Abstract
Fanconi anemia (FA) is a rare familial genome instability syndrome caused by mutations in FA genes that results in defective DNA crosslink repair. Activation of the FA pathway requires the FA core ubiquitin ligase complex-dependent monoubiquitination of 2 interacting FA proteins, FANCI and FANCD2. Although loss of either FANCI or FANCD2 is known to prevent monoubiquitination of its respective partner, it is unclear whether FANCI has any additional domains that may be important in promoting DNA repair, independent of its monoubiquitination. Here, we focus on an FA-I patient-derived FANCI mutant protein, R1299X (deletion of 30 residues from its C-terminus), to characterize important structural region(s) in FANCI that is required to activate the FA pathway. We show that, within this short 30 amino acid stretch contains 2 separable functional signatures, a nuclear localization signal and a putative EDGE motif, that is critical for the ability of FANCI to properly monoubiquitinate FANCD2 and promote DNA crosslink resistance. Our study enable us to conclude that, although proper nuclear localization of FANCI is crucial for robust FANCD2 monoubiquitination, the putative FANCI EDGE motif is important for DNA crosslink repair.
Introduction
Fanconi anemia (FA) is a rare, chromosome instability syndrome associated with developmental abnormalities, bone marrow failure, predisposition to cancer, and cellular hypersensitivity to DNA crosslinking agents.1,2 The disorder is genetically heterogeneous with at least 13 complementation groups currently defined. To date, at least 8 FA and other FA-associated proteins (FANCA, -B, -C, -E, -F, -G, -L, -M, and FAAP100) form a multisubunit nuclear core complex that contains a catalytic ubiquitin E3 ligase activity required for the activation of FANCD2 and its paralog FANCI by monoubiquitination.3,4 Monoubiquitinated and phosphorylated FANCD2 and FANCI5–9 are then targeted to BRCA1-containing DNA damage and repair sites in chromatin (nuclear foci) to assist in DNA crosslink repair along with at least 4 other downstream FA members, FANCJ (BRIP1, a DNA helicase),10–12 FANCD1 (BRCA2, a protein that mediates homologous recombination),13 FANCN (PALB2, partner and localizer of BRCA2),14,15 and RAD51C (a member of the RAD51-like gene family involved in HR-mediated DNA repair).16,17
Currently, it is thought that the key trigger in the activation of the FA pathway lies in the molecular events surrounding the DNA damage-inducible monoubiquitination of the FANCD2 and FANCI proteins. Insights into the mechanism of activation came with the discovery of the gene FANCI, responsible for the FA-I complementation group.6,7,18 The FANCI protein is thought to function similarly to that of FANCD2. Besides possessing regions of the protein with weak homology to FANCD2, FANCI also gets monoubiquitinated in a lysine site-specific and DNA damage-dependent manner by the FA core E3 ubiquitin ligase complex.6,7 A fraction of FANCI is found to be associated with FANCD2, forming the FANCI-FANCD2 (ID) complex. Interestingly, the monoubiquitination of FANCI and FANCD2 is dependent on each other. Loss of either FANCI or FANCD2 prevents the monoubiquitination of its other protein partner. As expected, negative regulation of the FA pathway can be achieved, in part, through FANCI and FANCD2 deubiquitination, which is mediated by the deubiquitinating enzyme USP1.6,19,20 More recently, it was shown that monoubiquitinated ID complex is required for replication-coupled interstrand crosslink repair in a cell-free system.21 However, it is still unclear how FANCI can contribute to both the upstream monoubiquitination of FANCD2 and subsequent downstream repair of DNA crosslinks.
FANCI is a relatively large protein with 1328 amino acids (Mr = 149 kDa) and, like other Fanconi proteins, contains little or no known domain(s) that may provide clues to possible protein function. To gain insight into potential structural region(s) on FANCI that may be important for mediating FANCD2 monoubiquitination and FA pathway function, we sought to analyze patient-derived FANCI mutant proteins and determine their functional defect in a reconstituted cell line system. From our previous study, we identified 7 different allelic mutations in the FANCI gene from 4 different FA-I patient-derived cell lines.6 Interestingly, cells from most, if not all, FA-I persons studied have retained expression of mutant FANCI proteins. Similar to what has been reported for patients harboring mutations in the FANCD2 gene,22 it is probable that complete null mutations of FANCI are lethal in humans.6 Studying the different hypomorphic mutations in FANCI may provide clues to determining important functional regions/domains in FANCI for promoting genome stability. In one particular patient cell line (F010191), we identified 2 truncating mutations in FANCI. The first results in a severe deletion of more than half of the coding sequence, whereas the other results in a deletion of the last 30 residues of FANCI (R1299X).6 Despite having similar levels of mutant FANCI compared with normal patient cells, the FA-I F010191 lymphoblasts cells have severely reduced levels of FANCD2 and FANCI monoubiquitination.6 This suggests that the extreme C-terminal region of FANCI may be critical for mediating the activation of the FA pathway.
The goal of the present study is to characterize the function of the C-terminal domain of FANCI in promoting FANCD2 monoubiquitination and DNA crosslink repair. We found that the last 30 residues of FANCI contain a functional nuclear localization signal (NLS) that is critical for proper nuclear localization of the FANCI protein. Mislocalization of FANCI resulting from the loss of the C-terminal NLS sequence results in defective FANCD2 and FANCI monoubiquitination and DNA crosslink sensitivity in human cells. We also suggest a DNA crosslink repair function for the putative EDGE motif6,7,23 that also resides within the C-terminal region of FANCI. Our present findings establish the importance of separable motifs, such as protein localization, as one type of mechanism that contributes to the molecular pathogenesis of FA.
Methods
Cell culture
U20S (ATCC) were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, glutamine, and antibiotics. Patient-derived FA-I F010191 lymphoblasts, as described,6,18 were propagated in RPMI 1640 medium supplemented with 15% fetal bovine serum, glutamine, and antibiotics. After informed consent, patient-derived FA-I F010191 fibroblasts were established from a small skin biopsy under local anesthesia and primary fibroblasts were grown out over several weeks in Dulbecco modified Eagle medium supplemented with 15% fetal bovine serum, glutamine, and antibiotics. Fibroblasts were immortalized by transduction with a retroviral vector that expresses the large and small T antigens from the SV40 viruses under an SFFV promoter (H.H., unpublished, July 2010). All cells were grown at 37°C.
Cell transfections
siRNA transfections were performed using HiPerfect (QIAGEN) and plasmid transfections were performed using Fugene6 (Roche Diagnostics) according to the manufacturer's protocol. To replace endogenous FANCI with siRNA-resistant FANCI constructs (WT FANCI, FANCI R1299X, FANCI NLS2, and FANCI NLS3), cells were first transfected with FANCI siRNA and followed by transfection of siRNA-resistant FANCI constructs 24 hours later.
Plasmids and siRNAs
FANCI and negative control small interfering RNA (siRNA) oligonucleotide sequences were purchased from QIAGEN: KIAA1794-3 (CACGGGCATCTGGGAGATATA) for FANCI siRNA and All-Stars Negative Control siRNA for control. Flag-tagged wild-type (WT) FANCI was constructed by cloning the full-length human FANCI cDNA into N-terminal pFLAG-CMV vector (Sigma-Aldrich). WT Flag-FANCI was mutagenized by polymerase chain reaction using QuikChange Site-Directed Mutagenesis Kit (Stratagene) to generate Flag-FANCI resistant to siRNA KIAA1794-3 (see Figure 4A). Other FANCI mutants, such as K523R, R1299X, D1301A, ΔNLS (K1323X), NLS2, and NLS3, were generated by polymerase chain reaction mutagenesis using the Flag-FANCI WT siRNA resistant clone as DNA template. Flag-FANCI-NLS2 and Flag-FANCI-NLS3 were amplified by polymerase chain reaction from N-terminal-pFLAG-CMV FANCI R1299X construct with the reverse primers containing FANCI NLS sequence for Flag-FANCI NLS2 (5-GAACGTGGATCCTCATTATTTTTTCCTTTTCTTCTTAAGAACCATGTCTAGGAT-3) and c-Myc NLS sequence for Flag-FANCI NLS3 (5-GAACGTGGATCCTTAGTCCAGTTTAACCCTTTTTGCTGCTGGAAGAACCATGTCTAGGAT-3).
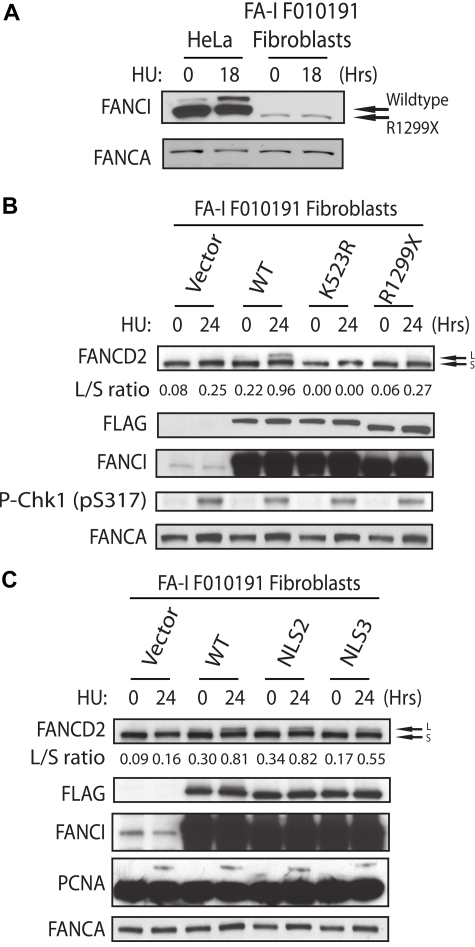
Figure 4.
Recovery of FANCD2 monoubiquitination in the FA-I F010191 fibroblasts. (A) HeLa and F010191 fibroblasts were either untreated or treated with HU (2mM for 18 hours). (B-C) F010191 fibroblasts were transfected with empty vector or the indicated FANCI expression constructs. Forty-eight hours after transfection, cells were treated with 2mM HU for 24 hours and FANCD2 monoubiquitination was assessed by Western blot analysis and probed with the indicated antibodies. The L/S ratio was calculated as described in Figure 3A.
Immunocytochemistry
Detection of FANCD2 and BRCA1 foci, U2OS cells, or F010191 fibroblasts were treated with 2mM hydroxyurea (HU) for indicated times. Cells were permeabilized with Scully buffer (0.5% Triton X-100, 20mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.4, 50mM NaCl, 3mM MgCl2, 300mM sucrose) for 2 minutes and fixed with 2% (vol/vol) paraformaldehyde in phosphate-buffered saline (PBS) for 20 minutes at room temperature. Cells were then blocked in blocking solution (5% goat serum 0.1% NP-40 in PBS) and incubated with anti FANCD2 (1:500; NB 100-182 Novus) and anti-BRCA1 (1:400; D9 Santa Cruz Biotechnology) antibodies diluted in blocking solution for 2 hours. Cells were washed 3 times with 0.1% NP-40 PBS and incubated with AlexaFluor 546-conjugated goat anti–rabbit immunoglobulin G (IgG) (Invitrogen) and AlexaFluor 488-conjugated goat anti–mouse IgG (Invitrogen) at a dilution of 1:500 in blocking solution for 1 hour. Cells were washed 3 times with 0.1% NP-40 PBS and mounted in Vectashield with 4,6-diamidino-2-phenylindole (Vector Laboratories). For WT FANCI, FANCI R1299X, FANCI ΔNLS, FANCI NLS2, and FANCI NLS3 localization, cells were first fixed with 2% (vol/vol) paraformaldehyde in PBS for 20 minutes and then permeabilized with Scully buffer for 2 minutes. Anti-FLAG (1:500, Sigma-Aldrich M2) antibody and AlexaFluor 546-conjugated goat anti–mouse IgG (Invitrogen) were used. Images were captured using a Deltavision personal DV system (Applied Precision) on a base Olympus IX71 microscope and a CoolSnap HQ camera (Photometrics). Images were deconvolved using Softworx Suite software (Applied Precision). The images were opened and then sized and placed into figures using Image J Version 1.43 software (available at http://rsb.info.nih.gov/ij) or Adobe Photoshop Version 7.0. Professional (Adobe Systems).
Western blotting
Western blots were performed with whole-cell extracts prepared in lysis buffer (0.1M Tris, pH 6.8, 2% [wt/vol] sodium dodecyl sulfate and 12% [vol/vol] β-mercaptoethanol) and separated on Nupage 3% to 8% Tris-acetate gels (Invitrogen). Immunoblotting was performed as previously described6 using anti-FANCD2, anti-FANCI (A301-254A, Bethyl Laboratories), antiphospho ATM (ab2888, Abcam), anti-ATM (ab17995, Abcam), antiphospho-Chk1 (ab2834, Abcam), antitubulin (ab4074, Abcam), anti-proliferating cell nuclear antigen (sc-56, Santa Cruz Biotechnology), anti-FANCA (A301-980A, Bethyl Laboratories), and anti-FLAG (M2, Sigma-Aldrich) antibodies.
Subcellular fractionation
FA-I F010191 lymphoblast cells were fractionated into cytoplasmic and nuclear fractions by hypotonic swelling with a protocol adapted from Mendez and Stillman.24 Cells were resuspended in buffer A (10mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.9, 10mM KCl, 1.5mM MgCl2, 0.34M sucrose, 10% [vol/vol] glycerol 1mM dithiothreitol, and 1mM phenylmethylsulfonyl fluoride) and Triton X-100 (0.1% [vol/vol] final concentration) was added for 5 minutes on ice. Nuclei were pelleted by low-speed centrifugation (1300g at 4°C for 5 minutes). The supernatant (cytoplasmic fraction) was transferred to a new tube and further clarified by high-speed centrifugation (20 000g at 4°C for 15 minutes). Nuclei were washed once with buffer A and lysed in lysis buffer (0.1M Tris, pH 6.8, 2% [wt/vol] sodium dodecyl sulfate and 12% [vol/vol] β-mercaptoethanol). Fractionation of the cell lysates was confirmed by Western blot with antitubulin antibody (ab6160, Abcam) and anti-LSD1 antibody (Reinberg Laboratory) as cytoplasmic and nuclear markers, respectively.
MMC sensitivity assay
F010191 fibroblasts were transfected with different FANCI constructs (WT FANCI, FANCI NLS2, FANCI NLS3, FANCI D1301A, and FANCI ΔNLS). Twenty-four hours after transfection, cells were replated at 750 cells per well in 96-well plates. Twenty-four hours after replating, cells were treated with mitomycin C (MMC; Sigma-Aldrich) at the desired final concentrations. Cells were incubated with the drug for 5 days and their survival rates were measured using a CellTiter Glo kit (Promega) following the manufacturer's instructions.
Results
DNA damage-induced monoubiquitination of FANCD2 is severely reduced in FA-I F010191 lymphoblasts
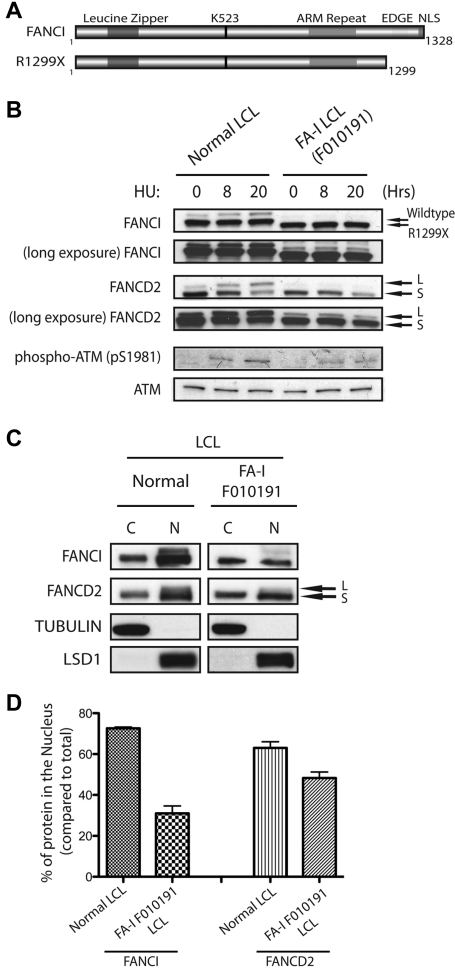
In our previous study, we showed that biallelic mutations in the FANCI gene were found in different FA-I patient-derived cells.6 Different allelic mutations of FANCI may provide clues to the requirement of different structural regions of FANCI toward their function in the FA pathway. Figure 1A shows a schematic diagram of predicted FANCI motifs and FANCI proteins expressed from normal or FA-I cell line (F010191). Interestingly, the mutant allele detected in the FA-I cell line (F010191) results in a deletion of the last 30 residues of FANCI (Figure 1A). As shown previously and in Figure 1B, the expression level of the truncated FANCI protein (R1299X) in F010191 lymphoblastoid cells (LCLs) is similar to that of full-length FANCI from normal LCLs. However, DNA damage-induced monoubiquitination of both FANCI and FANCD2 was inhibited after HU treatment in the F010191 LCLs expressing the FANCI R1299X protein. Unlike FA patient-derived cells harboring severe deletions of FA core ubiquitin ligase components, the levels of FANCI and FANCD2 monoubiquitination in the F010191 LCLs were still detectable with a long exposure of a Western blot, albeit extremely low (Figure 1B, long exposure). ATM (pSer1981) and Chk1 (pSer317) phosphorylation levels were comparable between the normal and F010191 lymphoblasts after HU treatment, implicating that the DNA damage response in the F010191 cells is functioning properly despite the defect in the FA pathway (Figure 1B; and data not shown). This suggests that the function of the extreme C-terminal region of FANCI may be critical for the efficiency or amplification of the monoubiquitination signal for the FA pathway.
Figure 1.
FANCI is mislocalized in FA-I F010191 lymphoblasts. (A) Schematic representation of WT FANCI and mutant FANCI proteins expressed in F010191 FA-I cells. In silico predicted leucine zipper, armadillo (ARM) repeat domain, EDGE motif and NLS, and lysine 523 (the monoubiquitination target of the FA core complex). (B) Time course of FANCI and FANCD2 monoubiquitination after 2mM HU treatment in normal or F010191 LCLs evaluated by Western blot analysis with the indicated antibodies. Note the slightly faster migrating FANCI protein in the F010191 LCLs resulting from the missing residues from the C-terminus. (C) Equal amounts of fractionated cytoplasmic (C) and nuclear (N) protein extracts from normal or FA-I lymphoblast lines were analyzed by Western blot with the indicated antibodies. Procedure for fractionation is described in “Subcellular fractionation.” (D) Nuclear accumulation of FANCI and FANCD2 proteins was quantified by determining the band intensity of underexposed Western blot film with ImageJ Version 1.43 software. Error bars represent SD from 3 independent experiments. The percentage of protein in the nucleus is compared with total from both cytoplasmic and nuclear fractions.
Reduced nuclear accumulation of endogenous FANCI R1299X mutant in F010191 LCLs
According to the schematic of predicted FANCI motifs, FANCI has a putative EDGE motif and NLS sequence that resides in the last 30 residues of the protein.6,7,25 Because these 2 putative motifs are missing in the FANCI R1299X mutant, we speculate that either one or both of these motifs are required for efficient FANCI and FANCD2 monoubiquitination after DNA damage. Interestingly, when cell fractionation studies were performed using normal and F010191 LCLs, the percentage of total FANCI protein in the nucleus of endogenous R1299X expressing cells was significantly reduced compared with WT cells (Figure 1C-D). It is known that FANCI and FANCD2 can associate with each other to form the ID complex.6,7 However, the mislocalization of R1299X did not dramatically alter the nuclear accumulation of endogenous FANCD2, suggesting that the defect is largely confined to suboptimal nuclear localization of FANCI (Figure 1C-D). It is probable that FANCD2 nuclear localization is not dependent on FANCI.
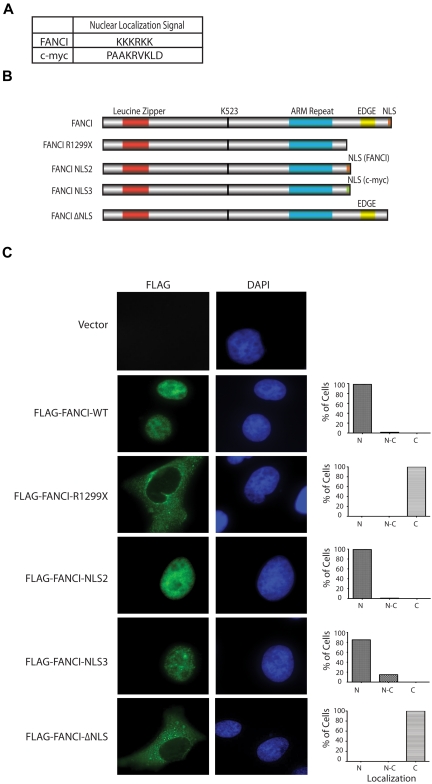
Ectopic expression of FANCI R1299X in U2OS cells reveals its defect in nuclear localization
To test whether the FANCI R1299X mutant protein is defective in nuclear localization, we generated and expressed different FANCI constructs in U2OS cells to determine their overall subcellular localization by indirect immunofluorescence (Figure 2B). As expected, we showed that WT Flag-FANCI localized predominantly in the nucleus. However, Flag-FANCI R1299X was found to be predominantly cytoplasmic (Figure 2C). To confirm whether the localization defect is the result of the loss of the C-terminal NLS on FANCI R1299X, the FANCI NLS sequence (Figure 2A, KKKRKK) was added back to the C-terminal end of the FANCI R1299X construct (Flag-FANCI-NLS2). We also used an unrelated NLS sequence from the c-Myc protein26,27 (Figure 2A, PAAKRVKLD) for control (Flag-FANCI-NLS3). The expression of both Flag-FANCI-NLS2 and -NLS3 were found to be predominantly nuclear, suggesting that the mislocalization of FANCI 1299X could be rescued by the addition of an NLS sequence (Figure 2C). We noted that the Flag-FANCI-NLS3 nuclear localization was not as complete or efficient as the -NLS2 construct (Figure 2C). This may explain the subtle FANCD2 monoubiquitination differences observed between these 2 mutant expression constructs described later in the paper (Figures 3A, 4C). We also mutated KKKRKK to RRRKRR in the Flag-FANCI-NLS2 construct and found that the expression of the mutant NLS2 was primarily in the cytoplasm, similar to that of R1299X (data not shown). In addition, the EDGE motif, which lies immediately upstream of the NLS and is present only in the FANCI WT construct but absent in the other 3 expression constructs tested (FANCI-R1299X, -NLS2, -NLS3) is probably not required for the nuclear localization of FANCI. To demonstrate that this is the case, we deleted the C-terminal NLS sequence in FANCI while keeping the EDGE motif intact (Flag-FANCI-ΔNLS or FANCI K1323X) and showed that the protein was primarily localized in the cytoplasm, similar to the R1299X mutant (Figure 2C).
Figure 2.
Exogenous expression of FANCI R1299X mutant localizes to cytoplasm in U2OS cells. (A) Table showing FANCI and c-Myc NLS amino acid sequence used in the study. NLS software (www.predictprotein.org) was used to identify the putative C-terminal NLS sequence for FANCI. (B) Schematic diagram of FANCI WT, FANCI R1299X, and FANCI R1299X proteins with the fusion of NLS sequences from FANCI (FANCI-NLS2) or c-Myc (FANCI-NLS3) at the C-terminus and C-terminal NLS deleted (FANCI-ΔNLS). (C) U2OS were transfected with the indicated Flag-tagged FANCI constructs. Forty-eight hours after transfection, cells were fixed and stained with anti-Flag antibody. 4,6-diamidino-2-phenylindole is used for DNA staining in the nucleus. A total of 200 cells were counted and scored for predominantly nuclear (N), both nuclear and cytoplasmic (N-C), or cytoplasmic (C) staining and graphed as percentage of total cells. Representative images of each expression constructs are shown.
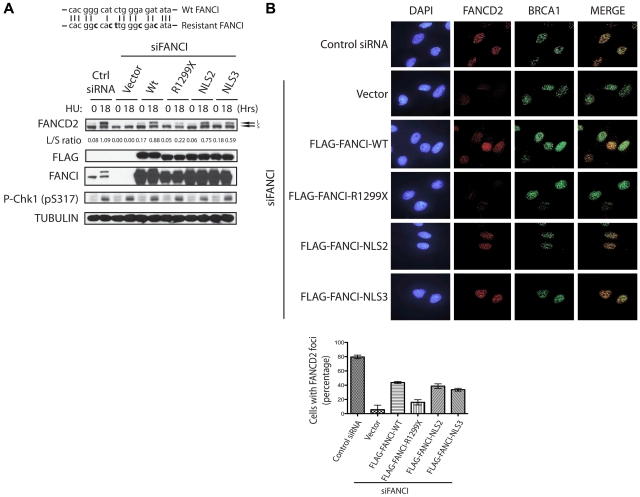
Figure 3.
Addition of an NLS sequence to FANCI R1299X can rescue FANCD2 monoubiquitination and nuclear foci formation after DNA damage. (A) Diagram of the 21-bp FANCI oligonucleotide siRNA target sequence (top line). Bold letters indicate silent mutations introduced in the siRNA-resistant FANCI cDNA (bottom line). U2OS cells were transfected with control or FANCI-specific siRNA for 24 hours, followed by transfection of empty vector or the indicated FANCI expression constructs. Twenty-four hours after the second transfection, cells were treated with 2mM HU for 18 hours and FANCD2 monoubiquitinatation was assessed by Western blot analysis with the indicated antibodies. The L/S ratio between the monoubiquitinated (L) and unmodified form of FANCD2 (S) is shown and measured using ImageJ Version 1.43 software. (B) Replacement of endogenous FANCI in U2OS with empty vector or the indicated FANCI expression constructs were performed as described in panel A. Cells were then treated with 2mM HU for 18 hours and FANCD2 and BRCA1 foci formation was analyzed by indirect immunofluorescence as indicated. FANCD2 nuclear foci after HU treatment were quantified as percentage of cells with 5 or more FANCD2 foci. Error bars represent the SD from 3 independent experiments.
The addition of an NLS sequence to the FANCI R1299X protein can rescue DNA damage-inducible FANCD2 monoubiquitination
To determine whether the loss of FANCI nuclear localization is the cause for the monoubiquitination defect observed in the F010191 FA-I cells, we decided to test, in a heterologous cell-based system, what effect the expression of different FANCI mutants had on FANCD2 monoubiquitination. To do this, we first developed a FANCI WT expression construct harboring silent mutations that could not be targeted by the oligonucleotide siRNA used in this study to knockdown human endogenous FANCI (Figure 3A). As shown in Figure 3A, siRNA knockdown of endogenous FANCI abolished FANCD2 monoubiquitination after HU treatment. However, the monoubiquitination of FANCD2 could be rescued with the expression of WT FANCI that is resistant to the FANCI siRNA sequence (Figure 3A). Importantly, the expression of FANCI R1299X could not restore FANCD2 monoubiquitination in FANCI siRNA-treated cells. This provided us an opportunity to independently validate the FANCI R1299X-mediated FA pathway defect in a heterologous cell-based system. Analysis of 2 different NLS sequences added to the FANCI R1299X mutant showed that they were both capable of correcting the FANCD2 monoubiquitination defect (Figure 3A). This suggests that the inability of FANCI to appreciably accumulate inside the nucleus results in the defect in FANCD2 monoubiquitination. This underscores the relevance and significance of proper protein localization as a mean to fully activate and engage a DNA damage response pathway.
It has been shown that monoubiquitination of FANCI and FANCD2 is required for its targeting to DNA damage/repair foci in the nucleus.5–7 As expected, the expression of FANCI constructs that were capable of supporting FANCD2 monoubiquitination, was also necessary to promote DNA damage-induced FANCD2 nuclear foci formation in cells treated with FANCI siRNA (Figure 3B). As shown previously, the loss of FANCI did not perturb BRCA1 nuclear foci formation6 (Figure 3B). These studies suggest that loss of the C-terminal NLS on FANCI can dramatically alter the cell's ability to mediate a DNA damage response through the FA pathway. Despite the loss of the EDGE motif, artificial fusion of NLS to FANCI R1299X mutant can rescue the FANCD2 monoubiquitination and localization to DNA damage/repair sites on chromatin.
Correction of the F010191 FA-I patient-derived fibroblasts with ectopic expression of FANCI
Despite the increasing number of FA-I patients being characterized, only one FA-I patient cell line has been functionally corrected with the introduction of WT FANCI.7 It is unclear why FA-I patient-derived cells have been difficult to functionally correct. Several laboratories, including ours, have used different methods of gene transduction to achieve FANCI WT expression into the different FA-I lymphoblasts, but with minimal success (data not shown). Instead of using patient lymphoblasts, we decided to obtain fibroblasts from the same F010191 FA-I patient to determine whether these cells would be easier to functionally correct. Interestingly, the level of FANCI R1299X protein expression is significantly lower than in other normal fibroblast or transformed epithelial cells compared with their FANCI levels (Figure 4A; and data not shown). It is unclear what causes the decrease in total FANCI protein in F010191 FA-I fibroblasts but not in the lymphoblasts. Nevertheless, the F010191 fibroblasts mimic a cell line with reduced FANCI protein levels, similar to the condition during FANCI siRNA knockdown treatment. Surprisingly, simple transient expression of WT FANCI was capable of increasing the levels of FANCD2 monoubiquitination (Figure 4B). As expected, expression of FANCI K523R (FANCI mutant that cannot be monoubiquitinated) and R1299X failed to increase FANCD2 monoubiquitination in the F010191 fibroblasts (Figure 4B). This is an important control to show that simple overexpression of FANCI mutants is insufficient to activate FANCD2 monoubiquitination. Importantly, expression of FANCI-NLS2 and -NLS3 was capable of increasing the monoubiquitination levels of FANCD2 (compare WT with NLS2 and NLS3 L/S ratios; Figure 4C). This was also true for the correction of FANCD2 nuclear foci formation in the F010191 fibroblasts with WT versus NLS2 and NLS3 mutants (Figure 5A-B). Thus, the molecular events associated with FA pathway activation appears to be operational after complementing the F010191 FA-I fibroblasts with WT FANCI or a FANCI mutant missing the EDGE motif that is still capable of localizing and accumulating to the nucleus.
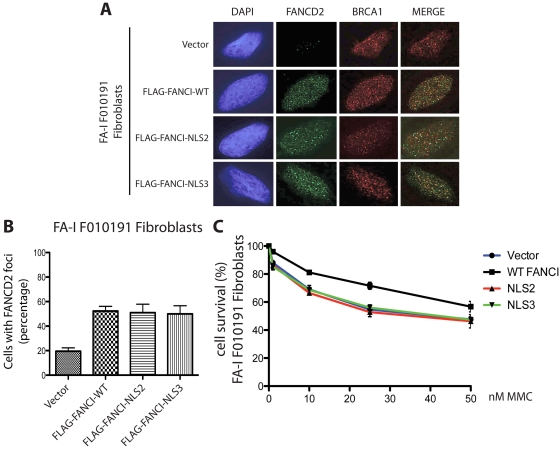
Figure 5.
FANCI missing the EDGE motif fails to correct MMC sensitivity in the FA-I F010191 fibroblasts. (A) F010191 fibroblasts were transfected with empty vector or the indicated FANCI expression constructs. Forty-eight hours after transfection, cells were treated with 2mM HU for 24 hours and processed for immunostaining. FANCD2 and BRCA1 nuclear foci formation was analyzed by immunofluorescence as previously described. (B) FANCD2 nuclear foci after HU treatment were quantified. Error bars represent SD from 3 independent experiments. (C) FA-I F010191 fibroblasts were transfected with either empty vector or the indicated FANCI expression constructs; and 48 hours after transfection, cells were treated with MMC at the indicated dose for 5 more days. Percentage cell survival is shown as an average of 3 independent experiments. Error bars represent the SD for the different experiments performed.
FANCI missing the EDGE motif fails to correct DNA crosslinker sensitivity
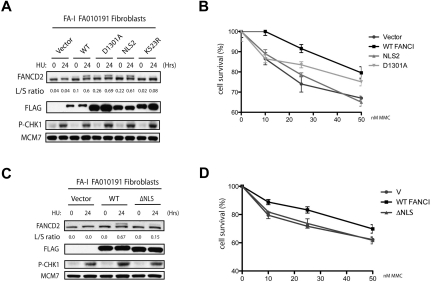
The FANCI R1299X mutant has a deletion of the last 30 amino acids. Within this short region contains 2 putative motifs: one is an NLS and the other is a lesser-known or characterized EDGE motif, which lies upstream of the NLS. Throughout this study, we have shown that the defect in FANCD2 monoubiquitination and nuclear foci localization can be attributed to the loss of the C-terminal NLS sequence in the naturally occurring R1299X FANCI mutant. However, it is unclear whether the EDGE motif plays any functional role in the FA pathway because it is not required for FANCI nuclear localization, FANCD2 monoubiquitination, and FANCD2 nuclear foci formation. Therefore, we tested whether expression of FANCI without the EDGE motif could still correct DNA crosslinker sensitivity in the F010191 fibroblasts. Surprisingly, expression of WT FANCI, but not the FANCI-NLS2 and -NLS3 mutants, was found to be more resistant to the DNA crosslinker MMC in the F010191 fibroblasts (Figure 5C). Thus, artificial nuclear accumulation of the FANCI R1299X mutant is not sufficient to fully correct FA-I cells. This opens the possibility that the EDGE motif is required for an unspecified function that is independent or downstream of FANCD2-FANCI DNA damage-induced monoubiquitination and foci formation. To further characterize the EDGE motif, we tested the D1301A point mutant of the EDGE motif that was first shown to modulate in vitro DNA-binding activity of FANCI.25 We found that the D1301A mutant of FANCI behaved similarly to the NLS2 construct, in terms of rescuing the FANCD2 monoubiquitination and foci formation defect in the F010191 fibroblasts (Figure 6A and data not shown). However, the D1301A mutant could not fully complement the MMC sensitivity to WT levels (Figure 6B). The slight increase in MMC resistance of the D1301A mutant compared with the NLS2 construct is probably the result of the milder mutation of the EDGE motif generated in the mutants. Although highly improbable, it is a possibility that the FANCI EDGE motif may mediate DNA crosslinker resistance outside of the nucleus. To rule out a potential EDGE motif function that is independent of its nuclear localization, we tested whether the FANCI-ΔNLS could rescue MMC sensitivity in the F010191 fibroblasts. As expected, FANCI-ΔNLS construct failed to functionally complement the FA-I fibroblasts for FANCD2 monoubiquitination and MMC resistance (Figure 6C-D), suggesting that the intact EDGE motif is not sufficient for its role in DNA crosslink repair without proper protein localization. The potential role for the EDGE motif in FANCI will be addressed in “Discussion.”
Figure 6.
A deletion of the C-terminal NLS of FANCI fails to correct MMC sensitivity in the FA-I F010191 fibroblasts. (A,C) F010191 fibroblasts were transfected with empty vector or the indicated FANCI expression constructs. Forty-eight hours after transfection, cells were treated with 2mM HU for 24 hours and FANCD2 monoubiquitination was assessed by Western blot analysis and probed with the indicated antibodies. The L/S ratio was calculated as described in Figure 3A. (B,D) FA-I F010191 fibroblasts were transfected with either empty vector or the indicated FANCI expression constructs; and 48 hours after transfection, cells were treated with MMC at the indicated dose for 5 more days. Percentage cell survival is shown as an average of 3 independent experiments. Error bars represent the SD for the different experiments performed.
Discussion
In this study, we provide evidence to explain the molecular defect in a patient-derived FA-I cell line (F010191) expressing a short C-terminal truncation mutant form of FANCI. Although only missing the last 30 amino acids from the C-terminal end of the FANCI protein, this FA-I cell line has been reported previously to have defective FANCD2 and FANCI monoubiquitination.6 Thus, it was unclear how the C-terminal region mechanistically contributes to the activation of the FA pathway. Here we show that mislocalization of the FANCI protein is responsible for the loss of monoubiquitination and nuclear foci formation in the FANCD2 effector protein. Predicted motifs in the C-terminal region of FANCI suggest an EDGE motif and NLS sequence. To test the contribution of the NLS motif, we artificially fused an NLS sequence (one from the original FANCI NLS, the other from c-Myc NLS) to the C-terminal region of the truncated FANCI protein (R1299X). Importantly, fusion proteins carrying the C-terminal NLS sequences were capable of rescuing the FANCD2 monoubiquitination and nuclear foci formation defect from the R1299X FANCI mutant in different cell types.
For the first time, we were able to functionally complement the FA-I fibroblast cell line (F010191) with WT FANCI expression. This enabled us to directly test the functional contribution of different FANCI mutants toward the monoubiquitination and nuclear foci formation of FANCD2 during DNA damage. We were able to confirm our heterologous cell-based studies and show that the addition of an NLS to the R1299X FANCI mutant was sufficient to overcome the localization defect of the R1299X FANCI mutant and rescue the monoubiquitination and nuclear foci formation of FANCD2 in the FA-I patient-derived fibroblasts (F010191). However, unlike the WT FANCI, the NLS fused R1299X FANCI could not correct MMC sensitivity in the patient fibroblasts. This suggests that the putative EDGE motif (which is still missing in the NLS fused R1299X FANCI mutant) may be important in mediating a function of FANCD2-FANCI independent of FANCI, monoubiquitination and foci formation, such as in DNA damage sensing or DNA crosslink repair.
As mentioned in “Results,” FANCI has a putative EDGE motif that resides in the last 30 residues of the protein. To study the role of different structural domains on FANCI, we initially focused our attention on patient-derived mutations of the FANCI gene that may shed some light on critical structural regions of the FANCI protein for normal FA pathway function. Fortunately, the FA-I F010191 patient-derived cells have one mutant expressing allele that results in the deletion of the last 30 residues of FANCI (the other mutant allele results in a more severe protein truncation).6 This gave us an excellent opportunity to dissect the structure and function of the C-terminal region containing the EDGE motif in the patient-derived cells because it was already known that the human patient missing this region results in the FA phenotype. The EDGE motif was previously defined by the EDGE sequence itself, containing a cluster of acidic residues (letters denote amino acids).23 It was originally discovered and characterized in a splice variant form of FANCD2 containing exon 44. Reconstitution of FANCD2 mutants with a defective EDGE motif failed to rescue MMC sensitivity in FANCD2-deficient fibroblasts, implying a crucial function of the EDGE motif in DNA damage sensing or DNA repair.23 Interestingly, expression of FANCD2 with defective EDGE motif in the FA-D2 cells displayed normal cellular phenotype regarding the levels observed for FANCD2 monoubiquitination and nuclear foci formation. This molecular signature is strikingly similar to the FANCI EDGE motif scenario portrayed in our present study.
In vitro studies have suggested that purified FANCI can directly bind to different types of DNA substrates.21,25,28 The intrinsic DNA-binding activity of FANCI does not require its monoubiquitination or association with FANCD2.25,28 Interestingly, it was shown that changing a single residue (D1301A) in the FANCI EDGE motif dramatically enhanced its DNA binding for different substrates, suggesting that the EDGE motif may be involved in regulating the dynamics of FANCI-FANCD2 DNA binding during interstrand crosslink repair.25 However, from these previous studies, it was still unclear whether the intrinsic DNA-binding activity of FANCI functions upstream or downstream of FANCI and FANCD2 monoubiquitination and nuclear foci formation in vivo. Evidence from our present study points to an independent function for the FANCI EDGE motif. This is in agreement with the current model suggesting that both FANCI and FANCD2 may be directly involved in binding to specific DNA structural intermediates to function in DNA repair.25,29 It will be interesting to test whether mutations in the EDGE motif prevent FANCI-FANCD2 from dissociating from DNA in a timely manner to insure proper recovery from DNA crosslink repair, in vivo. Recently, several studies suggest that the monoubiquitinated FANCI-FANCD2 pair is responsible for recruiting a novel FA-associated nuclease, FAN1, which is critical for DNA crosslink repair.30–33 It will be interesting to test whether the EDGE motif in FANCI is required for subsequent FAN1 binding after DNA damage. It has been suggested, although not proven, that the monoubiquitination of FANCI-FANCD2 provides a dock for the recruitment of FAN1 to FANCD2 via its UBZ domain (ubiquitin-binding zinc finger motif). Generally, ubiquitin-binding domain-containing substrates that interact with their ubiquitinated receptors have a secondary binding site with their cognate receptors. This appears to be the case for proliferating cell nuclear antigen and the translesion synthesis polymerases, whereby the translesion synthesis polymerases interact with monoubiquitinated proliferating cell nuclear antigen via their PIP box and ubiquitin-binding domains for a more specific interaction.34
Mutations in FA genes that cause protein mislocalization probably play an important role in the molecular pathogenesis of FA. For example, the 8 proteins forming the FA core ubiquitin ligase complex, and other FA-associated proteins are normally localized predominantly in the nucleus.35–42 Specific patient-derived mutations in FA core complex genes, such as in FANCA, can result in aberrant cytoplasmic localization of the FANCA protein, which can lead to defects in FA core complex assembly, FANCD2 monoubiquitination, and interstrand crosslink repair.43 Although it is possible that FA proteins may possess cytoplasmic function, the majority of the FA pathway function takes place in the nucleus and possibly on chromatin-bound DNA. Localization of FA proteins, including FANCI, to the nuclear compartment is probably critical for the efficient activation of DNA damage sensing, signaling, and repair of DNA lesions. In our present study, we provide evidence that a patient-derived mutant form of FANCI (R1299X) is mislocalized into the cytoplasmic compartment. This is the result of the loss of its C-terminal NLS sequence. We found that the localization defect in the FANCI mutant did not drastically affect or change the localization of FANCD2. This suggests that FANCI may be targeted to the nucleus independently of FANCD2 and that FANCD2 subcellular localization is not completely dependent on FANCI. Based on primary amino acid sequence analysis, human FANCD2 has 2 NLS sequences that are unrelated to the C-terminal NLS on FANCI.44 However, to the best of our knowledge, the function of these 2 FANCD2 NLS sequences has not been tested. Currently, the model for FA pathway activation involves the interaction between FANCI with FANCD2 on nuclear chromatin-bound DNA to facilitate their mutual monoubiquitination.6,7,21 In addition, it is probable that the nuclear PI3-like kinase, ATR, is important in stimulating this reaction through phosphorylation of the ID complex.8,45,46
Mislocalization of FA proteins may also play a role in carcinogenesis. In a recent study, FANCD2 was found to be largely cytoplasmic in more malignant human breast tumor tissues.47 Cytoplasmic localization of FANCD2 in cells, similar to that of FANCI, also probably prevents FA pathway activation, leading to genome instability. It is still unclear what is the precise mechanism that causes the FANCD2 protein mislocalization in these human breast cancer tissues.47 It will be interesting to determine whether FANCI may also be abnormally expressed and cytoplasmically localized in human breast cancers and whether this is the result of sporadic mutations that target the C-terminal NLS function of FANCI. It is known that defects in protein NLSs have been associated with human disease conditions, such as numerous cancers and developmental disorders.48 The increasing prevalence of FA gene mutations observed in human cancers1,49 underscores the importance of careful structure/function analyses of FA proteins and their role in facilitating genome stability.
Acknowledgments
The authors thank the affected FA-I persons and their families for providing tissue samples for these studies, the many physicians that have referred families for participation in the research, A. Auerbach and F. Lach for normal patient fibroblasts and lymphoblast control cells and for helping with the FA-I F010191 characterization and cell line work, Y. Deng and NYU Skirball Imaging core facility for assistance with microscopy, and members of the laboratories of T.T.H., D. Bar-Sagi, and D. Reinberg for their technical assistance, equipment, and helpful discussions.
This work was supported in part by the Wendy Will Case Foundation, National Institutes of Health (grant RO1GM084244), and New York University School of Medicine Start-up funding (T.T.H.).
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: L.C., M.J.K.J., X.M.C.-R., and T.T.H. designed and executed experiments; D.S. and H.H. provided patient-derived fibroblasts and other critical reagents for this study; and L.C. and T.T.H. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tony T. Huang, Department of Biochemistry, New York University School of Medicine, 522 First Ave, Smilow Bldg 210, New York, NY 10016; e-mail: tony.huang@nyumc.org.
References
- 1.D'Andrea AD. Susceptibility pathways in Fanconi's anemia and breast cancer. N Engl J Med. 2010;362(20):1909–1919. doi: 10.1056/NEJMra0809889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auerbach AD. Fanconi anemia and its diagnosis. Mutat Res. 2009;668(1):4–10. doi: 10.1016/j.mrfmmm.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moldovan GL, D'Andrea AD. How the Fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8(10):735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Higuera I, Taniguchi T, Ganesan S, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7(2):249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 6.Sims AE, Spiteri E, Sims RJ, III, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14(6):564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 7.Smogorzewska A, Matsuoka S, Vinciguerra P, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129(2):289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishiai M, Kitao H, Smogorzewska A, et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol. 2008;15(11):1138–1146. doi: 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhi G, Wilson JB, Chen X, et al. Fanconi anemia complementation group FANCD2 protein serine 331 phosphorylation is important for Fanconi anemia pathway function and BRCA2 interaction. Cancer Res. 2009;69(22):8775–8783. doi: 10.1158/0008-5472.CAN-09-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bridge WL, Vandenberg CJ, Franklin RJ, Hiom K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat Genet. 2005;37(9):953–957. doi: 10.1038/ng1627. [DOI] [PubMed] [Google Scholar]
- 11.Levran O, Attwooll C, Henry RT, et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat Genet. 2005;39(9):931–933. doi: 10.1038/ng1624. [DOI] [PubMed] [Google Scholar]
- 12.Levitus M, Waisfisz Q, Godthelp BC, et al. The DNA Helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat Genet. 2005;37(9):934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- 13.Howlett NG, Taniguchi T, Olson S, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297(5581):606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 14.Reid S, Schindler D, Hanenberg H, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39(2):162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 15.Xia B, Dorsman JC, Ameziane N, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39(2):159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 16.Vaz F, Hanenberg H, Schuster B, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42(5):406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 17.Meindl A, Hellebrand H, Wiek C, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42(5):410–414. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 18.Dorsman JC, Levitus M, Rockx D, et al. Identification of the Fanconi anemia complementation group I gene, FANCI. Cell Oncol. 2007;29(3):211–218. doi: 10.1155/2007/151968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nijman SM, Huang TT, Dirac AM, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17(3):331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Huang TT, Nijman SM, Mirchandani KD, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8(4):339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 21.Knipscheer P, Raschle M, Smogorzewska A, et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326(5960):1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalb R, Neveling K, Hoehn H, et al. Hypomorphic mutations in the gene encoding a key Fanconi anemia protein, FANCD2, sustain a significant group of FA-D2 patients with severe phenotype. Am J Hum Genet. 2007;80(5):895–910. doi: 10.1086/517616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montes de Oca R, Andreassen PR, Margossian SP, et al. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood. 2005;105(3):1003–1009. doi: 10.1182/blood-2003-11-3997. [DOI] [PubMed] [Google Scholar]
- 24.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20(22):8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan F, El Hokayem J, Zhou W, Zhang Y. FANCI protein binds to DNA and interacts with FANCD2 to recognize branched structures. J Biol Chem. 2009;284(36):24443–24452. doi: 10.1074/jbc.M109.016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dang CV, Lee WM. Identification of the human c-myc protein nuclear translocation signal. Mol Cell Biol. 1988;8(10):4048–4054. doi: 10.1128/mcb.8.10.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saphire AC, Bark SJ, Gerace L. All four homochiral enantiomers of a nuclear localization sequence derived from c-Myc serve as functional import signals. J Biol Chem. 1998;273(45):29764–29769. doi: 10.1074/jbc.273.45.29764. [DOI] [PubMed] [Google Scholar]
- 28.Longerich S, San Filippo J, Liu D, Sung P. FANCI binds branched DNA and is monoubiquitinated by UBE2T-FANCL. J Biol Chem. 2009;284(35):23182–23186. doi: 10.1074/jbc.C109.038075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moldovan GL, D'Andrea AD. FANCD2 hurdles the DNA interstrand crosslink. Cell. 2009;139(7):1222–1224. doi: 10.1016/j.cell.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacKay C, Declais AC, Lundin C, et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142(1):65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kratz K, Schopf B, Kaden S, et al. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142(1):77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 32.Smogorzewska A, Desetty R, Saito TT, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39(1):36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329(5992):693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- 34.Bienko M, Green CM, Crosetto N, et al. Ubiquitin-binding domains in y-family polymerases regulate translesion synthesis. Science. 2005;310(5755):1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 35.Savoia A, Garcia-Higuera I, D'Andrea AD. Nuclear localization of the Fanconi anemia protein FANCC is required for functional activity. Blood. 1999;93(11):4025–4026. [PubMed] [Google Scholar]
- 36.Taniguchi T, D'Andrea AD. The Fanconi anemia protein, FANCE, promotes the nuclear accumulation of FANCC. Blood. 2002;100(7):2457–2462. doi: 10.1182/blood-2002-03-0860. [DOI] [PubMed] [Google Scholar]
- 37.Yamashita T, Kupfer GM, Naf D, et al. The Fanconi anemia pathway requires FAA phosphorylation and FAA/FAC nuclear accumulation. Proc Natl Acad Sci U S A. 1998;95(22):13085–13090. doi: 10.1073/pnas.95.22.13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naf D, Kupfer GM, Suliman A, Lambert K, D'Andrea AD. Functional activity of the Fanconi anemia protein FAA requires FAC binding and nuclear localization. Mol Cell Biol. 1998;18(10):5952–5960. doi: 10.1128/mcb.18.10.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kupfer G, Naf D, Garcia-Higuera I, et al. A patient-derived mutant form of the Fanconi anemia protein, FANCA, is defective in nuclear accumulation. Exp Hematol. 1999;27(4):587–593. doi: 10.1016/s0301-472x(99)00022-3. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Higuera I, Kuang Y, Naf D, Wasik J, D'Andrea AD. Fanconi anemia proteins FANCA, FANCC, and FANCG/XRCC9 interact in a functional nuclear complex. Mol Cell Biol. 1999;19(7):4866–4873. doi: 10.1128/mcb.19.7.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kupfer GM, Naf D, Suliman A, Pulsipher M, D'Andrea AD. The Fanconi anaemia proteins, FAA and FAC, interact to form a nuclear complex. Nat Genet. 1997;17(4):487–490. doi: 10.1038/ng1297-487. [DOI] [PubMed] [Google Scholar]
- 42.Garcia-Higuera I, Kuang Y, Denham J, D'Andrea AD. The Fanconi anemia proteins FANCA and FANCG stabilize each other and promote the nuclear accumulation of the Fanconi anemia complex. Blood. 2000;96(9):3224–3230. [PubMed] [Google Scholar]
- 43.Adachi D, Oda T, Yagasaki H, et al. Heterogeneous activation of the Fanconi anemia pathway by patient-derived FANCA mutants. Hum Mol Genet. 2002;11(25):3125–3134. doi: 10.1093/hmg/11.25.3125. [DOI] [PubMed] [Google Scholar]
- 44.Dequen F, St-Laurent JF, Gagnon SN, Carreau M, Desnoyers S. The Caenorhabditis elegans FancD2 ortholog is required for survival following DNA damage. Comp Biochem Physiol B Biochem Mol Biol. 2005;141(4):453–460. doi: 10.1016/j.cbpc.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 45.Andreassen PR, D'Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18(16):1958–1963. doi: 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho GP, Margossian S, Taniguchi T, D'Andrea AD. Phosphorylation of FANCD2 on two novel sites is required for mitomycin C resistance. Mol Cell Biol. 2006;26(18):7005–7015. doi: 10.1128/MCB.02018-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rudland PS, Platt-Higgins AM, Davies LM, et al. Significance of the Fanconi anemia FANCD2 protein in sporadic and metastatic human breast cancer. Am J Pathol. 2010;176(6):2935–2947. doi: 10.2353/ajpath.2010.090779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McLane LM, Corbett AH. Nuclear localization signals and human disease. IUBMB Life. 2009;61(7):697–706. doi: 10.1002/iub.194. [DOI] [PubMed] [Google Scholar]
- 49.Kennedy RD, D'Andrea AD. DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes. J Clin Oncol. 2006;24(23):3799–3808. doi: 10.1200/JCO.2005.05.4171. [DOI] [PubMed] [Google Scholar]