GSH binding region of Clostridium selenoprotein Grx are indicated by arrowheads. Human and zebrafish Grx3s are represented as monothiol Grxs that contain the CGFS catalytic motif. GenBank accession numbers are as follows: Clostridium sp. OhILAs Grx1, 158321154; Clostridium Grx2, 158139258; E. coli Grx1, 146273; Saccharomyces cerevisiae Grx1, 5328; Drosophila melanogaster Grx1, 16226192; Mus musculus Grx1, 31981458; Homo sapiens Grx1, 531405; H. sapiens Grx2, 15077235; H. sapiens Grx3, 95113651; Danio rerio Grx3, 54400406.
Expression of Clostridium selenoprotein Grx in E. coli
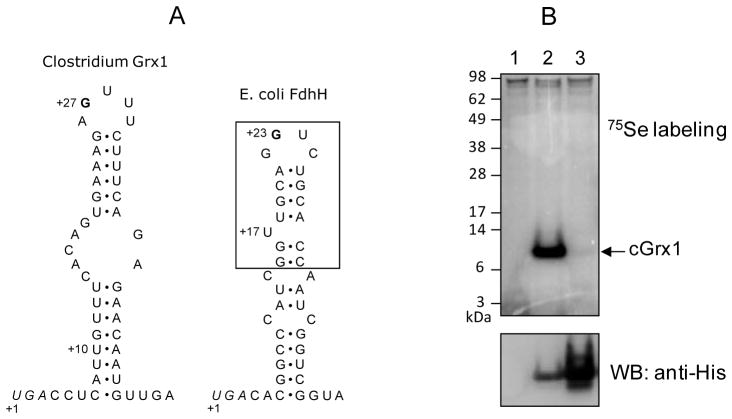
Consistent with the consensus bacterial SECIS structural model, the SECIS element of cGrx1 had a G in the apical loop, but lacked a bulged U in the stem, which is present in the E. coli SECIS element (Figure 2A). To test whether the SECIS element of cGrx1 can be recognized by the E. coli Sec insertion machinery, we metabolically labeled E. coli cells expressing cGrx1 with 75Se. Also, cells transformed with a Sec-to-Cys mutant construct were labeled to determine whether selenium incorporation is specific into Sec residue. As shown in upper panel of Figure 2B, a 10 kDa radioactive band corresponding in size to the calculated molecular mass of recombinant cGrx1 was detected (lane 2). In contrast, no apparent Sec-to-Cys mutant protein band was detected (lane 3) in the phosphor image, although its high expression level (Western blot of lower panel). We also verified the expression of selenoprotein cGrx1 with a C-terminal His tag by Western blot analysis using anti-His antibodies. Thus, Clostridium selenoprotein Grx can be expressed in E. coli with specific selenium incorporation into Sec, indicating that its SECIS element is functional in the E. coli Sec insertion system.
Figure 2. Structure of the SECIS element and expression of Clostridium selenoprotein Grx in E. coli.
(A) Structures of SECIS elements of Clostridium Grx1 (left) and E. coli fdhH (right). The Clostridium Grx1 SECIS contains a conserved G nucleotide in the apical loop, but lacks a bulged U, which is present in the minimal step-loop structure (boxed) in E. coli fdhH SECIS. Sec UGA codons are shown in italics and numbering of nucleotides is initiated at these Sec codons. The U at +10 position of Clostridium Grx1 SECIS is changed to A in monoselenol form of the protein. The SECIS structure of Clostridium Grx1 was predicted with RNAfold (Schuster et al., 1994). (B) 75Se metabolic labeling. E. coli BL21(DE3) cells transformed with an empty vector (lane 1), pET-CLOS-Grx1 (lane 2), or pET-CLOS-Grx1/U13C (lane 3) were metabolically labeled with 75Se. Proteins were separated by SDS-PAGE, transferred onto a PVDF membrane, and visualized with a PhosphorImager (upper panel). The same membrane was immunoblotted with anti-His antibody (lower panel).
Catalytic advantage provided by Sec in Grx
To investigate catalytic properties of cGrx1 and cGrx2, we purified these proteins from E. coli cells (Supplementary Figure S2A). We also cloned, expressed and purified E. coli Grx1 (eGrx1) for comparison. We performed the standard Grx activity assay using hydroxyethyl disulfide (HED) as substrate as described in Experimental Procedures. Notably, a significant Trp insertion in cGrx1 (83% of total protein) mediated by opal UGA suppression was identified by latter mass spectrometry analysis. To be sure whether Trp-inserted cGrx1 form is enzymatically inactive, we generated a Trp mutant cGrx1/U13W, purified this protein from E. coli cells, and assayed its activity. As expected, this Trp mutant was completely inactive (Table 1). Thus, for determining kcat value of cGrx1, the protein concentration of cGrx1 was normalized by subtracting the concentration of Trp-inserted form. The kcat value of cGrx1 was approximately 200-fold higher than that of its Cys mutant (cGrx1/U13C) while the Km value was similar (Table 1). These data show that Sec is essential for catalysis by cGrx1. Interestingly, the Km values of both selenoprotein cGrx1 and its Cys mutant were 10-fold lower than those of Cys homologs, cGrx2 and eGrx1. The kcat value of selenoprotein cGrx1 was 2-fold lower than that of Cys-containing cGrx2, but the catalytic efficiency, kcat/Km, was 5-fold higher that of cGrx2. The kinetic parameters of cGrx2 appeared to be similar to those of eGrx1.
Table 1.
Kinetic parameters of Clostridium Grx forms
| Protein | kcat (min−1) | Km (HED) (mM) | kcat/Km (mM−1 min−1) |
|---|---|---|---|
| cGrx1 | 87 ± 15 | 1.5 ± 0.4 | 58 |
| cGrx1/U13C | 0.5 ± 0.1 | 1.5 ± 0.8 | 0.3 |
| cGrx1/U13W | ND | - | - |
| cGrx1/C16S | 83 ± 8 | 0.9 ± 0.2 | 92 |
| cGrx2 | 186 ± 28 | 17.6 ± 5.6 | 11 |
| cGrx2/C15S | 396 ± 67 | 18.1 ± 4.5 | 22 |
| eGrx1 | 126 ± 8 | 11.2 ± 2.2 | 11 |
Purified proteins were assayed in the standard Grx reaction using HED as a substrate. Km and kcat values were determined by fitting the experimental data to the Michaelis-Menten equation. The protein concentrations of selenoprotein Grxs (cGrx1 and cGrx1/C16S) were normalized by subtracting the concentrations of Trp-inserted forms and subjected to calculating kcat values. cGrx1, wild-type selenoprotein Clostridium Grx1; cGrx1/U13C, Sec-to-Cys mutant of cGrx1; cGrx1/U13W, Sec-to-Trp mutant of cGrx1; cGrx1/C16S, monoselenol mutant of cGrx1; cGrx2, wild-type Cys-containing Clostridium Grx2; cGrx2/C15S, monothiol mutant of cGrx2; eGrx1, E. coli Grx1. ND, activity not detectable.
We also generated a monoselenol form of cGrx1 (cGrx1/C16S) in which Cys16 was replaced with Ser and a monothiol form of cGrx2 (cGrx2/C15S) in which Cys15 was substituted to Ser. The mutation that led to cGrx1/C16S included a single nucleotide change (U to A) in the lower stem of the SECIS element (Figure 2A, left), and therefore should not have interfered with Sec insertion in E. coli. Indeed, this protein was expressed as verified by Western blot with anti-His antibodies and mass spectrometry analyses.
A Trp insertion by opal suppression was also analyzed in the cGrx1/C16S form (the ratio of Trp to Sec was similar to that in wild-type cGrx1). Similarly, the protein concentration of cGrx1/C16S was normalized by subtracting the concentration of Trp-inserted form and subjected to determining kcat value. The kinetic parameters of cGrx1/C16S appeared to be similar to those of wild-type cGrx1 with a slightly higher catalytic efficiency (Table 1). We also obtained kinetic properties of monothiol cGrx2/C15S and compared them with the wild-type dithiol cGrx2. cGrx2/C15S showed a 2-fold increased kcat, compared to the wild-type dithiol form, while the Km value remained unchanged. Thus, the kcat/Km value of the monothiol cGrx2/C15S was 2-fold higher than that of the wild-type dithiol cGrx2. These data suggest that conversion of dithiol to monothiol in cGrx2 somewhat enhances catalytic efficiency.
Redox potentials of Clostridium Grxs
We further determined redox potentials of Clostridium Grxs using an AMS/gel-shift method: the proteins were equilibrated with a GSH/GSSG buffer, followed by alkylation with AMS and mobility of oxidized (non-modified) and reduced (modified; slower migration) forms were analyzed using non-reducing SDS-PAGE. The redox potentials of Cys mutant of cGrx1 (cGrx1/U13C) and cGrx2 were calculated as −215 mV and −210 mV, respectively. These values are within the range of redox potentials of Grxs characterized previously (Aslund et al., 1997, Su & Gladyshev, 2004). We could not determine the redox potential of selenoprotein cGrx1.
It has been reported that a chemically engineered selenoprotein form of E. coli Grx3 exhibits lower (more negative) redox potential than Cys-containing wild-type Grx3 (−260 mV vs −194 mV) (Metanis et al., 2006). Thus, the redox potential of our selenoprotein cGrx1 would be expected to be lower than that of the Cys mutant form. However, unexpectedly, only reduced forms of selenoprotein cGrx1 were observed in GSH redox buffers ranging from −240 mV to −193 mV of redox potentials. It is possible that Sec insertion may be low and instead Trp is inserted by opal suppression in a significant fraction of protein molecules expressed in E. coli. Then, the protein would appear as the redox-insensitive form at any GSH/GSSG ratio. Indeed, the insertion ratio of Trp to Sec in selenoprotein cGrx1 was semi-quantitatively determined to be 5:1 by counting the number of identified peptides with tandem mass spectrometry analysis (Supplementary Figure S3).
No apparent peroxidase activity of selenothiol and monoselenol Clostridium Grxs
Previous reports showed that engineered selenoprotein Grxs displayed strong GSH peroxidase (Gpx) activity (Casi et al., 2008, Ge et al., 2009). In particular, the artificial monoselenol Grx domain from mouse Trx/GSH reductase exhibited a high Gpx activity comparable to that of Gpx1 from rabbit liver (Ge et al., 2009). Thus, it was of interest to examine whether cGrx1 also exhibits Gpx activity. We measured the Gpx activity with H2O2 and used Gpx1 from bovine erythrocytes as a positive control. As shown in Figure 3, the Gpx activity of selenothiol cGrx1 was not detected while the activity of the control Gpx1 enzyme was 39,000 nmol·min−1·mg protein−1. Moreover, the monoselenol cGrx1/C16S also was inactive. No Gpx activity was also detected in all Cys-containing dithiol and monothiol Grxs tested (cGrx2, cGrx2/C15S, and eGrx1).
Figure 3. Lack of peroxidase activity by Clostridium selenoprotein Grx forms.
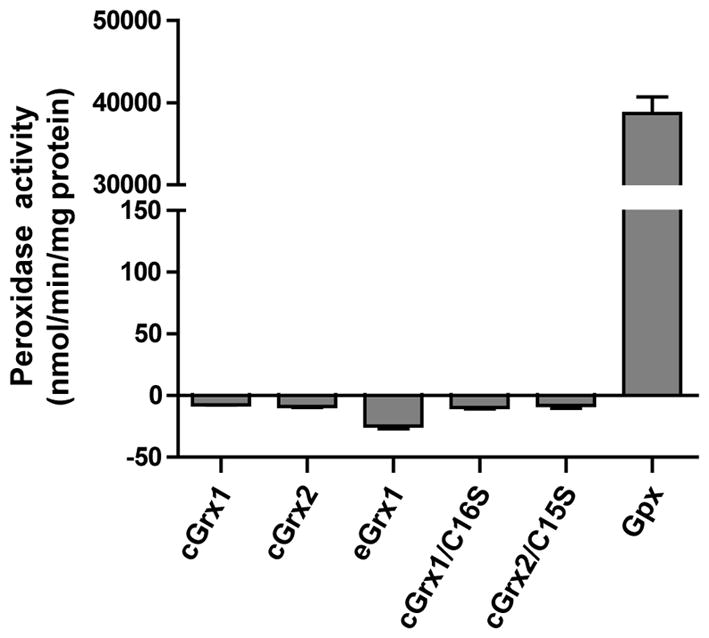
Peroxidase activity of Sec- and Cys-containing Grxs was measured with H2O2. Bovine erythrocyte Gpx1 was used as positive control for this assay. The decreased absorbance values of samples at 340 nm were subtracted from that of the background without the enzyme. Enzyme activity was calculated using a molar extinction coefficient of NADPH oxidation. Negative activity values were derived from all Grxs samples, indicating no apparent peroxidase activity.
Specific reduction of selenoprotein MsrA by Clostridium selenoprotein Grx
Methionine sulfoxide reductases (Msrs) are thiol-dependent enzymes that catalyze the reduction of oxidized methionines back to methionines in a stereospecific manner (Kim & Gladyshev, 2007). MsrA reduces free and protein-based methionine-S-sulfoxides. We previously characterized selenoprotein MsrAs from Clostridium sp. OhILAs and Chlamydomonas reinhardtii (Kim et al., 2006, Kim et al., 2009), which lack a resolving Cys. Interestingly, the Clostridium and Chlamydomonas selenoprotein MsrAs and their Cys mutant forms were not reducible by Trx, suggesting an alternative in vivo reductant for these enzymes.
The Cys-containing cGrx2 appeared to be an efficient catalyst comparable to selenoprotein cGrx1 (Table 1). This raised the question of why Sec is needed in a Clostridium Grx. Trx is generally considered as the in vivo reductant for MsrA. However, we have recently found that Grxs also can reduce Cys-containing MsrAs, and as mentioned above, the Clostridium and Chlamydomonas selenoprotein MsrAs could not be reduced by Trx. Thus, we speculated that Grxs might be natural reducing agents for selenoprotein MsrAs.
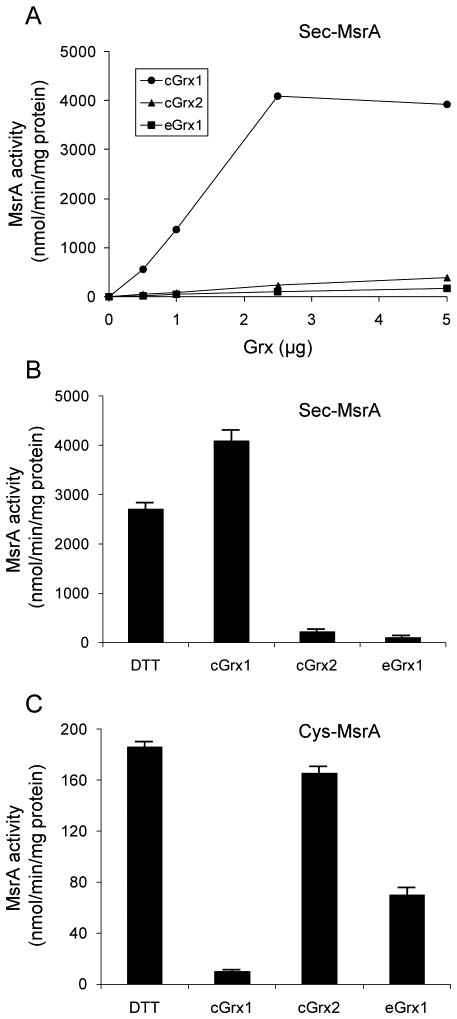
We assessed Grx-dependent Clostridium selenoprotein MsrA activities using selenoprotein cGrx1 and Cys-containing cGrx2 and eGrx1 (Figure 4A and B). Most remarkably, only selenoprotein cGrx1 could efficiently reduce the selenoprotein Clostridium MsrA, whereas cGrx2 and eGrx1 poorly reduced it; the selenoprotein cGrx1 had >20-fold higher reduction ability for this selenoprotein MsrA than the Cys-containing cGrx2 and eGrx1 (Figure 4B). We also tested whether the selenoprotein cGrx1 reduces a Cys version of Clostridium MsrA, in which Sec is replaced with Cys. Interestingly, cGrx1 poorly reduced this Cys mutant (Figure 4C). In contrast, cGrx2 and eGrx1 efficiently reduced this protein, consistent with our previous findings that Grxs are capable of reducing this protein. Thus, our data demonstrated that selenoprotein cGrx1 could efficiently reduce the selenoprotein MsrA but poorly reduce its Cys mutant, whereas cGrx2 or eGrx1, which efficiently reduce the Cys mutant, poorly reduced this selenoprotein. Overall, both selenoprotein and Cys-containing Grxs displayed high substrate specificities for reduction of MsrA: Sec-containing MsrA reduction by a Sec-containing Grx vs Cys-containing MsrA reduction by a Cys-containing Grx. This unexpected preference for MsrA reduction could explain why Sec is utilized in Clostridium Grx.
Figure 4. Selenoprotein and non-selenoprotein Grxs reduce MsrAs with high substrate specificity.
(A) Reduction of selenoprotein Clostridium MsrA by Grxs. A. MsrA activity was measured according to concentrations of selenoprotein (cGrx1) and non-selenoprotein Grxs (cGrx2 and eGrx1) indicated. (B) Comparison of Grx-dependent selenoprotein MsrA activities with the corresponding DTT-dependent activity. The Grx-dependent activities were measured using 2.5 μg Grxs. (C) Reduction of the Cys mutant of selenoprotein Clostridium MsrA by Grxs. Activities of the Cys mutant of selenoprotein Clostridium MsrA, in which Sec is replaced with Cys, were analyzed in DTT- and Grx-dependent reactions. 2.5 μg Grxs were used in the Grx-dependent assays.
Discussion
This study features the first characterization of a naturally-occurring selenoprotein Grx. Our data show that in the Sec-containing Grx, Sec confers a higher catalytic activity compared to its Cys mutant, as it does in the unnatural engineered selenoprotein Grxs characterized previously (Casi et al., 2008, Metanis et al., 2006). Interestingly, the Km value was dramatically reduced (>10-fold) in selenoprotein cGrx1 compared with Cys homologs, cGrx2 and eGrx1. This lower Km feature, i.e., higher affinity for the substrate, provides an advantage for the selenoprotein cGrx1 in the reduction of selenoprotein MsrA in cells because the expression levels of selenoproteins are normally much lower compared to those of Cys-containing forms. However, Sec residue alone is not the crucial determinant for affinity of the substrate in cGrx1.
To our knowledge, there have been no recombinant selenoproteins that contained 100% Sec insertion when expressed in E. coli. Trp insertion, so called opal suppression, always occurs during translation of selenoproteins in E. coli cells (Su et al., 2005, Sambol & Dubes, 1984, Sandman & Noren, 2000). To increase Sec insertion, we co-transformed cells with the plasmid that expresses the components of the Sec insertion machinery (selA, selB, and selC). Nevertheless, the insertion ratio of Trp to Sec in selenoprotein cGrx1 was 5:1. Such high ratio of Trp insertion is probably due to the relatively inefficient SECIS element of cGrx1 in E. coli system.
MsrAs are important antioxidant protein repair enzymes that reduce free and protein-bound methionine-S-sulfoxides back to methionines. The catalytic mechanism of MsrA is based on a common sulfenic acid chemistry (Boschi-Muller et al., 2000, Lowther et al., 2000, Tete-Favier et al., 2000), and reducing agents are required to regenerate the enzyme for the next catalytic cycle. Trx is generally considered as the in vivo reductant while DTT can be used as the in vitro reductant. However, it was previously shown that Trx could not reduce Clostridium selenoprotein MsrA (and also its Cys mutant), suggesting an alternative in vivo reductant for this selenoprotein. Our recent finding was that Grxs can reduce MsrAs. Thus, in the present study, we tested whether Clostridium Grxs can serve as reductants for Clostridium selenoprotein MsrA and whether selenoprotein and non-selenoprotein Grxs display substrate specificity for Sec- and Cys-containing MsrAs. Most surprisingly, only selenoprotein cGrx1 was an efficient catalyst for the reduction of Clostridium selenoprotein MsrA, whereas Cys-containing cGrx2 and eGrx1 poorly reduced it. In contrast, cGrx1 could not efficiently reduce the Cys mutant of selenoprotein MsrA, whereas Cys-containing Grxs could. Thus, the reduction of MsrA by Grxs is highly specific: only Sec-containing Grx efficiently reduces Sec-containing MsrA, while only Cys-containing Grxs reduce Cys-containing MsrA with efficiency.
To explore these higher specificities in the reduction of MsrAs by Grxs, we attempted to measure redox potentials of selenoprotein and Cys forms of Grxs using a GSH/GSSG redox buffer. The redox potentials of Cys mutant cGrx1 and cGrx2 were determined as −215 mV and −210 mV, respectively, fitting the range of previously characterized Grxs redox potentials. The redox potential of selenoprotein cGrx1 could not be determined but would be expected to be lower than that of Cys mutant form. We also attempted to measure the redox potentials of Clostridium selenoprotein MsrA and its Cys mutant using the AMS/gel-shift method, but failed to determine them. For such substrate specificities, the redox potential of selenoprotein MsrA is assumed to be higher than that of selenoprotein cGrx1 but lower than that of Cys-containing cGrx2, while the redox potential of the Cys mutant MsrA is likely higher than that of cGrx2. However, this assumption for redox potential values would not explain why the selenoprotein Grx was unable to efficiently reduce the Cys version of MsrA. In addition to determining redox potentials of MsrA and selenoprotein Grx, further studies, such as physiological relevance of selenoprotein Grx-selenoprotein MsrA interaction and structural analyses, will be needed, in order to elucidate the molecular basis of the higher specificity in the reduction of MsrA by Grx.
Among selenoproteins characterized so far, the majority are oxidoreductases, such as Trx reductase, Gpxs, and Msrs. Sec provides selenoprotein oxidoreductases with higher catalytic efficiency compared to their Cys mutant forms (Kim & Gladyshev, 2004, Lee et al., 2000). However, there are also Cys-containing orthologs of selenoproteins with catalytic efficiencies comparable to those of selenoproteins. The majority of selenoproteins evolved by the replacement of Cys with Sec, but the evolution of selenoproteins is a complex process that balances enhanced catalysis provided by Sec with limitations imposed by the dependence on selenium, its toxicity, prior occurrence of the Sec insertion system, and availability of electron donors (or acceptors) for selenoproteins. We and other groups have made efforts to address the fundamental question of why Sec is used in biological systems (Gromer et al., 2003, Kim & Gladyshev, 2005), but as of yet there is little direct evidence to answer this question. Gromer et al. suggested that Sec is not necessarily needed for efficient catalysis by Trx reductase (Gromer et al., 2003). Instead, it provides advantages with regard to a broader range of substrates and increased flexibility in the active sites. We previously suggested that Sec is assisted by a different set of features in the active site of mammalian selenoprotein MsrB and operates different catalytic mechanisms from the Cys-containing mammalian MsrBs (Kim & Gladyshev, 2005). A recent review discussed chemical reasons for the use of Sec in enzymes and suggested that Sec can confer resistance to inactivation by irreversible oxidation (Hondal & Ruggles, 2010). The present study provided an interesting reason for Sec utilization, i.e., the specific reduction of MsrA by Grx requiring a tandem of selenoproteins.
Experimental Procedures
Cloning, expression and purification of the selenoprotein and Cys-containing forms of Clostridium Grx
A coding region of the selenoprotein grx1 gene was PCR-amplified using Clostridium sp. OhILAs genomic DNA and forward (5′-GCGCCATATGGCAAAAGAAGTAATCGTG-3′) and reverse (5′-GCGCCTCGAGTCCTAAAAGTTCCTCAATT-3′) primers. The amplified DNA fragment digested with NdeI and XhoI was cloned into pET21b (Novagen). The resulting construct, designated pET-CLOS-Grx1, coded for full-length selenoprotein Grx1 (cGrx1) with a C-terminal His-tag (LEHHHHHH). The Cys-containing homolog grx2 gene was also PCR-cloned into NdeI/XhoI sites of pET21b using forward (5′-GCGCCATATGAAGAATATTACGATTTAC-3′) and reverse (5′-GCGCCTCGAGTTTTAGGCCAAGCTTTTTA-3′) primers. The resulting plasmid pET-CLOS-Grx2 coded for full-length Grx2 (cGrx2) with a C-terminal His-tag (LEHHHHHH).
Cys mutant (cGrx1/U13C) and Trp mutant (cGrx1/U13W) of selenoprotein Grx1 were generated in which Sec13 was replaced with Cys and Trp, respectively, by site-directed mutagenesis. We also generated a monoselenol mutant of Grx1 (cGrx1/C16S), in which Cys16 was replaced with Ser, and a monothiol mutant of Grx2 (cGrx2/C15S), in which Cys15 was substituted to Ser.
To express cGrx1 or cGrx1/C16S in E. coli, the corresponding plasmids were transformed into BL21(DE3) cells also containing a plasmid pSUABC (Arner et al., 1999), which encodes E. coli selA, selB, and selC. The transformed cells were grown in LB media containing 2 μM sodium selenite, 100 μg/ml ampicillin, and 25 μg/ml chloramphenicol at 37°C. IPTG (0.1 mM) was added when an optical density at 600 nm of the culture reached ~0.6–0.8, and the cells were further cultured for 6 h at 30°C. The cell pellets were resuspended in an extraction buffer (50 mM sodium phosphate, pH 7.0, 300 mM NaCl, 10 mM imidazole, and 1 mM phenylmethylsulfonyl fluoride) and lysed by sonication. The supernatant of the lysate was loaded on a Talon-metal affinity resin (Clontech), washed with the extraction buffer, and eluted with a buffer containing 50 mM sodium phosphate (pH 7.0), 50 mM NaCl, and 150 mM imidazole. The eluted proteins were dialyzed against 50 mM sodium phosphate (pH 7.5) and 50 mM NaCl. Typical yield of the purified selenoprotein Grx1 forms was 30–60 μg from 500 ml culture broth, as estimated by Western blot assays.
To express the Cys-containing Grxs (cGrx1/U13C, cGrx2 or cGrx2/C15S) or Trp mutant cGrx1/U13W, BL21(DE3) cells transformed with the corresponding plasmid were cultivated in LB media containing 100 μg/ml ampicillin at 37°C until optical density at 600 nm reached ~0.6–0.8, IPTG was added with a concentration of 0.1 mM, and the cells were further cultured for 6 h at 30°C. The cells were harvested and stored at −20°C until use. Procedures for purification of the proteins were as described above for the selenoprotein Grx forms. The samples were analyzed for purity by SDS-PAGE and consisted almost exclusively of the ectopic protein. We also cloned E. coli Grx1 (eGrx1, GI 146273) into pET21b vector and purified this protein by a Talon-metal affinity chromatography as described above.
Metabolic labeling with 75Se
To verify expression of Clostridium selenoprotein Grx1 in E. coli, we performed metabolic labeling with 75Se. Briefly, E. coli BL21(DE3) cells transformed with an empty vector, pET-CLOS-Grx1, or pET-CLOS-Grx1/U13C were grown at 37°C in 5 ml LB media containing ampicillin until optical density at 600 nm reached ~0.6. Then, 0.05 mCi of freshly neutralized [75Se]selenous acid and 1 mM IPTG were added to the cell culture. The cells were further cultured at 37°C for 5 h, harvested, washed with PBS buffer and lysed. A total of 30 μg protein was separated by SDS-PAGE and transferred onto a PVDF membrane. The 75Se radioactivity pattern on the membrane was analyzed by using a PhosphorImager (GE Health Care).
Determination of protein concentration
Due to small amounts of purified recombinant selenoprotein Grxs, concentrations of these selenoproteins were determined by Western blot analysis using anti-His antibodies, followed by quantifying the blot signals with a densitometric analysis (ImageJ) (Supplementary Figure S2B). The cGrx1/U13C protein was used as an internal standard. Concentrations of purified Cys-containing Grxs and Trp mutant cGrx1/U13W were determined by the Bradford method using a Bio-Rad protein assay reagent and bovine serum albumin as a standard.
Grx enzyme assay and analysis of kinetics
Grx activity was measured by using HED as a substrate. The reaction mixture (200 μl) contained 50 mM sodium phosphate (pH 7.5), 50 mM NaCl, 0.2 mM NADPH, 0.5 mM GSH, 0.5 unit yeast GSH reductase (Sigma), 2 mM HED, and purified Grx. The amount of Grx enzymes used was 1 μg except for 10 μg of cGrx1/U13C and 20 μg of cGrx1/U13W. The reaction was carried out at 25oC for 5 min following a 1-min preincubation. The decrease in absorbance of NADPH at 340 nm was monitored using a spectrophotometer. Enzyme activity was defined as nmole of oxidized NADPH per min using a molar extinction coefficient of 6220 M−1 cm−1. The protein concentrations of selenoprotein Grxs (cGrx1 and cGrx1/C16S) were normalized by subtracting the concentrations of Trp-inserted forms determined by mass spectrometry analysis and subjected to calculating their kcat values. Km and kcat values were determined by non-linear regression using GraphPad Prism 5 software.
Determination of peroxidase activity
The peroxidase activity of Grxs was measured with H2O2. The reaction mixture (200 μl) contained 50 mM sodium phosphate (pH 7.5), 50 mM NaCl, 0.2 mM NADPH, 0.5 mM GSH, 0.5 unit yeast GSH reductase, 0.4 mM H2O2, and 5 μg purified Grx. GSH peroxidase (Gpx) from bovine erythrocytes (Sigma) was used as positive control. The reaction was carried out at 25oC for 5 min following a 2-min preincubation and the NADPH oxidation was analyzed at A340. The decreased absorbance values of samples were subtracted from that of the background without the enzyme. Activity was defined as nmole of oxidized NADPH per min using a molar extinction coefficient of 6220 M−1 cm−1.
MsrA enzyme assay
Selenoprotein Clostridium MsrA and its Cys mutant proteins were purified from E. coli as described previously (Kim et al., 2009). The reaction mixture (100 μl) contained 50 mM sodium phosphate (pH 7.5), 50 mM NaCl, 0.2 mM NADPH, 10 mM GSH, 0.5 unit GSH reductase, 0.5–5 μg purified Grx, 200 μM dabsyl-methionine-S-sulfoxide, and purified Clostridium MsrA. The reactions were carried out at 37oC for 30 min and analyzed by HPLC as described previously (Kumar et al., 2002). DTT-dependent assay was also performed for comparison.
Analysis of redox potentials
To determine the redox potential of Grxs, a previously reported gel-shift method was used (Regeimbal & Bardwell, 2002, Su & Gladyshev, 2004). Briefly, purified Grx (2–10 μg) was incubated at room temperature in redox buffer containing 100 mM sodium phosphate (pH 7.0), 1 mM EDTA, and variable GSH/GSSG concentrations and ratios for 4 h. The proteins were then precipitated with 15% trichloroacetic acid by 15 min incubation on ice and pelleted by centrifugation at 13,000 rpm for 10 min. The pellets were washed with ice-acetone, dried, and resuspended in 100 mM sodium phosphate (pH 7.0), 1 mM EDTA, 1% SDS, and 15 mM 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS; Molecular Probes). The alkylation was performed at 37oC for 2 h, and the proteins were subjected to non-reducing SDS-PAGE and visualized by Coomassie staining. Protein band intensities were quantified using ImageJ software and the redox potentials were calculated using the Nernst equation as described (Aslund et al., 1997).
Mass spectrometry analysis
To determine the insertion ratio of Trp to Sec in cGrx1, semi-quantitative analysis was performed by counting the number of identified peptides containing Trp or Sec by mass spectrometry. The separated cGrx1 band was destained and digested with trypsin and peptides were extracted. The peptide extracts were evaporated to dryness in SpeedVac for mass spectrometry analysis, and dissolved in 10% acetonitrile solution containing 1% formic acid. Dissolved peptides were desalted prior to separation using a trap column (ID 180 μm × 20 mm, Symmetry® C18, Waters) cartridge. Peptides were separated using a C18 reversed-phase 75 μm i. d. × 250 mm analytical column (1.7 μm particle size, BEH130 C18, Waters) with an integrated electrospray ionization SilicaTip™ (± 10 μm i.d., New Objective, USA). Chromatography was performed by nanoUPLC-ESI-q-TOF (SYNAPT™ HDMS™, Waters Co. USA) as described previously (Choi et al., 2010).
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation of Korea grants 2009-0084101 and 2010-0001240 (to HYK), by National Institutes of Health grants AG021518 and GM061603 (to VNG), by the World Class University grant R31-2008-000-10010-0 (to VNG), and by Center for Cell Signaling & Drug Discovery Research grant R15-2006-020 (to KJL).
Abbreviations
- Grx
glutaredoxin
- Gpx
glutathione peroxidase
- HED
hydroxyethyl disulfide
- Msr
methionine sulfoxide reductase
- Sec
selenocysteine
- SECIS
Sec insertion sequence
- Trx
thioredoxin
References
- Arner ES, Sarioglu H, Lottspeich F, Holmgren A, Bock A. High-level expression in Escherichia coli of selenocysteine-containing rat thioredoxin reductase utilizing gene fusions with engineered bacterial-type SECIS elements and co-expression with the selA, selB and selC genes. J Mol Biol. 1999;292:1003–1016. doi: 10.1006/jmbi.1999.3085. [DOI] [PubMed] [Google Scholar]
- Aslund F, Berndt KD, Holmgren A. Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein-protein redox equilibria. J Biol Chem. 1997;272:30780–30786. doi: 10.1074/jbc.272.49.30780. [DOI] [PubMed] [Google Scholar]
- Bock A, Forchhammer K, Heider J, Leinfelder W, Sawers G, Veprek B, Zinoni F. Selenocysteine: the 21st amino acid. Mol Microbiol. 1991;5:515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- Boschi-Muller S, Azza S, Sanglier-Cianferani S, Talfournier F, Van Dorsselear A, Branlant G. A sulfenic acid enzyme intermediate is involved in the catalytic mechanism of peptide methionine sulfoxide reductase from Escherichia coli. J Biol Chem. 2000;275:35908–35913. doi: 10.1074/jbc.M006137200. [DOI] [PubMed] [Google Scholar]
- Casi G, Roelfes G, Hilvert D. Selenoglutaredoxin as a glutathione peroxidase mimic. Chembiochem. 2008;9:1623–1631. doi: 10.1002/cbic.200700745. [DOI] [PubMed] [Google Scholar]
- Choi S, Jeong J, Na S, Lee HS, Kim HY, Lee KJ, Paek E. New algorithm for the identification of intact disulfide linkages based on fragmentation characteristics in tandem mass spectra. J Proteome Res. 2010;9:626–635. doi: 10.1021/pr900771r. [DOI] [PubMed] [Google Scholar]
- Cone JE, Del Rio RM, Davis JN, Stadtman TC. Chemical characterization of the selenoprotein component of clostridial glycine reductase: identification of selenocysteine as the organoselenium moiety. Proc Natl Acad Sci U S A. 1976;73:2659–2663. doi: 10.1073/pnas.73.8.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll DM, Copeland PR. Mechanism and regulation of selenoprotein synthesis. Annu Rev Nutr. 2003;23:17–40. doi: 10.1146/annurev.nutr.23.011702.073318. [DOI] [PubMed] [Google Scholar]
- Fourmy D, Guittet E, Yoshizawa S. Structure of prokaryotic SECIS mRNA hairpin and its interaction with elongation factor SelB. J Mol Biol. 2002;324:137–150. doi: 10.1016/s0022-2836(02)01030-6. [DOI] [PubMed] [Google Scholar]
- Ge Y, Qi Z, Wang Y, Liu X, Li J, Xu J, Liu J, Shen J. Engineered selenium-containing glutaredoxin displays strong glutathione peroxidase activity rivaling natural enzyme. Int J Biochem Cell Biol. 2009;41:900–906. doi: 10.1016/j.biocel.2008.08.032. [DOI] [PubMed] [Google Scholar]
- Gladyshev VN, Jeang KT, Stadtman TC. Selenocysteine, identified as the penultimate C-terminal residue in human T-cell thioredoxin reductase, corresponds to TGA in the human placental gene. Proc Natl Acad Sci U S A. 1996;93:6146–6151. doi: 10.1073/pnas.93.12.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromer S, Johansson L, Bauer H, Arscott LD, Rauch S, Ballou DP, Williams CH, Jr, Schirmer RH, Arner ES. Active sites of thioredoxin reductases: why selenoproteins? Proc Natl Acad Sci U S A. 2003;100:12618–12623. doi: 10.1073/pnas.2134510100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol Cell Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. Hydrogen donor system for Escherichia coli ribonucleoside-diphosphate reductase dependent upon glutathione. Proc Natl Acad Sci U S A. 1976;73:2275–2279. doi: 10.1073/pnas.73.7.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A, Johansson C, Berndt C, Lonn ME, Hudemann C, Lillig CH. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem Soc Trans. 2005;33:1375–1377. doi: 10.1042/BST0331375. [DOI] [PubMed] [Google Scholar]
- Hondal RJ, Ruggles EL. Differing views of the role of selenium in thioredoxin reductase. Amino Acids. 2010 doi: 10.1007/s00726-010-0494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Fomenko DE, Yoon YE, Gladyshev VN. Catalytic advantages provided by selenocysteine in methionine-S-sulfoxide reductases. Biochemistry. 2006;45:13697–13704. doi: 10.1021/bi0611614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Gladyshev VN. Methionine sulfoxide reduction in mammals: characterization of methionine-R-sulfoxide reductases. Mol Biol Cell. 2004;15:1055–1064. doi: 10.1091/mbc.E03-08-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Gladyshev VN. Different catalytic mechanisms in mammalian selenocysteine- and cysteine-containing methionine-R-sulfoxide reductases. PLoS Biol. 2005;3:e375. doi: 10.1371/journal.pbio.0030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Gladyshev VN. Methionine sulfoxide reductases: selenoprotein forms and roles in antioxidant protein repair in mammals. Biochem J. 2007;407:321–329. doi: 10.1042/BJ20070929. [DOI] [PubMed] [Google Scholar]
- Kim HY, Zhang Y, Lee BC, Kim JR, Gladyshev VN. The selenoproteome of Clostridium sp. OhILAs: characterization of anaerobic bacterial selenoprotein methionine sulfoxide reductase A. Proteins. 2009;74:1008–1017. doi: 10.1002/prot.22212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RA, Koc A, Cerny RL, Gladyshev VN. Reaction mechanism, evolutionary analysis, and role of zinc in Drosophila methionine-R-sulfoxide reductase. J Biol Chem. 2002;277:37527–37535. doi: 10.1074/jbc.M203496200. [DOI] [PubMed] [Google Scholar]
- Lee SR, Bar-Noy S, Kwon J, Levine RL, Stadtman TC, Rhee SG. Mammalian thioredoxin reductase: oxidation of the C-terminal cysteine/selenocysteine active site forms a thioselenide, and replacement of selenium with sulfur markedly reduces catalytic activity. Proc Natl Acad Sci U S A. 2000;97:2521–2526. doi: 10.1073/pnas.050579797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillig CH, Berndt C, Holmgren A. Glutaredoxin systems. Biochim Biophys Acta. 2008;1780:1304–1317. doi: 10.1016/j.bbagen.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Lillig CH, Holmgren A. Thioredoxin and related molecules--from biology to health and disease. Antioxid Redox Signal. 2007;9:25–47. doi: 10.1089/ars.2007.9.25. [DOI] [PubMed] [Google Scholar]
- Liu Z, Reches M, Groisman I, Engelberg-Kulka H. The nature of the minimal ‘selenocysteine insertion sequence’ (SECIS) in Escherichia coli. Nucleic Acids Res. 1998;26:896–902. doi: 10.1093/nar/26.4.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowther WT, Brot N, Weissbach H, Matthews BW. Structure and mechanism of peptide methionine sulfoxide reductase, an “anti-oxidation” enzyme. Biochemistry. 2000;39:13307–13312. doi: 10.1021/bi0020269. [DOI] [PubMed] [Google Scholar]
- Metanis N, Keinan E, Dawson PE. Synthetic seleno-glutaredoxin 3 analogues are highly reducing oxidoreductases with enhanced catalytic efficiency. J Am Chem Soc. 2006;128:16684–16691. doi: 10.1021/ja0661414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regeimbal J, Bardwell JC. DsbB catalyzes disulfide bond formation de novo. J Biol Chem. 2002;277:32706–32713. doi: 10.1074/jbc.M205433200. [DOI] [PubMed] [Google Scholar]
- Sambol AR, Dubes GR. Phenotypic suppression of opal mutants by L-tryptophan. Microbios. 1984;39:19–27. [PubMed] [Google Scholar]
- Sandman KE, Noren CJ. The efficiency of Escherichia coli selenocysteine insertion is influenced by the immediate downstream nucleotide. Nucleic Acids Res. 2000;28:755–761. doi: 10.1093/nar/28.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandman KE, Tardiff DF, Neely LA, Noren CJ. Revised Escherichia coli selenocysteine insertion requirements determined by in vivo screening of combinatorial libraries of SECIS variants. Nucleic Acids Res. 2003;31:2234–2241. doi: 10.1093/nar/gkg304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster P, Fontana W, Stadler PF, Hofacker IL. From sequences to shapes and back: a case study in RNA secondary structures. Proc Biol Sci. 1994;255:279–284. doi: 10.1098/rspb.1994.0040. [DOI] [PubMed] [Google Scholar]
- Stadtman TC. Selenocysteine. Annu Rev Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- Su D, Gladyshev VN. Alternative splicing involving the thioredoxin reductase module in mammals: a glutaredoxin-containing thioredoxin reductase 1. Biochemistry. 2004;43:12177–12188. doi: 10.1021/bi048478t. [DOI] [PubMed] [Google Scholar]
- Su D, Li Y, Gladyshev VN. Selenocysteine insertion directed by the 3′-UTR SECIS element in Escherichia coli. Nucleic Acids Res. 2005;33:2486–2492. doi: 10.1093/nar/gki547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tete-Favier F, Cobessi D, Boschi-Muller S, Azza S, Branlant G, Aubry A. Crystal structure of the Escherichia coli peptide methionine sulphoxide reductase at 1.9 A resolution. Structure. 2000;8:1167–1178. doi: 10.1016/s0969-2126(00)00526-8. [DOI] [PubMed] [Google Scholar]
- Tormay P, Bock A. Barriers to heterologous expression of a selenoprotein gene in bacteria. J Bacteriol. 1997;179:576–582. doi: 10.1128/jb.179.3.576-582.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gladyshev VN. An algorithm for identification of bacterial selenocysteine insertion sequence elements and selenoprotein genes. Bioinformatics. 2005;21:2580–2589. doi: 10.1093/bioinformatics/bti400. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Romero H, Salinas G, Gladyshev VN. Dynamic evolution of selenocysteine utilization in bacteria: a balance between selenoprotein loss and evolution of selenocysteine from redox active cysteine residues. Genome Biol. 2006;7:R94. doi: 10.1186/gb-2006-7-10-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.