Abstract
Inflammatory bowel disease (IBD) is a group of idiopathic, chronic and relapsing inflammatory conditions of the gastrointestinal tract. Familial and epidemiological studies have stressed the involvement of genetic factors and have also shown the critical role of environmental factors such as sanitation and hygiene in the development of IBD. However, the molecular mechanisms of intestinal inflammation in IBD have long remained unknown. In recent years, the study of susceptibility genes involved in the detection of bacterial components and in the regulation of the host immune response has shed light onto the potential role of intestinal pathogens and gut flora in IBD immunobiology. This review presents current knowledge on intestinal epithelial barrier alterations and on dysfunction of mucosal innate and acquired immune responses in IBD. The data support the etiological hypothesis which argues that pathogenic intestinal bacteria and/or infectious agents initiate and perpetuate the inflammation of the gut through disruption of tolerance towards the commensal microbiota in an individual with genetic vulnerability.
Key words: IBD, Crohn disease, ulcerative colitis, immunobiology, defensins, intestinal bacteria, T cell, dendritic cell
Definition and Epidemiology
Inflammatory bowel disease (IBD) is characterized by severe inflammation of the small bowel and/or the colon leading to recurrent diarrhea and abdominal pain. Crohn disease (CD) and ulcerative colitis (UC) are the two main clinicopathological subtypes of IBD. Despite both being chronic and relapsing inflammatory diseases of the bowel, they can be differentiated by the location of the inflammation in the gastrointestinal tract and by the nature of the histological alterations in the intestinal wall. Anatomically, CD can affect the entire gastrointestinal tract from mouth to anus, although it commonly affects the terminal ileum and colon. UC is restricted to the rectum, colon and caecum. Microscopically, CD is transmural and often discontinuous while UC affects only the intestinal mucosa in a continuous pattern.1
IBD is a very disabling disease due to the fatigue associated with the inflammatory symptoms and due to the chronic pain suffered by patients. IBD affects quality of life but not life span: the mortality rate of patients is not different from the normal population. The prevalence of IBD is similar in men and women and peaks in young adults. The highest incidences are reported in northern Europe and North America where they range from 12 to 19/100,000/year and from 5 to 29/100,000/year, respectively. Nearly 1.4 million Americans and 2.2 million Europeans are affected.2 The prevalence of IBD varies depending on ethnicity or race. Caucasians and African Americans are most affected while IBD is rare among Hispanics and Asians. Ashkenazi Jews have a much higher risk of developing IBD with a 2–4 times higher incidence when compared to non-Jewish Caucasians.3 However, there is evidence to suggest that the prevalence of IBD increases in populations and regions with industrialization. For example, a study demonstrated a similar incidence rate of IBD in African Americans compared to that observed in Americans of European ancestry.4
IBD Etiology
The pathogenesis of IBD is only partially understood; various environmental and host (e.g., genetic, epithelial, immune and nonimmune) factors are involved. Complex interactions between the immune system, enteric commensal bacteria/pathogens and host genotype are thought to underlie the development of IBD.5–7 These relapsing chronic inflammatory disorders appear to be caused by overly aggressive T-cell responses directed against environmental factors and/or a subset of commensal bacteria/pathogens that inhabit the distal ileum and colon of genetically susceptible hosts.
Environmental contribution to IBD.
The lower incidence of IBD in Asia and Africa compared with North America and Europe probably reflects both genetic and environmental factors. The hygiene hypothesis has been invoked to account for the rising prevalence of various autoimmune and inflammatory conditions, which are thought to result from the lack of early exposure to select microbial agents due to stringent sanitation conditions.8 Although the precise environmental factors that account for changing IBD prevalence have not been defined, changes in dietary and intestinal microbial milieu probably play a key pathogenic role that is common to CD and UC. One important environmental factor is smoking. The effect of smoking is discordant between CD and UC.9 Smoking was found to be protective against UC and, after the onset of the disease, might improve its course, decreasing the need for colectomy.10 In contrast, smoking increases the risk of developing CD and aggravates its course by approximately twofold in western countries.10–12 Patients with a high lifetime tobacco exposure (>150 cigarettes/year) suffer small bowel disease more often than do nonsmokers. Smokers also have a higher accumulated number of fistulae and/or abscesses than nonsmokers and are also more vulnerable to CD flare-ups.13 In particular, heavy smokers (>10 cigarettes/day) are more likely to require gastric surgery as a result of IBD and suffer a significantly decreased quality of life.14,15
Susceptibility genes in IBD.
Considering that IBD incidence seems linked to race, a strong genetic component in IBD etiology is very likely. Some epidemiological data support this hypothesis: (1) the heterogeneity in the geographical distribution of IBD, (2) the existence of familial forms of IBD and (3) the high rate of concordance in monozygotic twins. Indeed, population-based studies find that 5–10% of patients have a first-degree family member with IBD, with the relative risk to siblings of affected individuals estimated to be 30- to 40-fold higher for CD and 10- to 20-fold higher for UC, suggesting that some susceptibility genes should be specific for CD and UC.16,17 Twin studies provide additional evidence for genetic contribution in IBD: the concordance rate is significantly greater in monozygotic than dizygotic twins for both CD (50–58% versus 0–12%) and UC (6–14% versus 0–5%).18,19 However, the significantly higher monozygotic twin concordance rates for CD than UC suggest that there is a greater contribution of genetic factors in CD.
In recent years, genome-wide association (GWA) studies have provided a broad view of the relative contributions of various genomic loci and have substantially improved the understanding of the molecular pathways leading to IBD. GWA have stressed an association between IBD and specific chromosomal loci20,21 and thereby have led to the identification of candidate genes which are involved in the development of inflammatory processes (Table 1). In general, the main genetic associations can be divided into genes that contribute to the innate immune response and those that contribute to the acquired immune response. In the innate immune arm, the association of CD with polymorphisms in NOD222,23 and the two autophagy-related genes ATG16L1 and IRGM24–26 suggests that alterations in the recognition and intracellular processing of bacterial components may have a role in the immunopathogenesis of the disease.27 A link between nucleotide-binding oligomerization domain (NOD) protein and the autophagy protein ATG16L1 has been recently reported by showing that these two proteins form a complex at the cytoplasmic membrane.28 This highlights the hypothesis of autophagy deficiency in IBD patients since mutations in NOD or the ATG16L1 protein result in diminished and ineffective autophagic response to intracellular pathogens.
Table 1.
Chromosomal loci of susceptibility for IBD
| Genetic marker | Locus | Candidate gene(s) | Proposed role(s) in inflammation | Related disease | Associated polymorphism(s) | Reference(s) | PMID |
| IBD1 | 16q12 | NOD2/CARD15, CD19, sialophorin, CD11 integrin cluster, IL4R | Detection of cytosolic bacterial components | CD; UC? | SNPs D16S409 and D16S419 | Hugot, et al. 1996 | 11385576 |
| SNPs D16S409 and D16S753 | Cavanaugh, et al. 1998 | 9924607 | |||||
| SNP D16S3117 | Zouali, et al. 2001 | 11781683 | |||||
| IBD2 | 12p13.2-q24.1 | STAT6, VDR, IFNγ | Development of T-cell response | CD + UC | SNP D12S83 | Duerr, et al. 1998 | 9634527 |
| IBD3 | 6p21.3 | MHC HLA-DQA1, HLA-DRA, HLA-DRB5 and HLA-DRB1, TNFalpha, TNFbeta, IL10, BTLN2 | Detection of non-self components | CD + UC | SNPs D6S289 and D6S276 | Hampe, et al. 1999 | 10053016 |
| IBD4 | 14q11-q12 | MMP14, DAD1, TCR | Development of T-cell response | CD | SNP D14S261 | Duerr, et al. 2000 | 10747815 |
| IBD5 | 5q31 | IL3, IL4, IL5 and IL13, OCTN1, OCTN2, CSF2, SLC22A5 | Regulation of inflammation | CD | SNPs rs6596075 and rs2188962 | Rioux, et al. 2000–2001 | 11586304, 10777714 |
| IBD6 | 19p13 | ICAM-1, C3, TBXA2R, LTB4H | Diapedesis, stimulation of neutrophil response | CD + UC | ns | Van Heel, et al. 2003 | 12928481 |
| IBD7 | 1p36 | PADI4 | ns | CD + UC | SNPs D1S1597, D1S507 and D1S1628 | Cho, et al. 1998–2000 | 9636179, 10814724 |
| IBD8 | 16p | ns | ns | UC | SNP D16S408 | Annese, et al. 2003 | 14627676 |
| IBD9 | 3p26 | DEC1 | Anti-neoplasia | CD + UC | SNP D3S1297 | Duerr, et al. 2002 | 12354785 |
| IBD10 | 2q37.1 | ATG16L1 | Autophagy | CD | SNP rs2241880 | Hampe, et al. 2007 | 17200669 |
| IBD11 | 7q22 | MUC3A | Protection of the epithelial barrier | CD + UC | SNP D7S669 | Satsangi, et al. 1996 | 8841195 |
| IBD12 | 3p21 | MST1, BSN, GNAI2 | Regulation of proinflammatory mediators expression | CD + UC | SNP D3S2432 | Paavola, et al. 2001 | 11378820 |
| IBD13 | 7q21.1 | ABCA1 | Regulation of glucocorticosteroids production? | CD + UC | SNP D7S669 | Satsangi, et al. 1996 | 8841195 |
| IBD14 | 7q32 | IRF5 | Regulation of proinflammatory mediators expression | CD + UC | CGGGG insertion in the promoter of the gene IRF5 | Dideberg, et al. 2007 | 17881657 |
| IBD15 | 10q21 | ZNF365 | Regulation of proinflammatory mediators expression? | CD + UC | SNP rs224136 | Rioux, et al. 2007 | 17435756 |
| SNP rs10761659 | Fisher, et al. 2008 | 18438406 | |||||
| SNP rs10995271 | Barrett, et al. 2008 | 18587394 | |||||
| IBD16 | 9q32 | TNFSF15 | Costimulation of activated T CD4+ cells | CD + UC | SNP D9S2157 | Cho, et al. 1998 | 9636179 |
| SNPs rs3810936, rs6478108, rs6478109, rs7848647 and rs7869487 | Yamakasi, et al. 2005 | 16221758 | |||||
| SNP rs4263839 | Barrett, et al. 2008 | 18587394 | |||||
| IBD17 | 1p31.1 | IL23R, PTGER4 | Generation and maintenance of Th17 cells; prostaglandins signaling | CD + UC | R381Q substitution in IL23R (rs11209026) | Libioulle, et al. 2007 | 17447842 |
| SNP rs7517847 | Rioux, et al. 2007 | 17435756 | |||||
| SNP rs4613763 | Barrett, et al. 2008 | 18587394 | |||||
| IBD18 | 5p13.1 | Gene modulating PTGER4? | Prostaglandins signaling? | CD + UC | SNP rs1373692 | Libioulle, et al. 2007 | 17447842 |
| SNP rs4613763 | Barrett, et al. 2008 | 18587394 | |||||
| SNP rs1992660 | Franke, et al. | 2007 18438405 | |||||
| IBD19 | 5q33.1 | IRGM, IL12B | Autophagy | CD | SNP IL12B rs6887695 | Parkes, et al. 2007 | 17554261 |
| SNP IL12B rs6556416 | Fisher, et al. 2008 | 18438406 | |||||
| SNPs IRGM rs4958847 and rs4958427 | Franke, et al. 2007 | 18438405 | |||||
| SNPs IRGM rs11747270 and rs10045431 | Barrett, et al. 2008 | 18587394 | |||||
| IBD20 | 10q23-q24 | DLG5, NKX2-3 | Maintaining epithelial integrity | CD + UC | SNPs rs11190140 and rs10883365 | Franke, et al. 2008 | 18438405 |
| SNPs D10S547 and D10S20 | Hampe, et al. 1999 | 10053016 | |||||
| SNP rs10883365 | Parkes, et al. 2007 | 17554261 | |||||
| IBD21 | 18p11 | NDUFV2, VAPA, PACAP, PTPN2 | TCR signaling | CD + UC | SNP PTPN2 rs2542151 | Parkes, et al. 2007 | 17554261 |
| IBD22 | 17q21 | STAT3, ORMDL3 | Development of T cell response | CD | SNP ORMDL3 rs2872507 | Barrett, et al. 2008 | 18587394 |
| SNP STAT3 rs744166 | Franke, et al. 2008 | 18438405 | |||||
| IBD23 | 1q32 | IL10 | Regulation of inflammatory mediators production | CD + UC | SNP-1082G | Fowler, et al. 2005 | 15937090 |
| SNP rs17419032 | Franke, et al. 2008 | 18438405 | |||||
| SNP rs3024505 | Franke, et al. 2008 | 18836448 | |||||
| IBD24 | 20q13 | TNFRSF6B or DCR3 | Costimulation of activated T CD4+ cells | CD + UC | SNPs rs2315008 and rs4809330 | Kugathasan, et al. 2008 | 18758464 |
| IBD25 | 21q22 | PSMG1 | Autophagy | CD + UC | SNP rs2836878 | Kugathasan, et al. 2008 | 18758464 |
| IBD26 | 12q15 | ns | ns | UC | SNPs rs1558744, rs7134599, rs2870946 | Silverberg, et al. 2009 | 19122664 |
| IBD27 | 13q13.3 | ns | ns | CD | SNPs rs20411 | Shugart, et al. 2008 | 18246054 |
| UC | SNPs rs17085007 | Asano, et al. 2009 | 19915573 | ||||
| IBD28 | 21q22 | IL10RA | Regulation of inflammation | CD + UC | SNPs rs2836878 | Glocker, et al. 2009 | 19890111 |
ns, not specified; ABC, ABC transporter; ATG16L1, autophagy-related protein 16-1; BSN, Bsn nuclease; C3, complement C3; CD, cluster of differentiation or Crohn disease; CSF, colony stimulating factor; DAD, daughters against ddp protein; DCR, decoy receptor; DEC, differentially expressed in chondrocytes; DLG, disc large homolog; GNAI, guanine nucleotide binding protein, alpha inhibiting; IBD, inflammatory bowel disease; ICAM, intercellular adhesion molecule; IL, interleukin; IFN, interferon; IRF, interferon regulatory factor; IRGM, immunity-related GTPase family, M protein; HLA, human leukocyte antigen; LTB4, leukotriene B4; MHC, major histocompatibility complex; MMP, matrix metalloprotease; NFκB, nuclear factor kappaB; MST, microtubule stabilizing; MUC, mucin; NDUFV, NADH dehydrogenase ubiquinone flavoprotein; NKX, homeodomain protein NKX; NOD2/CARD15, nucleotide-binding oligomerization domain/caspase-activating recruitment domain 15; OCTN, organic cation transporter; ORMDL, orosomucoid (ORM)-like protein family; PACAP, pituitary adenylate cyclase-activating polypeptide; PADI, peptidylarginine deiminase; PSMG, proteasome assembly chaperone; PTGER, G-protein coupled, E-series prostaglandin receptor; PTPN, protein tyrosine-phosphatase; SLC, chemokine CCL21; SNP, single nucleotide polymorphism; STAT, signal transducers and activator of transcription; TBXA2, thromboxane receptor A2; TCR, T-cell receptor; TGF, transforming growth factor; TLR, toll-like receptor; TNF, tumor necrosis factor; TNFSF, tumor necrosis factor superfamily; UC, ulcerative colitis; VAPA, vesicle-associated protein A; VDR, vitamin D receptor; ZNF, zinc finger protein. Reference publication: Wellcome Trust Case Control Consortium: Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007; 447: 661–78; PMID: 17554300.
Among a myriad of genes pinpointed by genetic linkage studies, alterations in genes of the innate immune system, such as NOD2, ATG16L1 and IRGM are specific to patients with CD and are not observed in those with UC. By contrast, multiple genes implicated in the IL-23 pathway, including IL23R (encoding a subunit for IL-23 receptor) and IL12B (encoding the p40 subunit of IL-12 and IL-23), STAT3 (encoding signal transducer and activator of transcription 3) and the homeodomain-containing transcription factor NKX2-3 have been shown to be associated with both UC and CD.25,29–33 On the other hand, mutations in ECM1 (extracellular matrix protein 1) are specific to patients with UC.21,31,32
Microbiota in IBD.
The importance of gut flora in IBD is directly supported by studies in a variety of murine models in which ‘spontaneous’ chronic colitis seems to be entirely dependent on the presence of a luminal flora.34,35 Thus, colitis cannot develop when these lines are maintained in a germ-free gnotobiotic state, but rapidly emerges when they are reconstituted with bacteria that are considered normal constituents of luminal flora.36,37 At least three not necessarily mutually exclusive theories can be proposed concerning the implication of bacteria in the aetiopathogenesis of IBD: (1) an involvement of a persistent pathogen; (2) an abnormally permeable mucosal barrier leading to excessive bacterial translocation; and (3) a breakdown in the balance between putative “protective” versus “harmful” intestinal bacteria (“dysbiosis”) which can promote inflammation.38
The possibility that IBD is an infection has been debated since the first description of CD. Many infectious agents (viral, bacterial or parasitic agents; Table 2) have been suspected in IBD etiology but the strongest evidence comes from studies pointing to the role of bacteria. In the intestinal mucosa of patients with IBD, bacteria that can potentially be pathogenic such as Escherichia coli, Listeria monocytogenes, Yersinia enterocolitica and Mycobacterium avium paratuberculosis are found in excess.39 Mycobacterium avium paratuberculosis was the first organism to be suggested as an IBD pathogen, and it has been argued that it fulfils Koch's postulates40 and could be designated the cause of CD. Other organisms have been postulated as possible IBD pathogens, including various Helicobacter species, one of which has been identified in primate colitis while others are widely used in animal models of IBD.
Table 2.
Targeted search for micro-organisms in tissues of patients with IBD
| Virus | Bacteria | Parasite |
| Adenovirus | Brucella spp. | Borrelia spp. |
| Epstein-Barr virus | Chlamidia spp. | Toxoplasma gondii |
| Cytomegalovirus | Campylobacter spp. | Treponema spp. |
| Flu virus | Escherichia coli | |
| Coronavirus | Helicobacter spp. | |
| Rotavirus | Legionella spp. | |
| Measles virus | Mycobacterium spp. | |
| Respiratory syncytial virus | Pseudomonas spp. | |
| Para-influenza type 2 and 3 virus | Salmonella spp. | |
| Bovine viral diarrhea virus | Staphylococcus spp. | |
| Streptococcus spp. | ||
| Shigella spp. | ||
| Yersinia spp. | ||
| Enterococcus | ||
| Listeria monocytogenes | ||
| Bacteroides vulgatus | ||
| Bacteroides burgdorferi |
Ssp, subspecies.
Escherichia coli appear specific to ileal CD and have been shown to induce the release of TNFα, a key cytokine in IBD inflammation.41,42 Increased numbers of mucosa-associated E. coli forming a biofilm on the surface of the gut mucosa are observed in patients with IBD.43–49 Bacteria colonizing the gut mucosa have the ability (1) to strongly adhere to intestinal epithelial cells (IEC),44,46 (2) to invade IEC by a mechanism involving actin polymerisation and microtubule recruitment,50,51 (3) to survive and replicate inside macrophages inducing the release of large amounts of TNFα and (4) to induce granuloma formation in vitro.42,52,53 Based on the pathogenic characteristics of CD-associated E. coli, an additional pathogenic group of E. coli was defined and named AIEC for Adherent-Invasive E. coli. Such pathogenic strains are achieving increasing relevance since various studies performed in France,54 the United Kingdom,46 Spain55 and the USA56–58 have reported them to be more prevalent in CD patients than in controls. The presence of AIEC in healthy subjects suggests that AIEC are facultative pathogens that cause disease in susceptible hosts. AIEC strains were found to be highly associated with ileal mucosa in CD patients, suggesting that there are specific alterations to the ileal epithelial cells in patients with CD that allow AIEC to adhere to a greater extent. The receptor involved in AIEC colonization and abnormally expressed on ileal mucosa in 35% of CD patients was characterized as being the carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6).59,60 Recently, the endoplasmic reticulum (ER) stress protein Gp96 was found to be overexpressed on the apical surface of ileal epithelial cells in CD patients and to act as a host cell receptor for outer membrane vesicles, promoting AIEC invasion.61
Dysbiosis could be a key factor in the immunopathogenesis of IBD by disrupting the host immune defences to commensal flora microbes at the mucosal border. Arguments in favour of dysbiosis are: (1) the low proportion of Faecalibacterium prausnitzii, i.e., bacteria with anti-inflammatory properties, on ileal CD mucosa,39,62 and (2) the abnormal colonization of the ileal mucosa by AIEC. In line with this hypothesis, a recent twin study revealed specific imbalances in the mucosa-associated microbiota.63 Large interindividual variation in the composition of the intestinal microbiota between unrelated individuals has made it challenging to identify specific aspects of dysbiosis that lead to CD. The microbial compositions at all biopsy locations for each individual were similar, regardless of disease state, but differences between individuals were observed. In particular, individuals with predominantly ileal CD had a dramatically lower abundance of Faecalibacterium prausnitzii and increased abundance of Escherichia coli compared to healthy co-twins and those with CD localized in the colon. This dysbiosis was significantly correlated to the disease phenotype rather than genotype.63 Dysbiosis can promote the growth of invasive pathogenic bacteria and also can facilitate bacterial translocation through the intestinal mucosal barrier to the mesenteric lymph nodes. These two phenomena contribute to the permeabilization process of the epithelial barrier, which is the prerequisite for the activation of the mucosal immune response.
Immunopathogenesis of IBD
In IBD, the immune defense against intestinal microbes fails at two levels: (1) the epithelial mucosal barrier is impaired and (2) the innate and acquired host immune responses are altered. The immunopathogenesis of IBD occurs in three temporally distinct stages: (1) penetration of luminal contents into underlying tissues which may be facilitated by environmental factors such as infection or inherent defects in mucosal barrier, (2) impaired clearance of foreign material from the bowel wall which may be due to defective secretion of pro-inflammatory cytokines by macrophages and (3) a compensatory acquired immune response which leads to a chronic inflammatory response and gives rise to characteristic IBD lesions.
Alterations of the mucosal epithelial barrier.
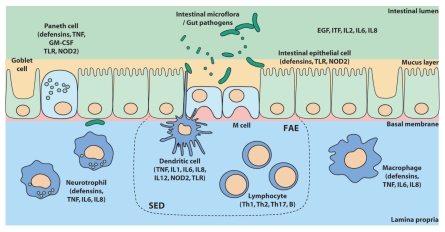
The intestinal barrier (Fig. 1) consists of a bacterial biofilm, a layer of mucus and an intestinal epithelium in which lie the innate immune system (dendritic cells, Paneth cells, macrophages and neutrophils). The intestinal epithelial barrier normally protects the body from potential bacterial threats but in IBD each of these defences is impaired. Indeed, the number of goblet cells secreting mucins, which constitute the protective mucus in the intestinal epithelium, is decreased in IBD. In addition, the use of microarrays has revealed that genes encoding mucins are underexpressed in the ileum and colon of patients.64 The cohesion and tightness of the intestinal mucosa depend on the cell junctions of IEC. Defects in the structure and function of apical junctional complexes (AJC) are implicated in both animal models of IBD and in IBD patients.65,66 For example, deregulated epithelial barrier function, specifically tight junctions, precedes histological evidence of ileitis in SAMP mice.67 Moreover, barrier dysfunction precedes relapse of CD in asymptomatic patients as proteins forming the tight junctions of enterocytes (occludin, cadherins and catenins) are underexpressed68 and studies demonstrate a specific distribution pattern of the tight junction proteins claudin 2, 3, 4, 5 and 8 in IBD patients.69–71
Figure 1.
Most important mechanisms of defence in the intestinal epithelial barrier. Invasion of the mucosa is prevented by physical defences (mucus layer secreted by goblet cells, cellular barrier formed by epithelial cells) and by epithelial immune cells (Paneth cells and M cells). Recognition of pathogenic bacterial components is performed via TLR and NOD receptors. The inflammatory reaction involves mucosal immune system activation (mononuclear cells, lymphocytes and dendritic cells of the subepithelial dome). Inflammation is mediated by cytokines (TNF, IL) secreted by these different immune cells. EGF, epithelial growth factor; GM-CSF, granulocyte macrophage-colony stimulating factor; FAE, follicle-associated epithelium; ITF, intestinal trefoil factor; IL, interleukin; NOD2, nucleotide oligomerization domain 2; SED, subepithelial dome of the Peyer patch; Th, T helper lymphocyte; TLR, toll-like receptor; TNF, tumor necrosis factor. Source: Med Sci (Paris) 2010; 26:405–10.
Intestinal permeability is significantly increased in 36% of CD patients72 and bacterial translocation of E. coli, Enterococcus spp., Clostridium perfringens has been observed in mesenteric nodes in 30–50% of CD patients versus 5–15% in healthy controls.73,74 This could be due to: (1) bacterial uptake via Peyer's patches and (2) bacterial translocation due to abnormal tight junctions. Increased intestinal permeability has been shown in CD patients carrying NOD2 polymorphisms.75 Consistently, abnormal development of Peyer's patches as well as increased permeability and bacterial translocation were observed in NOD2 deficient mice.76 The findings that increased E. coli translocation occurs in follicle-associated epithelium or in Peyer's patches isolated from CD patients compared with non-IBD patients77,78 and that increased numbers of Peyer's patches are found in CD, represent novel insights into CD pathophysiology by a previously unrecognized defect of intestinal barrier function in ileal CD.
Moreover, epithelial cells are the first line of defence against invading pathogens. In constant interaction with the intraluminal flora, they are able to identify pathogenic bacterial components via extracellular receptors to bacterial products like TLR (toll-like receptors) and via intracellular receptors like NLR (NOD-like receptors), in particular NOD2 receptor (or NOD2/CARD15 receptor, Nucleotide-binding Oligomerization Domain 2/Caspase-Activating Recruitment Domain 15 receptor). Once activated through TLR and NOD2 receptors, epithelial cells produce antimicrobial peptides (human β-defensins, HBD) and express MHC molecules to trigger the mucosal acquired immune response. In IBD, there is a functional defect of the immune barrier formed by antimicrobial peptides.79 Indeed, UC is associated with a very high production of HBD2, 3 and 4.80 However, the induction of these HBD is low in CD. The lack of induction of HBD2 in patients suffering from colonic CD could be explained by a reduced copy number of the HBD gene.81 Paneth cells, which are specialized cells located in intestinal crypts, also secrete antimicrobial peptides (human α-defensins, HD) following the binding of pathogenic bacterial components to TLR and NOD receptors. In UC, the production of HD5 and 6 is greatly increased whereas in ileal CD, a sharp decrease in the production of HD is observed.82–84 The lack of HD5 synthesis in ileal CD could result from a defect in the biology of Paneth cells caused by mutations in NOD2. Besides, NOD2-deficient animals are susceptible to Listeria monocytogenes infection via the oral route but not through intravenous or peritoneal delivery. Expression of defensin-related cryptdin 4 (Defcr4) and Defcr-related sequence 10 (Defcr-rs10) was very low in NOD2-/- mice and was further reduced after infection in NOD2-/- animals relative to wild-type mice.85 Cryptdins are antimicrobial peptides that are preferentially produced in intestinal Paneth cells, and their antimicrobial activity is important in suppressing infection with pathogenic bacteria, including L. monocytogenes86 and Mycobacterium paratuberculosis, an organism implicated in CD.87,88 Overall, current data suggest that, in UC, changes in expression of defensins are secondary to inflammation while they could cause impairment of innate mucosal immunity in ileal CD.
Innate immune response deregulation.
The weakening of mucosal defences promotes intestinal epithelium permeabilization resulting in more frequent contacts between the commensal flora and the mucosal immune system. In CD, these interactions would still be facilitated through a lack of bacterial clearance by macrophages in which secretion of inflammatory cytokines is defective.89 An excess of such interactions would cause a loss of tolerance to commensal flora by activating mucosal dendritic cells, the sentinel cells of innate immunity (Fig. 1).
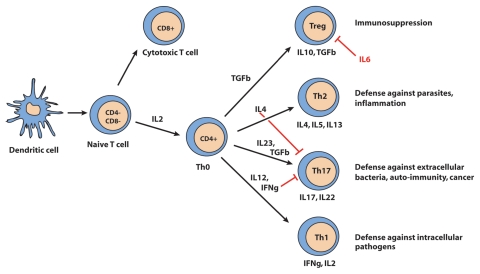
Dendritic cells (DC) are at the interface between IEC and T cells. They present antigens to naive CD4+ helper T cells (Th0) and ensure tolerance to commensal flora by promoting their differentiation into regulatory T cells (Treg). In case of infection, DC activated by their TLR and NOD2 receptors produce pro-inflammatory cytokines and promote the differentiation of effector T cells Th1, Th2 and Th17 (Fig. 2), which triggers a local persistent inflammation.90 The interplay between secreted cytokines determines the balance between the different types of CD4+ effector T cells.91 In IBD, an overactivation of DC was observed at sites of inflammation.92 DC overactivation induces a strong differentiation of effector lymphocytes (CD4+ and CD8+) and of other effector cells such as natural killer (NK) and NK T (NKT) cells while abolishing the production of regulatory cells. The absence of Treg lymphocytes contributes to the development of an immune response to commensal bacteria normally tolerated by the mucosal immune system (peripheral tolerance). Loss of peripheral tolerance then perpetuates inflammation.
Figure 2.
Differentiation of naive T cells (Th0). CD, cluster of differentiation; Th, T helper lymphocyte; IFNγ, interferon gamma; IL, interleukin; TGFβ, transforming growth factor β. Source: Med Sci (Paris) 2010; 26:405–10.
DC overactivation is mainly the consequence of abnormalities in TLR and NOD receptor function and the consecutive inability to detect bacterial components. Indeed, in IBD, alterations in epithelial mechanisms of innate immunity could be caused by changes in TLR expression patterns. The hypothesis of a TLR-dependent dysfunction of the immune response to bacteria in the intestinal lumen is supported by the description of polymorphisms in genes coding TLR1, TLR2, TLR4 and TLR6 which are associated with increased risk of developing IBD.93 The existence of polymorphisms of the NOD2 gene is another argument which suggests a defective mechanism in antigen recognition by TLR and NOD2. In fact, 10–15% of CD patients are homo or heterozygous carriers of one of the three major NOD2 mutations (Gly908Arg, Arg702Trp and Leu1007fs).22,23 “Knock-in” mice were used to evaluate the functions of the NOD2 Leu1007fs variant and showed that the susceptibility to develop colitis induced by dextran sodium sulphate (DSS) is higher in mice with such variant to the mouse NOD2 locus than in wild-type mice. DSS-induced colitis is associated with an overactivation of the nuclear factor kappaB (NFκB) pathway and increased secretion of IL1β.94 These results, which are contradictory with clinical data that correlate NOD2 mutations to decreased NFκB activity in patients, could be explained by the existence of a negative regulation exerted by NOD2 on the TLR2 signalling pathway. When the NOD2 mutation Leu1007fs is present, inhibition of the TLR-NFκB pathway by NOD2 would not take place, leading to increased production of pro-inflammatory cytokines in response to contact with commensal bacteria and/or pathogens. The TLR2 signalling disruption and the development of a Th1 immune response with excessive production of pro-inflammatory cytokines reported in these “knock-in” mice give weight to this hypothesis.95 Thus, in patients who carry these NOD2 variants, the production of proinflammatory cytokines following immune cell stimulation by bacterial components and the elimination of intracellular microbes are very likely to be altered.96
Acquired immune response deregulation.
In IBD, alterations in innate immune responses disrupt antigen recognition and presentation to effector cells (Fig. 1). Consequently, the epithelial barrier is more vulnerable to infections and activation of the mucosal immune system is deregulated. When IBD is active, there is an imbalance in the number of Th and Treg cells.97 In CD, Th1 lymphocytes, characterized by elevated production of IL2, IL12 and IFNγ, are predominant.98 In contrast, in the mucosa of patients with UC, there is mostly an infiltration of atypical Th2 lymphocytes which are T cells characterized by the production of IL5, IL13 and TGFβ.99
MICA, a MHC-related class Ib molecule expressed on IEC, could be involved in the activation of mucosal lymphocytes. Under normal conditions, MICA expression is restricted to intestinal and thymic epithelium. This expression can be induced by stress in different epithelial cells and is upregulated upon exposure to intracellular pathogens,100–102 in particular E. coli.103 MICA is a ligand of the NKG2D activating receptor preferentially expressed on CD8+ T cells, γδ T cells and NK cells.104,105 A subset of CD4+ T cells expressing NKG2D was increased in the lamina propria from patients with CD compared to controls or UC.106 CD4+NKG2D+ T cells with Th1 cytokine profile and expressing perforin were increased in the periphery and in the mucosa in CD and mediate inflammatory and cytotoxic responses through MICA interaction.106
Moreover, others studies have pointed to a new T cell population defined as Th17 lymphocytes that contribute to the shift towards effector T cells over regulator T cells populations in IBD.107 Th17 lymphocytes (CD4+ CD25−) are T cells which produce the proinflammatory cytokine IL17, especially in response to the presence of extracellular bacteria. Th17 differentiation of naive Th0 cells is induced by the coexpression of IL23 and TGFβ108 whose role in determining the balance between Treg lymphocytes (anti-inflammatory) and Th17 lymphocytes (proinflammatory) is essential (Fig. 2). Genetic association studies showing an involvement of the IL23R gene in IBD pathogenesis29,31 have thus led naturally to suspicions that effector subpopulation of T cells which possess strong inflammatory activity promote the activation and local accumulation of neutrophils at the site of tissue inflammation and induce the production of antimicrobial peptides as HBD.109 The current assumption is that there is a contribution of the IL23R signaling pathway to inflammation via impaired synthesis of antimicrobial peptides and by promotion of the Th17 pro-inflammatory state. IL23 stimulates the development and expansion of pathogenic memory T cells and ensures the survival and clonal expansion of Th17.110
Besides the role of cytokines such as IL23, changes in the pattern of expression of chemokines (or chemo-attractive cytokines) can also explain the abnormal influx of immune effector cells in the intestinal mucosa. The expression of many chemokines (in particular IL8; granulocyte chemotactic protein-2, GCP2; growth regulated protein α and β, GROα and GROβ; epithelial neutrophil activating protein 78, ENA-78; etc.,) and the expression of their respective receptors (CXCR1, CXCR2, etc.) are increased during the active phase of IBD (Table 3).111 In IBD, a defect in chemokine production or a defect in the regulation of chemokine signal transduction pathways could contribute to the loss of epithelial integrity by inducing local production of free radicals112,113 and by inducing an influx of leukocytes in the mucosa by means of neo-angiogenesis.114
Table 3.
Chemokines and receptors with a suspected role in IBD pathophysiology
| Name | Gene | Other names | Receptor | Uniprot | Related disease | Target cells |
| CC chemokines | ||||||
| CCL2 | Scya2 | MCP1 | CCR2 | P13500 | CD + UC | Th/mono/baso |
| CCL3 | Scya3 | MIP1a | CCR1 | P10147 | CD + UC | Tc |
| CCL4 | Scya4 | MIP1b | CCR1, CCR5 | P13236 | CD + UC | Th |
| CCL5 | Scya5 | RANTES | CCR5 | P13501 | CD + UC | Th/mono/baso |
| CCL7 | Scya7 | MARC, MCP3 | CCR2 | P80098 | CD + UC | mono/baso |
| CCL8 | Scya8 | MCP2 | CCR1, CCR2B, CCR5 | P80075 | CD + UC | neutro/baso/mono |
| CCL25 | Scya25 | TECK, Ckb15 | CCR9 | O15444 | CD + UC | Th17 |
| CXC chemokines | ||||||
| CXCL1 | Scyb1 | Gro-a, GRO1, NAP3, KC | CXCR2 | P09341 | CD + UC | neutro/baso |
| CXCL2 | Scyb2 | Gro-b, GRO2, MIP2a | CXCR2 | P19875 | CD; UC? | neutro/baso |
| CXCL5 | Scyb5 | ENA78 | CXCR2 | P42830 | CD + UC | neutro |
| CXCL6 | Scyb6 | GCP2 | CXCR1, CXCR2 | P80162 | CD + UC | neutro/baso |
| CXCL8 | Scyb8 | IL8, NAP1, MDNCF, GCP1 | CXCR1, CXCR2 | P10145 | CD + UC | neutro/baso |
| CXCL10 | Scyb10 | IP10, CRG2 | CXCR3 | P02778 | CD + UC | T/mono |
| CXCL12 | Scyb12 | SDF1, PBSF | CXCR4 | P48061 | CD + UC | T/B |
| CX3C chemokines | ||||||
| CX3CL1 | Scyd1 | Fractalkine, neurotactin, ABCD3 | CX3CR1 | P78423 | CD + UC | mono/DC |
CD, Crohn disease; DC, dendritic cell; Th, T helper cell; Tc, T cytotoxic cell; mono, monocyte; baso, basophil; neutro, neutrophil; UC, ulcerative colitis.
Finally, UC and CD are associated with a humoral response characterized by infiltration of plasma B cells.115 In the mucosa, the levels of G1 immunoglobulins (IgG1), IgG2, IgM and IgE are increased while the secretory IgA concentration is decreased.116–118 In addition, overactivation of B cells in IBD results in a high production of mucosal IgG auto-antibodies directed against commensal bacterial antigens from the intestinal lumen.118–120 These data suggest a shift from antibodies of the protector subtype (IgA) to the aggressive subtype (IgG) leading to mucosal inflammation.
Conclusion and New Therapies
In IBD, the existence of a genetic vulnerability leads to disrupted identification and presentation of intestinal antigens to effector cells. The subsequent inappropriate adaptive immune response results in loss of tolerance to commensal flora and to amplification and maintenance of the inflammatory response to intestinal pathogens, especially in CD where there is a weakness of the immune system. In parallel to inflammation, infiltration of immune cells in the intestinal mucosa and in the proximity of nerve endings leads to enteric neuro-immune direct contacts. These interactions cause the activation of visceral afferents which is the first step to the development of chronic abdominal pain consecutive to inflammation.
The main goal of therapy for IBD is to induce a clinical remission and then maintain it for a long period of time, in order to realize the best attainable quality of life. Choice of therapy depends on the severity and location of disease, as well as side effects and other adverse events. Recent discoveries have led to the development of novel therapeutic drugs that regulate activation of the cellular immune response and inflammation (antibodies against cytokines and their receptors, antibodies blocking T-cell costimulatory molecules, etc.,) as a complement to the standard anti-inflammatory treatments based on corticosteroids and 5-amino-salicylic acids. In CD, new therapeutic avenues with immunostimulation strategies are now envisaged to overcome the primary immunodeficiency in patients and to reduce complications caused by immunosuppressive drugs and by antibodies against pro-inflammatory cytokines.91 Several new drugs have been introduced in the last decade or are under investigation for the treatment of IBD. They include anti-TNFα agents, anti-adhesion molecules, anti-IL12/23, anti-IL6R and others. A novel therapeutic target for IBD could be the interaction between MICA and NKG2D since blockade of NKG2D signaling prevents the development of murine CD4+ T-cell mediated colitis121 and inhibition of NKG2D receptor function using antibodies attenuates transfer-induced colitis in SCID mice.122 To conclude, current therapies generally involve a combination of pharmacologic agents and dietary manipulation. Newer agents including monoclonal antibodies targeted to specific pro-inflammatory cytokines and adhesion molecules as well as the induction of anti-inflammatory cytokines and T-cell activation, have emerged and provided clinical benefit in the treatment and relapse of IBD.
Abbreviations
- AIEC
adherent-invasive E. coli
- CEACAM6
carcinoembrionic antigen-related cell adhesion molecule 6
- CD
Crohn disease
- DC
dendritic cell
- HBD
human β-defensin
- HD
human α-defensin
- IBD
inflammatory bowel disease
- IL
interleukin
- MHC
major histocompatibility complex
- NFκB
nuclear factor kappaB
- NOD2/CARD15
nucleotide-binding oligomerization domain 2/caspase-activating recruitment domain 15
- TLR
toll-like receptor
- UC
ulcerative colitis
Footnotes
Previously published online: www.landesbioscience.com/journals/selfnonself/article/13560
Authorship
Wrote the paper: J.M. and N.B. Made the tables and the illustrations: J.M. Supervision and final approval of the manuscript: D.A.
References
- 1.Baumgart DC, Sandborn WJ. Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet. 2007;369:1641–1657. doi: 10.1016/S0140-6736(07)60751-X. [DOI] [PubMed] [Google Scholar]
- 2.Loftus E., Jr Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence and environmental influences. Gastroenterology. 2004;126:1504–1517. doi: 10.1053/j.gastro.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen GC, Torres EA, Regueiro M, Bromfield G, Bitton A, Stempak J, et al. Inflammatory bowel disease characteristics among African Americans, Hispanics and non-Hispanic Whites: characterization of a large North American cohort. Am J Gastroenterol. 2006;101:1012–1023. doi: 10.1111/j.1572-0241.2006.00504.x. [DOI] [PubMed] [Google Scholar]
- 4.Ogunbi SO, Ransom JA, Sullivan K, Schoen BT, Gold BD. Inflammatory bowel disease in African-American children living in Georgia. J Pediatr. 1998;133:103–107. doi: 10.1016/s0022-3476(98)70187-8. [DOI] [PubMed] [Google Scholar]
- 5.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–429. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 6.Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J Clin Invest. 2007;117:514–521. doi: 10.1172/JCI30587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 8.Gent AE, Hellier MD, Grace RH, Swarbrick ET, Coggon D. Inflammatory bowel disease and domestic hygiene in infancy. Lancet. 1994;343:766–767. doi: 10.1016/s0140-6736(94)91841-4. [DOI] [PubMed] [Google Scholar]
- 9.Lakatos PL, Szamosi T, Lakatos L. Smoking in inflammatory bowel diseases: good, bad or ugly? World J Gastroenterol. 2007;13:6134–6139. doi: 10.3748/wjg.v13.i46.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calkins BM. A meta-analysis of the role of smoking in inflammatory bowel disease. Dig Dis Sci. 1989;34:1841–1854. doi: 10.1007/BF01536701. [DOI] [PubMed] [Google Scholar]
- 11.Boyko EJ, Koepsell TD, Perera DR, Inui TS. Risk of ulcerative colitis among former and current cigarette smokers. N Engl J Med. 1987;316:707–710. doi: 10.1056/NEJM198703193161202. [DOI] [PubMed] [Google Scholar]
- 12.Odes HS, Fich A, Reif S, Halak A, Lavy A, Keter D, et al. Effects of current cigarette smoking on clinical course of Crohn's disease and ulcerative colitis. Dig Dis Sci. 2001;46:1717–1721. doi: 10.1023/a:1010609722315. [DOI] [PubMed] [Google Scholar]
- 13.Cosnes J, Carbonnel F, Carrat F, Beaugerie L, Cattan S, Gendre J. Effects of current and former cigarette smoking on the clinical course of Crohn's disease. Aliment Pharmacol Ther. 1999;13:1403–1411. doi: 10.1046/j.1365-2036.1999.00630.x. [DOI] [PubMed] [Google Scholar]
- 14.Lindberg E, Jarnerot G, Huitfeldt B. Smoking in Crohn's disease: effect on localisation and clinical course. Gut. 1992;33:779–782. doi: 10.1136/gut.33.6.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russel MG, Nieman FH, Bergers JM, Stockbrugger RW. Cigarette smoking and quality of life in patients with inflammatory bowel disease. South Limburg IBD Study Group. Eur J Gastroenterol Hepatol. 1996;8:1075–1081. doi: 10.1097/00042737-199611000-00009. [DOI] [PubMed] [Google Scholar]
- 16.Binder V, Orholm M. Familial occurrence and inheritance studies in inflammatory bowel disease. Neth J Med. 1996;48:53–56. doi: 10.1016/0300-2977(95)00093-3. [DOI] [PubMed] [Google Scholar]
- 17.Peeters M, Nevens H, Baert F, Hiele M, de Meyer AM, Vlietinck R, et al. Familial aggregation in Crohn's disease: increased age-adjusted risk and concordance in clinical characteristics. Gastroenterology. 1996;111:597–603. doi: 10.1053/gast.1996.v111.pm8780562. [DOI] [PubMed] [Google Scholar]
- 18.Binder V. Genetic epidemiology in inflammatory bowel disease. Dig Dis. 1998;16:351–355. doi: 10.1159/000016891. [DOI] [PubMed] [Google Scholar]
- 19.Tysk C, Lindberg E, Jarnerot G, Floderus-Myrhed B. Ulcerative colitis and Crohn's disease in an unselected population of monozygotic and dizygotic twins. A study of heritability and the influence of smoking. Gut. 1988;29:990–996. doi: 10.1136/gut.29.7.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wellcome, author. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 23.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 24.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 25.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 28.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 29.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Einarsdottir E, Koskinen LL, Dukes E, Kainu K, Suomela S, Lappalainen M, et al. IL23R in the Swedish, Finnish, Hungarian and Italian populations: association with IBD and psoriasis and linkage to celiac disease. BMC Med Genet. 2009;10:8. doi: 10.1186/1471-2350-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fisher SA, Tremelling M, Anderson CA, Gwilliam R, Bumpstead S, Prescott NJ, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn's disease. Nat Genet. 2008;40:710–712. doi: 10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franke A, Balschun T, Karlsen TH, Hedderich J, May S, Lu T, et al. Replication of signals from recent studies of Crohn's disease identifies previously unknown disease loci for ulcerative colitis. Nat Genet. 2008;40:713–715. doi: 10.1038/ng.148. [DOI] [PubMed] [Google Scholar]
- 33.Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hata K, Andoh A, Sato H, Araki Y, Tanaka M, Tsujikawa T, et al. Sequential changes in luminal microflora and mucosal cytokine expression during developing of colitis in HLA-B27/beta2-microglobulin transgenic rats. Scand J Gastroenterol. 2001;36:1185–1192. doi: 10.1080/00365520152584824. [DOI] [PubMed] [Google Scholar]
- 35.Rath HC, Schultz M, Freitag R, Dieleman LA, Li F, Linde HJ, et al. Different subsets of enteric bacteria induce and perpetuate experimental colitis in rats and mice. Infect Immun. 2001;69:2277–2285. doi: 10.1128/IAI.69.4.2277-2285.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elson CO, Cong Y, McCracken VJ, Dimmitt RA, Lorenz RG, Weaver CT. Experimental models of inflammatory bowel disease reveal innate, adaptive and regulatory mechanisms of host dialogue with the microbiota. Immunol Rev. 2005;206:260–276. doi: 10.1111/j.0105-2896.2005.00291.x. [DOI] [PubMed] [Google Scholar]
- 37.Onderdonk AB, Hermos JA, Bartlett JG. The role of the intestinal microflora in experimental colitis. Am J Clin Nutr. 1977;30:1819–1825. doi: 10.1093/ajcn/30.11.1819. [DOI] [PubMed] [Google Scholar]
- 38.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 39.Tamboli CP, Neut C, Desreumaux P, Colombel JF. Dysbiosis as a prerequisite for IBD. Gut. 2004;53:1057. [PMC free article] [PubMed] [Google Scholar]
- 40.Falkow S. Molecular Koch's postulates applied to microbial pathogenicity. Rev Infect Dis. 1988;10:274–276. doi: 10.1093/cid/10.supplement_2.s274. [DOI] [PubMed] [Google Scholar]
- 41.Hansen R, Thomson JM, El-Omar EM, Hold GL. The role of infection in the aetiology of inflammatory bowel disease. J Gastroenterol. 45:266–276. doi: 10.1007/s00535-009-0191-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glasser AL, Boudeau J, Barnich N, Perruchot MH, Colombel JF, Darfeuille-Michaud A. Adherent invasive Escherichia coli strains from patients with Crohn's disease survive and replicate within macrophages without inducing host cell death. Infect Immun. 2001;69:5529–5537. doi: 10.1128/IAI.69.9.5529-5537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conte MP, Schippa S, Zamboni I, Penta M, Chiarini F, Seganti L, et al. Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut. 2006;55:1760–1767. doi: 10.1136/gut.2005.078824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology. 1998;115:1405–1413. doi: 10.1016/s0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- 45.Kotlowski R, Bernstein CN, Sepehri S, Krause DO. High prevalence of Escherichia coli belonging to the B2 + D phylogenetic group in inflammatory bowel disease. Gut. 2007;56:669–675. doi: 10.1136/gut.2006.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, et al. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology. 2004;127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 47.Mylonaki M, Rayment NB, Rampton DS, Hudspith BN, Brostoff J. Molecular characterization of rectal mucosa-associated bacterial flora in inflammatory bowel disease. Inflamm Bowel Dis. 2005;11:481–487. doi: 10.1097/01.mib.0000159663.62651.4f. [DOI] [PubMed] [Google Scholar]
- 48.Neut C, Bulois P, Desreumaux P, Membre JM, Lederman E, Gambiez L, et al. Changes in the bacterial flora of the neoterminal ileum after ileocolonic resection for Crohn's disease. Am J Gastroenterol. 2002;97:939–946. doi: 10.1111/j.1572-0241.2002.05613.x. [DOI] [PubMed] [Google Scholar]
- 49.Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 50.Boudeau J, Barnich N, Darfeuille-Michaud A. Type 1 pili-mediated adherence of Escherichia coli strain LF82 isolated from Crohn's disease is involved in bacterial invasion of intestinal epithelial cells. Mol Microbiol. 2001;39:1272–1284. doi: 10.1111/j.1365-2958.2001.02315.x. [DOI] [PubMed] [Google Scholar]
- 51.Boudeau J, Glasser AL, Masseret E, Joly B, Darfeuille-Michaud A. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease. Infect Immun. 1999;67:4499–4509. doi: 10.1128/iai.67.9.4499-4509.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bringer MA, Glasser AL, Tung CH, Meresse S, Darfeuille-Michaud A. The Crohn's disease-associated adherent-invasive Escherichia coli strain LF82 replicates in mature phagolysosomes within J774 macrophages. Cell Microbiol. 2006;8:471–484. doi: 10.1111/j.1462-5822.2005.00639.x. [DOI] [PubMed] [Google Scholar]
- 53.Meconi S, Vercellone A, Levillain F, Payre B, Al Saati T, Capilla F, et al. Adherent-invasive Escherichia coli isolated from Crohn's disease patients induce granulomas in vitro. Cell Microbiol. 2007;9:1252–1261. doi: 10.1111/j.1462-5822.2006.00868.x. [DOI] [PubMed] [Google Scholar]
- 54.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 55.Martinez-Medina M, Aldeguer X, Lopez-Siles M, Gonzalez-Huix F, Lopez-Oliu C, Dahbi G, et al. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E.coli (AIEC) in Crohn's disease. Inflamm Bowel Dis. 2009;15:872–882. doi: 10.1002/ibd.20860. [DOI] [PubMed] [Google Scholar]
- 56.Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. Isme J. 2007;1:403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 57.Eaves-Pyles T, Allen CA, Taormina J, Swidsinski A, Tutt CB, Jezek GE, et al. Escherichia coli isolated from a Crohn's disease patient adheres, invades and induces inflammatory responses in polarized intestinal epithelial cells. Int J Med Microbiol. 2008;298:397–409. doi: 10.1016/j.ijmm.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 58.Sasaki M, Sitaraman SV, Babbin BA, Gerner-Smidt P, Ribot EM, Garrett N, et al. Invasive Escherichia coli are a feature of Crohn's disease. Lab Invest. 2007;87:1042–1054. doi: 10.1038/labinvest.3700661. [DOI] [PubMed] [Google Scholar]
- 59.Barnich N, Carvalho FA, Glasser AL, Darcha C, Jantscheff P, Allez M, et al. CEACAM6 acts as a receptor for adherent-invasive E.coli, supporting ileal mucosa colonization in Crohn disease. J Clin Invest. 2007;117:1566–1574. doi: 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carvalho FA, Barnich N, Sivignon A, Darcha C, Chan CH, Stanners CP, et al. Crohn's disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J Exp Med. 2009;206:2179–2189. doi: 10.1084/jem.20090741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rolhion N, Barnich N, Bringer MA, Glasser AL, Ranc J, Hebuterne X, et al. Abnormally expressed ER stress response chaperone Gp96 in CD favours adherent-invasive Escherichia coli invasion. Gut. 2010;59:1355–1362. doi: 10.1136/gut.2010.207456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Willing B, Halfvarson J, Dicksved J, Rosenquist M, Jarnerot G, Engstrand L, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn's disease. Inflamm Bowel Dis. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 64.Moehle C, Ackermann N, Langmann T, Aslanidis C, Kel A, Kel-Margoulis O, et al. Aberrant intestinal expression and allelic variants of mucin genes associated with inflammatory bowel disease. J Mol Med. 2006;84:1055–1066. doi: 10.1007/s00109-006-0100-2. [DOI] [PubMed] [Google Scholar]
- 65.Bruewer M, Samarin S, Nusrat A. Inflammatory bowel disease and the apical junctional complex. Ann NY Acad Sci. 2006;1072:242–252. doi: 10.1196/annals.1326.017. [DOI] [PubMed] [Google Scholar]
- 66.Weber CR, Turner JR. Inflammatory bowel disease: is it really just another break in the wall? Gut. 2007;56:6–8. doi: 10.1136/gut.2006.104182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reuter BK, Pizarro TT. Mechanisms of tight junction dysregulation in the SAMP1/YitFc model of Crohn's disease-like ileitis. Ann NY Acad Sci. 2009;1165:301–307. doi: 10.1111/j.1749-6632.2009.04035.x. [DOI] [PubMed] [Google Scholar]
- 68.Hill KA, Wang KL, Stryker SJ, Gupta R, Weinrach DM, Rao MS. Comparative analysis of cell adhesion molecules, cell cycle regulatory proteins, mismatch repair genes, cyclooxygenase-2 and DPC4 in carcinomas arising in inflammatory bowel disease and sporadic colon cancer. Oncol Rep. 2004;11:951–956. [PubMed] [Google Scholar]
- 69.Prasad S, Mingrino R, Kaukinen K, Hayes KL, Powell RM, MacDonald TT, et al. Inflammatory processes have differential effects on claudins 2, 3 and 4 in colonic epithelial cells. Lab Invest. 2005;85:1139–1162. doi: 10.1038/labinvest.3700316. [DOI] [PubMed] [Google Scholar]
- 70.Wyatt J, Vogelsang H, Hubl W, Waldhoer T, Lochs H. Intestinal permeability and the prediction of relapse in Crohn's disease. Lancet. 1993;341:1437–1439. doi: 10.1016/0140-6736(93)90882-h. [DOI] [PubMed] [Google Scholar]
- 71.Zeissig S, Burgel N, Gunzel D, Richter J, Mankertz J, Wahnschaffe U, et al. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn's disease. Gut. 2007;56:61–72. doi: 10.1136/gut.2006.094375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Benjamin J, Makharia GK, Ahuja V, Kalaivani M, Joshi YK. Intestinal permeability and its association with the patient and disease characteristics in Crohn's disease. World J Gastroenterol. 2008;14:1399–1405. doi: 10.3748/wjg.14.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ambrose NS, Johnson M, Burdon DW, Keighley MR. Incidence of pathogenic bacteria from mesenteric lymph nodes and ileal serosa during Crohn's disease surgery. Br J Surg. 1984;71:623–625. doi: 10.1002/bjs.1800710821. [DOI] [PubMed] [Google Scholar]
- 74.Takesue Y, Ohge H, Uemura K, Imamura Y, Murakami Y, Yokoyama T, et al. Bacterial translocation in patients with Crohn's disease undergoing surgery. Dis Colon Rectum. 2002;45:1665–1671. doi: 10.1007/s10350-004-7256-z. [DOI] [PubMed] [Google Scholar]
- 75.D'Inca R, Annese V, di Leo V, Latiano A, Quaino V, Abazia C, et al. Increased intestinal permeability and NOD2 variants in familial and sporadic Crohn's disease. Aliment Pharmacol Ther. 2006;23:1455–1461. doi: 10.1111/j.1365-2036.2006.02916.x. [DOI] [PubMed] [Google Scholar]
- 76.Barreau F, Meinzer U, Chareyre F, Berrebi D, Niwa-Kawakita M, Dussaillant M, et al. CARD15/NOD2 is required for Peyer's patches homeostasis in mice. PLoS ONE. 2007;2:523. doi: 10.1371/journal.pone.0000523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Keita AV, Salim SY, Jiang T, Yang PC, Franzen L, Soderkvist P, et al. Increased uptake of non-pathogenic E. coli via the follicle-associated epithelium in longstanding ileal Crohn's disease. J Pathol. 2008;215:135–144. doi: 10.1002/path.2337. [DOI] [PubMed] [Google Scholar]
- 78.Salim SY, Silva MA, Keita AV, Larsson M, Andersson P, Magnusson KE, et al. CD83+CCR7− dendritic cells accumulate in the subepithelial dome and internalize translocated Escherichia coli HB101 in the Peyer's patches of ileal Crohn's disease. Am J Pathol. 2009;174:82–90. doi: 10.2353/ajpath.2009.080273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramasundara M, Leach ST, Lemberg DA, Day AS. Defensins and inflammation: the role of defensins in inflammatory bowel disease. J Gastroenterol Hepatol. 2009;24:202–208. doi: 10.1111/j.1440-1746.2008.05772.x. [DOI] [PubMed] [Google Scholar]
- 80.Wehkamp J, Schmid M, Stange EF. Defensins and other antimicrobial peptides in inflammatory bowel disease. Curr Opin Gastroenterol. 2007;23:370–378. doi: 10.1097/MOG.0b013e328136c580. [DOI] [PubMed] [Google Scholar]
- 81.Fellermann K, Stange DE, Schaeffeler E, Schmalzl H, Wehkamp J, Bevins CL, et al. A chromosome 8 genecluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am J Hum Genet. 2006;79:439–448. doi: 10.1086/505915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simms LA, Doecke JD, Walsh MD, Huang N, Fowler EV, Radford-Smith GL. Reduced alpha-defensin expression is associated with inflammation and not NOD2 mutation status in ileal Crohn's disease. Gut. 2008;57:903–910. doi: 10.1136/gut.2007.142588. [DOI] [PubMed] [Google Scholar]
- 83.Wehkamp J, Harder J, Weichenthal M, Schwab M, Schaffeler E, Schlee M, et al. NOD2 (CARD15) mutations in Crohn's disease are associated with diminished mucosal alpha-defensin expression. Gut. 2004;53:1658–1664. doi: 10.1136/gut.2003.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wehkamp J, Salzman NH, Porter E, Nuding S, Weichenthal M, Petras RE, et al. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc Natl Acad Sci USA. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 86.Eisenhauer PB, Harwig SS, Lehrer RI. Cryptdins: antimicrobial defensins of the murine small intestine. Infect Immun. 1992;60:3556–3565. doi: 10.1128/iai.60.9.3556-3565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Naser SA, Ghobrial G, Romero C, Valentine JF. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn's disease. Lancet. 2004;364:1039–1044. doi: 10.1016/S0140-6736(04)17058-X. [DOI] [PubMed] [Google Scholar]
- 88.Selby WS. Mycobacterium avium subspecies paratuberculosis bacteraemia in patients with inflammatory bowel disease. Lancet. 2004;364:1013–1014. doi: 10.1016/S0140-6736(04)17071-2. [DOI] [PubMed] [Google Scholar]
- 89.Smith AM, Rahman FZ, Hayee B, Graham SJ, Marks DJ, Sewell GW, et al. Disordered macrophage cytokine secretion underlies impaired acute inflammation and bacterial clearance in Crohn's disease. J Exp Med. 2009;206:1883–1897. doi: 10.1084/jem.20091233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Niess JH. Role of mucosal dendritic cells in inflammatory bowel disease. World J Gastroenterol. 2008;14:5138–5148. doi: 10.3748/wjg.14.5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baumgart DC, Carding SR. Inflammatory bowel disease: cause and immunobiology. Lancet. 2007;369:1627–1640. doi: 10.1016/S0140-6736(07)60750-8. [DOI] [PubMed] [Google Scholar]
- 92.Hart AL, Al-Hassi HO, Rigby RJ, Bell SJ, Emmanuel AV, Knight SC, et al. Characteristics of intestinal dendritic cells in inflammatory bowel diseases. Gastroenterology. 2005;129:50–65. doi: 10.1053/j.gastro.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 93.Cario E, Podolsky DK. Differential alteration in intestinal epithelial cell expression of toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect Immun. 2000;68:7010–7017. doi: 10.1128/iai.68.12.7010-7017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maeda S, Hsu LC, Liu H, Bankston LA, Iimura M, Kagnoff MF, et al. Nod2 mutation in Crohn's disease potentiates NFkappaB activity and IL-1beta processing. Science. 2005;307:734–738. doi: 10.1126/science.1103685. [DOI] [PubMed] [Google Scholar]
- 95.Watanabe T, Kitani A, Murray PJ, Strober W. NOD2 is a negative regulator of Toll-like receptor 2-mediated T helper type 1 responses. Nat Immunol. 2004;5:800–808. doi: 10.1038/ni1092. [DOI] [PubMed] [Google Scholar]
- 96.Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 97.Martin B, Banz A, Bienvenu B, Cordier C, Dautigny N, Becourt C, et al. Suppression of CD4+ T lymphocyte effector functions by CD4+CD25+ cells in vivo. J Immunol. 2004;172:3391–3398. doi: 10.4049/jimmunol.172.6.3391. [DOI] [PubMed] [Google Scholar]
- 98.Bamias G, Sugawara K, Pagnini C, Cominelli F. The Th1 immune pathway as a therapeutic target in Crohn's disease. Curr Opin Investig Drugs. 2003;4:1279–1286. [PubMed] [Google Scholar]
- 99.Targan SR, Karp LC. Defects in mucosal immunity leading to ulcerative colitis. Immunol Rev. 2005;206:296–305. doi: 10.1111/j.0105-2896.2005.00286.x. [DOI] [PubMed] [Google Scholar]
- 100.Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci USA. 1996;93:12445–12450. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Groh V, Rhinehart R, Secrist H, Bauer S, Grabstein KH, Spies T. Broad tumor-associated expression and recognition by tumor-derived gamma delta T cells of MICA and MICB. Proc Natl Acad Sci USA. 1999;96:6879–6884. doi: 10.1073/pnas.96.12.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Groh V, Steinle A, Bauer S, Spies T. Recognition of stress-induced MHC molecules by intestinal epithelial gammadelta T cells. Science. 1998;279:1737–1740. doi: 10.1126/science.279.5357.1737. [DOI] [PubMed] [Google Scholar]
- 103.Tieng V, Le Bouguenec C, du Merle L, Bertheau P, Desreumaux P, Janin A, et al. Binding of Escherichia coli adhesin AfaE to CD55 triggers cell-surface expression of the MHC class I-related molecule MICA. Proc Natl Acad Sci USA. 2002;99:2977–2982. doi: 10.1073/pnas.032668099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li P, Morris DL, Willcox BE, Steinle A, Spies T, Strong RK. Complex structure of the activating immunoreceptor NKG2D and its MHC class I-like ligand MICA. Nat Immunol. 2001;2:443–451. doi: 10.1038/87757. [DOI] [PubMed] [Google Scholar]
- 105.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3:781–790. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 106.Allez M, Tieng V, Nakazawa A, Treton X, Pacault V, Dulphy N, et al. CD4+NKG2D+ T cells in Crohn's disease mediate inflammatory and cytotoxic responses through MICA interactions. Gastroenterology. 2007;132:2346–2358. doi: 10.1053/j.gastro.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 107.Fujino S, Andoh A, Bamba S, Ogawa A, Hata K, Araki Y, et al. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 2003;52:65–70. doi: 10.1136/gut.52.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mangan PR, Harrington LE, O'Quinn DB, Helms WS, Bullard DC, Elson CO, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 109.Matsuzaki G, Umemura M. Interleukin-17 as an effector molecule of innate and acquired immunity against infections. Microbiol Immunol. 2007;51:1139–1147. doi: 10.1111/j.1348-0421.2007.tb04008.x. [DOI] [PubMed] [Google Scholar]
- 110.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 111.Kraneveld AD, Rijnierse A, Nijkamp FP, Garssen J. Neuro-immune interactions in inflammatory bowel disease and irritable bowel syndrome: future therapeutic targets. Eur J Pharmacol. 2008;585:361–374. doi: 10.1016/j.ejphar.2008.02.095. [DOI] [PubMed] [Google Scholar]
- 112.Keshavarzian A, Banan A, Farhadi A, Komanduri S, Mutlu E, Zhang Y, et al. Increases in free radicals and cytoskeletal protein oxidation and nitration in the colon of patients with inflammatory bowel disease. Gut. 2003;52:720–728. doi: 10.1136/gut.52.5.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zimmerman NP, Vongsa RA, Wendt MK, Dwinell MB. Chemokines and chemokine receptors in mucosal homeostasis at the intestinal epithelial barrier in inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:1000–1011. doi: 10.1002/ibd.20480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Koutroubakis IE, Tsiolakidou G, Karmiris K, Kouroumalis EA. Role of angiogenesis in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12:515–523. doi: 10.1097/00054725-200606000-00012. [DOI] [PubMed] [Google Scholar]
- 115.Bitton A, Peppercorn MA, Antonioli DA, Niles JL, Shah S, Bousvaros A, et al. Clinical, biological and histologic parameters as predictors of relapse in ulcerative colitis. Gastroenterology. 2001;120:13–20. doi: 10.1053/gast.2001.20912. [DOI] [PubMed] [Google Scholar]
- 116.Bossuyt X. Serologic markers in inflammatory bowel disease. Clin Chem. 2006;52:171–181. doi: 10.1373/clinchem.2005.058560. [DOI] [PubMed] [Google Scholar]
- 117.Fahmy OA, Abdel-Fattah SA, Fahmy IA, Mokhtar I, el-Maragy MM. Estimation of serum and tissue immunoglobulins level in some colonic disorders. J Egypt Soc Parasitol. 1991;21:503–512. [PubMed] [Google Scholar]
- 118.Macpherson A, Khoo UY, Forgacs I, Philpott-Howard J, Bjarnason I. Mucosal antibodies in inflammatory bowel disease are directed against intestinal bacteria. Gut. 1996;38:365–375. doi: 10.1136/gut.38.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Adams RJ, Heazlewood SP, Gilshenan KS, O'Brien M, McGuckin MA, Florin TH. IgG antibodies against common gut bacteria are more diagnostic for Crohn's disease than IgG against mannan or flagellin. Am J Gastroenterol. 2008;103:386–396. doi: 10.1111/j.1572-0241.2007.01577.x. [DOI] [PubMed] [Google Scholar]
- 120.Holstein A, Burmeister J, Plaschke A, Rosemeier D, Widjaja A, Egberts EH. Autoantibody profiles in microscopic colitis. J Gastroenterol Hepatol. 2006;21:1016–1020. doi: 10.1111/j.1440-1746.2005.04027.x. [DOI] [PubMed] [Google Scholar]
- 121.Ito Y, Kanai T, Totsuka T, Okamoto R, Tsuchiya K, Nemoto Y, et al. Blockade of NKG2D signaling prevents the development of murine CD4+ T cell-mediated colitis. Am J Physiol Gastrointest Liver Physiol. 2008;294:199–207. doi: 10.1152/ajpgi.00286.2007. [DOI] [PubMed] [Google Scholar]
- 122.Kjellev S, Haase C, Lundsgaard D, Urso B, Tornehave D, Markholst H. Inhibition of NKG2D receptor function by antibody therapy attenuates transferinduced colitis in SCID mice. Eur J Immunol. 2007;37:1397–1406. doi: 10.1002/eji.200636473. [DOI] [PubMed] [Google Scholar]