Abstract
In eukaryotes transcription is complicated by the DNA being packed in nucleosomes and by supercoils induced by opening of the DNA double helix during elongation. Here we discuss our recent genome-wide work regarding topoisomerases and their role in chromatin remodeling during the transcription cycle and we report a novel function for topoisomerases in transcription termination.
Key words: topoisomerases, gene expression, nucleosome, transcription termination, nucleosome free region
Introduction
DNA topoisomerases solve the topological problems associated with DNA replication, transcription and recombination by introducing transient nicks in DNA and thereby relieving the helical torsion.1,2 Topoisomerases relax the structure of DNA by generating single-strand (type I subfamily) or double-strand (type II subfamily) breaks.1 The fundamental need for topoisomerases originates from the DNA double helix, as (1) during DNA replication, the two strands of the DNA must be untangled and as (2) during transcription, the opening of DNA structure to allow RNA elongation, generates DNA supercoils that must be relaxed in order for the polymerase to move rapidly along the template.1,3–6 In eukaryotic cells, even though the bulk of the genomic DNA is relaxed,7 supercoiled microdomains may appear at sites of active transcription.8–10 This has been demonstrated by studies of transcription at divergent promoters, shown to induce local negative supercoiling in human cells.11
In multicellular organisms, Top1 is essential for early development. However, the yeast deletion mutant is viable, likely through compensation from Top2.1,2,12 Inactivation of either Top1 or Top2 does not significantly affect transcription, whereas inactivation of both enzymes clearly reduces RNA levels and, to a lesser extent, mRNA synthesis.13–15 Although both type I and II topoisomerases have been implicated in transcription, Top1 seems to be the main actor in releasing transcription-generated helical stress.1,2,16 Recently, systematic mapping of 53 different chromosomal proteins combined with principal component analysis in Drosophila showed that Top1 is found in two of five principal chromatin types, and these two types are also characterized by having the active chromatin mark H3K4me and high levels of transcription.17
DNA dependent processes including transcription, replication, recombination and repair are complicated by the fact that chromosomes are compacted by DNA being wrapped around histone octamers, forming nucleosomes. Nucleosomes can form on both negatively and positively supercoiled circular DNA templates in vitro and, in the latter case, this increases the superhelical stress of the template molecule.18–20 In contrast, when nucleosomes are formed on negatively coiled circular templates this relaxes the superhelical tension by one turn per nucleosome.21 Negative supercoiling of DNA templates facilitates nucleosome formation in vitro.18–20 According to the ‘twin supercoiling model’ a transcribing RNA polymerase generates positive supercoils ahead of the enzyme and negative coils behind assuming that the DNA ends are not free to rotate.2 As a consequence nucleosome assembly behind the polymerase and disassembly ahead of the polymerase should be favored.
However, the relationships between topoisomerases and chromatin assembly and disassembly mechanisms are poorly understood and the existing data are mostly based on in vitro or single gene studies. In our recent genome-wide study,22 we provided a holistic view of the changes in transcription and nucleosome occupancy that occur in topoisomerase mutants. The results support a role for Top1 in facilitating the eviction of nucleosomes to create 5′NFR regions at gene promoters and stimulate transcription initiation. The data also suggest that Top1 and Top2 are required for efficient elongation of transcription, presumably by removing positive supercoiling ahead of the polymerase. Here, we review these findings and bring novel insights into topoisomerase functions linked chromatin dynamics at 3′NFR regions and control of transcription termination.
Topoisomerases Broadly Contribute to the Control of Gene Expression and Chromatin Structures
We have carried out a genome-wide analysis of topoisomerases of type I and II in the fission yeast Schizosaccharomyces pombe.22 We found that both topoisomerases occupy intergenic regions and Top1, but not Top2, is linked to actively transcribed gene promoters. In contrast, both topoisomerase enzymes were found in 3′ coding regions and 3′ intergenic regions. In vitro studies have suggested that nucleosome assembly is tightly coupled to DNA topology.23,24 To determine the influence of topoisomerase activity on chromatin organization as suggested in earlier studies,25 we used the double mutant top1Δ top2-191ts carrying a gene deletion for top1 and a temperature sensitive allele of top2. This strain displays reduced topoisomerase II activity when grown at the semi-permissive temperature.12 We showed that topoisomerase activity influences histone density and detected higher histone H3 occupancy in intergenic regions in top1Δ top2ts, especially in genes that are highly transcribed in wild-type cells. The same set of genes also showed reduced expression and Pol II occupancy in the top1Δtop2ts mutant. These results suggested that topoisomerase activity maintains a low histone density in promoters to allow for efficient Pol II recruitment and high transcription levels. Additionally, we found that Pol II accumulates on longer genes in the absence of topoisomerase activity. In longer genes, Top2 occupancy is markedly increased in 3′ coding regions. These findings support an essential role for topoisomerases in vivo to facilitate transcription elongation (and especially Top2 at longer genes) through the removal of positive DNA supercoils generated ahead of Pol II.1,2
Topoisomerases and the Chd1 Chromatin Remodeler Hrp1 are Both Needed for Nucleosome Disassembly
In vitro studies have shown that the activity of the ATP-dependent chromatin remodeler Snf2 generates superhelical torsion during nucleosome assembly and requires DNA topoisomerases to relieve the DNA tension.26 In S. pombe, the Snf2 remodeler and Chd1 homolog Hrp1 interacts with the histone chaperone Nap1 and stimulates nucleosome disassembly in vivo.27 Our analysis showed a significant overlap between Hrp1, Nap1 and Top1/Top2 occupancy where histone H3 density is changed in the respective mutants.22 It is conceivable that the remaining intergenic regions (unaffected by hrp1 and where H3 density is increased in top1Δ top2-191ts) require other Snf2 chromatin remodelers that evict nucleosomes together with Top1 and Top2.
Topoisomerases are Required for Efficient Transcription Termination
Genome-wide mapping of yeast nucleosomes show that promoters (5′NFR) and sites of transcript termination (3′NFR) are depleted in nucleosomes.28,29 In budding yeast the 3′ NFR was shown to be dependent on transcription by Pol II and it was hypothesized to be linked to some aspect of RNA 3′ end formation.28 An interesting aspect of topoisomerases is their occupancy at the 3′ end of genes.22 By revisiting our genome-wide data, we set out to test whether topoisomerase activity could play a role in modifying the histone density at the 3′ end of genes.
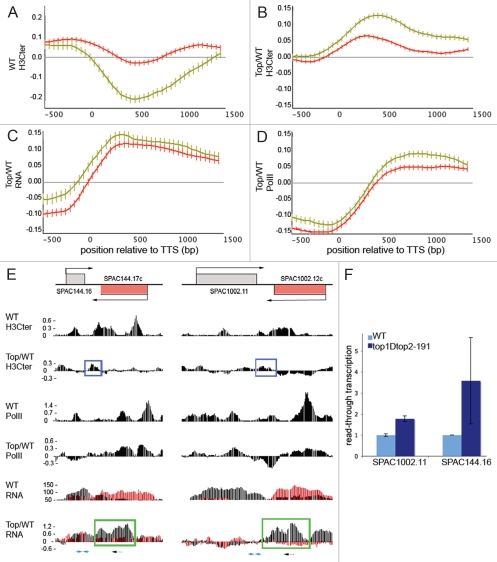
We determined the average H3 density in the region surrounding the 3′ TTS (using TTS annotations obtained from ref. 29). We filtered the data to include only genes transcribed above average and partitioned the data by the distance from a gene's stop codon to the start/stop codon of the next 3′ gene. With this step, we avoid interference by adjacent transcription (next gene within 500–1,000 bp are orange and next gene >1,000 bp away are green in Fig. 1). In wild-type cells, longer intergenic regions between transcribed genes show the most pronounced depletion of histone H3 (Fig. 1A). By comparing the high-resolution H3 maps for top1Δ top2-191ts and wild-type cells, we observed that there is a significant increase of histone occupancy regardless of the distance to adjacent genes (Fig. 1B). This indicates that topoisomerase activity participates in histone disassembly in the 3′ end of the gene.
Figure 1.
Topoisomerases are required for the global formation of 3′NFR and transcription termination. (A) Histone H3 occupancy in wild-type cells is dependent on the proximity to the adjacent gene, (B) H3 occupancy in top1Δ top2-191ts mutant relative to levels in wild-type cells, (C) RNA levels in top1Δ top2-191ts mutant relative to levels in wild-type cells, (D) Pol II occupancy in top1Δ top2-191ts mutant relative to levels in wild-type cells. Genes making up the green lines have no other transcript <1 kb downstream, genes making up the orange line have another transcript between 500 and 1,000 bp away. The genomic data are aligned at the transcript termination site (TTS). Error bars represent the 99% confidence intervals. Graphs were generated in Podbat (www.podbat.org). (E) Transcription from the forward strand is shown in black and transcription from the reverse strand is shown in red. The green boxes highlight the aberrant transcript in the top1Δ top2-191ts mutant. Blue boxes indicate the increased level of H3 in the intergenic regions. Dotted arrows indicate cDNA primer position and blue arrows indicate the qRT-PCR primer positions used in (F). Browser views are generated in Podbat. (F) qRT-PCR reveals aberrant transcript elongation in top1Δ top2-191ts cells. The primers specifically detect read-through transcripts from the indicated gene. Data are normalized to endogenous control.
Levels of histone H3 is generally linked to regulation of transcription, therefore we asked whether transcription termination defects could be observed at the 3′ end of genes. Using our transcriptome data,22 we performed the same analysis for wild-type cells and the top1Δ top2-191ts mutant at regions encompassing the 3′ TTS. As expected, we found increased transcription levels in the 3′ TTS regions of the top1Δ top2-191ts mutant compared to wild-type cells (Fig. 1C). Also the elevated RNA levels were accompanied by an increase of Pol II occupancy in the top1Δ top2-191ts mutant (Fig. 1D). Together, the data presented here suggest that topoisomerase activity is involved in controlling the supercoil structures at the 3′ end of genes and thereby contributing to nucleosome disassembly and efficient transcription termination.
We illustrate our findings with two pairs of convergent genes. By comparing top1Δ top2-191ts and wild-type cells, we observed an increased H3 occupancy at intergenic regions (Fig. 1E). In addition, the antisense transcription in top1Δ top2-191ts is increased whereas the sense transcription seems unaffected. The Pol II occupancy in wild type is slightly increased at the end of the transcript and, consistent with run-on transcription, Pol II occupancy in top1Δ top2-191ts cells is increased and shifted 3′, within the coding region of the neighboring gene. Detection by quantitative 5′ nested RT-PCR shows an increase of SPAC144.17c antisense transcripts initiated from SPAC144.16 in the top1Δ top2-191ts mutant (Fig. 1F), as well as a SPAC1002.12c antisense transcription from SPAC1002.11. This suggests that the defects in transcription termination in the neighboring converging gene causes Pol II read-through.
Normally, transcription termination is linked to a shift in phosphorylation levels of the C-terminal domain (CTD) of Pol II largest subunit. As the polymerase approaches the end of the open reading frame, serine 2 phosphorylation increases and serine 5 gets dephosphorylated. Factors such as Pcf11, needed for mRNA polyadenylation, bind to the serine 2 phosphorylated CTD, and the nascent RNA is cleaved at the polyA site. Pol II continues to transcribe the DNA but the resulting RNA is rapidly degraded by the Rat1/Xrn2 exonuclease.30 Both transcription initiation and termination are highly regulated processes and are associated with a checkpoint pause. This pause is controlled by the chromatin remodeler Isw1 in Saccharomyces cerevisiae.31 Mutation of Isw1 results in a denser nucleosome positioning around the 5′NFR as well as transcription termination defects. It has also been shown that Isw1 interacts physically with Top1.32 However, S. pombe has no Isw1 homolog and these functions could have been taken over by other Snf2 remodelers acting together with Top1/Top2. In the case of both Isw1 and the topoisomerases, the effect of decreased activity leads to a less accessible chromatin structure. We speculate that a possible consequence of the altered chromatin structure we observe at the 3′NFR obstructs the signals for transcript termination. For example this could happen through reduced availability for interactions with the CTD. The increase of read-through RNA that we observe could be from parts of the RNA molecules that normally are degraded by the Rat1/Xrn2 exonuclease.
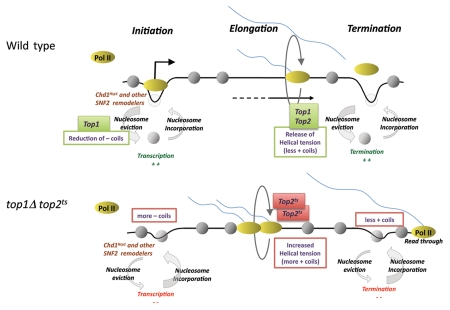
We propose a model for the function of Top1 and Top2 in controlling DNA supercoiling states and chromatin structure during all stages the transcription cycle: initiation, elongation and termination (Fig. 2, top part). First, in the initiation phase and pre-initiation complex formation, Top1 has a key role in removing negative coils to facilitate nucleosome disassembly by remodelers at the 5′NFR. Then, during transcription elongation Top1 and Top2 cooperate to decrease helical tension introduced by Pol II activity, as predicted by the ‘twin supercoiling’ model. Finally, in the process of transcription termination, topoisomerases are required to maintain the 3′NFR to allow for proper termination. It is possible that the positive supercoils that presumably accumulate in the 3′ region of the gene destabilize nucleosomes in the 3′ NFR region. Chromatin remodelers could potentially also stimulate 3′NFR formation. Interestingly, the Chd1 remodeler Hrp1 has also been implicated in facilitating transcription termination of individual genes in S. pombe.33 In the top1Δtop2ts mutant we assume that negative coils accumulate in the 5′ region leading to a reduced 5′NFR (Fig. 2: bottom part). Furthermore, the partially inactive Top2ts enzyme is not sufficient to allow for proper elongation by Pol II leading to a ‘piling up’ of Pol II 500–1,000 bp into the coding region, and the accumulation of extra positive supercoils in the same region. As a consequence of this elongation defect the positive supercoiling is reduced in the 3′ region thus inhibiting the nucleosome eviction process and the 3′NFR disappears and thereby obstructing the signals for transcript termination.
Figure 2.
A model of transcriptional regulation by Top1, Top2 and chromatin remodelers. All stages of the transcription cycle are affected by Top1 and Top2.
Acknowledgements
M.D.D. is funded by a Biotechnology and Biological Sciences Research Council (BBSRC) fellowship grant. We apologize to authors that could not be cited due to place restriction. This work was supported by grants to K.E. from the Swedish Cancer Society, Swedish Research Council (VR) and the Göran Gustafssons Foundation for Research in Natural Sciences. We thank Ulrika Norman for advice.
Abbreviations
- ChIP
chromatin immunoprecipitation
- Top1
topoisomerase I
- Top2
topoisomerase II
- Pol II
RNA polymerase II
- CTD
carboxy terminal domain
- TSS
transcription start site
- TTS
transcription termination site
- NFR
nucleosome free region
References
- 1.Champoux JJ. DNA topoisomerases: structure, function and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 3.Brill SJ, Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988;54:403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- 4.Kim RA, Wang JC. Function of DNA topoisomerases as replication swivels in Saccharomyces cerevisiae. J Mol Biol. 1989;208:257–267. doi: 10.1016/0022-2836(89)90387-2. [DOI] [PubMed] [Google Scholar]
- 5.Mondal N, Zhang Y, Jonsson Z, Dhar SK, Kannapiran M, Parvin JD. Elongation by RNA polymerase II on chromatin templates requires topoisomerase activity. Nucleic Acids Res. 2003;31:5016–5024. doi: 10.1093/nar/gkg705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu HY, Shyy SH, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 7.Sinden RR, Carlson JO, Pettijohn DE. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: analogous measurements in insect and human cells. Cell. 1980;21:773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]
- 8.Jupe ER, Sinden RR, Cartwright IL. Stably maintained microdomain of localized unrestrained supercoiling at a Drosophila heat shock gene locus. EMBO J. 1993;12:1067–1075. doi: 10.1002/j.1460-2075.1993.tb05748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kramer PR, Sinden RR. Measurement of unrestrained negative supercoiling and topological domain size in living human cells. Biochemistry. 1997;36:3151–3158. doi: 10.1021/bi962396q. [DOI] [PubMed] [Google Scholar]
- 10.Ljungman M, Hanawalt PC. Localized torsional tension in the DNA of human cells. Proc Natl Acad Sci USA. 1992;89:6055–6059. doi: 10.1073/pnas.89.13.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 12.Uemura T, Yanagida M. Isolation of type I and II DNA topoisomerase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J. 1984;3:1737–1744. doi: 10.1002/j.1460-2075.1984.tb02040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavalli G, Bachmann D, Thoma F. Inactivation of topoisomerases affects transcription-dependent chromatin transitions in rDNA but not in a gene transcribed by RNA polymerase II. EMBO J. 1996;15:590–597. [PMC free article] [PubMed] [Google Scholar]
- 14.Collins I, Weber A, Levens D. Transcriptional consequences of topoisomerase inhibition. Mol Cell Biol. 2001;21:8437–8451. doi: 10.1128/MCB.21.24.8437-8451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schultz MC, Brill SJ, Ju Q, Sternglanz R, Reeder RH. Topoisomerases and yeast rRNA transcription: negative supercoiling stimulates initiation and topoisomerase activity is required for elongation. Genes Dev. 1992;6:1332–1341. doi: 10.1101/gad.6.7.1332. [DOI] [PubMed] [Google Scholar]
- 16.Kouzine F, Levens D. Supercoil-driven DNA structures regulate genetic transactions. Front Biosci. 2007;12:4409–4423. doi: 10.2741/2398. [DOI] [PubMed] [Google Scholar]
- 17.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark DJ, Felsenfeld G. Formation of nucleosomes on positively supercoiled DNA. EMBO J. 1991;10:387–395. doi: 10.1002/j.1460-2075.1991.tb07960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hizume K, Yoshimura SH, Takeyasu K. Atomic force microscopy demonstrates a critical role of DNA superhelicity in nucleosome dynamics. Cell Biochem Biophys. 2004;40:249–261. doi: 10.1385/CBB:40:3:249. [DOI] [PubMed] [Google Scholar]
- 20.Negri R, Costanzo G, Buttinelli M, Venditti S, Di Mauro E. Effects of DNA topology in the interaction with histone octamers and DNA topoisomerase I. Biophys Chem. 1994;50:169–181. doi: 10.1016/0301-4622(94)85029-1. [DOI] [PubMed] [Google Scholar]
- 21.Germond JE, Hirt B, Oudet P, Gross-Bellark M, Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci USA. 1975;72:1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durand-Dubief M, Persson J, Norman U, Hartsuiker E, Ekwall K. Topoisomerase I regulates open chromatin and controls gene expression in vivo. EMBO J. 2010;29:2126–2134. doi: 10.1038/emboj.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blank TA, Becker PB. The effect of nucleosome phasing sequences and DNA topology on nucleosome spacing. J Mol Biol. 1996;260:1–8. doi: 10.1006/jmbi.1996.0377. [DOI] [PubMed] [Google Scholar]
- 24.Levchenko V, Jackson B, Jackson V. Histone release during transcription: displacement of the two H2A-H2B dimers in the nucleosome is dependent on different levels of transcription-induced positive stress. Biochemistry. 2005;44:5357–5372. doi: 10.1021/bi047786o. [DOI] [PubMed] [Google Scholar]
- 25.Almouzni G, Mechali M. Assembly of spaced chromatin promoted by DNA synthesis in extracts from Xenopus eggs. EMBO J. 1988;7:665–672. doi: 10.1002/j.1460-2075.1988.tb02861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gavin I, Horn PJ, Peterson CL. SWI/SNF chromatin remodeling requires changes in DNA topology. Mol Cell. 2001;7:97–104. doi: 10.1016/s1097-2765(01)00158-7. [DOI] [PubMed] [Google Scholar]
- 27.Walfridsson J, Khorosjutina O, Matikainen P, Gustafsson CM, Ekwall K. A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly. EMBO J. 2007;26:2868–2879. doi: 10.1038/sj.emboj.7601728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan XC, Moqtaderi Z, Jin Y, Zhang Y, Liu XS, Struhl K. Nucleosome depletion at yeast terminators is not intrinsic and can occur by a transcriptional mechanism linked to 3′-end formation. Proc Natl Acad Sci USA. 2010;107:17945–17950. doi: 10.1073/pnas.1012674107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol. 2010;17:251. doi: 10.1038/nsmb.1741. [DOI] [PubMed] [Google Scholar]
- 30.West S, Gromak N, Proudfoot NJ. Human 5′→3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 31.Morillon A, Karabetsou N, O'Sullivan J, Kent N, Proudfoot N, Mellor J. Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell. 2003;115:425–435. doi: 10.1016/s0092-8674(03)00880-8. [DOI] [PubMed] [Google Scholar]
- 32.Krogan NJ, Cagney G, Yu HY, Zhong GQ, Guo XH, Ignatchenko A, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 33.Alen C, Kent NA, Jones HS, O'Sullivan J, Aranda A, Proudfoot NJ. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol Cell. 2002;10:1441–1452. doi: 10.1016/s1097-2765(02)00778-5. [DOI] [PubMed] [Google Scholar]