Abstract
Histone acetylation modulates histone occupancy both at promoters and in coding sequences. Based on our recent observation that HDACs in the budding yeast, Saccharomyces cerevisiae, are co-transcriptionally recruited to coding regions by elongating polymerases, we propose a model in which Pol II facilitates recruitment of chromatin remodeling complexes as well as other factors required for productive elongation.
Key words: transcription, yeast, RNA polymerase II, RSC, histone deacetylases, HDACs, HATs, histone acetyltransferase, Kin28, GCN4
Introduction
The complexity of transcription regulation is underscored by evidence indicating that multiple mechanisms function at each stage of the process. Initially, the majority of regulation was thought to occur at initiation, which was presumed to be the most energetically favorable time point for the cell. The current view is that regulation occurs at every stage of transcription. Although the exact factors and mechanisms involved have not been fully characterized, the regulatory effect of chromatin structure on multiple stages of transcription has been well established. The packaging of eukaryotic DNA into chromatin creates a physical barrier for the transcription machinery both at initiation and throughout the elongation process. The dynamic disassembly and reassembly of nucleosomes observed at the promoter and coding regions of genes thus facilitates accurate transcription while preventing inappropriate transcription from or damage to accessible DNA.1–8 Despite the discovery of many factors involved in histone disassembly and reassembly, the exact mechanisms by which these processes occur during transcription, especially during elongation, are not well understood.
The initial studies giving rise to the histone code hypothesis led to the prevailing view that particular histone modifications or combinations of modifications serve to recruit various chromatin modifying and remodeling complexes to regulate chromatin structure and consequently transcription.9,10 This view is supported by the presence in these complexes of protein domains such as bromodomains, chromodomains (CHD) and plant homeobox domains (PHD) that are capable of recognizing specific histone modifications.11 Recent studies providing evidence for crosstalk between modifications, such as ubiquitination and methylation,12,13 and for combinatorial mechanisms distinguishing recruitment from function,4,14,15 reveal a far more complex code. In this article, we will focus on the application of one such combinatorial mechanism to the modulation of histone occupancy by co-transcriptionally recruited histone modifying and chromatin-remodeling complexes during transcription elongation. More specifically, we hypothesize that interactions with elongating RNA Polymerase II and histone acetylation are important for the co-transcriptional recruitment and subsequent function of chromatin remodeling complexes such as SWI/SNF or RSC.
HDACs are Recruited to Coding Regions by Elongating RNA Polymerase II
The Set2 methyltransferase is targeted to coding regions during elongation to methylate histone H3K36 through an interaction with the Pol II CTD, phosphorylated at Ser2 (Ser2P). Several studies have shown in the budding yeast Saccharomyces cerevisiae that the smaller of the two Rpd3 containing complexes, Rpd3C(S) recognizes and binds H3K36me2/3 through its PHD and CHD containing subunits, Rco1 and Eaf3, respectively.8,16 Since Set2 travels with elongating Pol II,17–19 the H3K36me2/3 mark was proposed to serve as a signal for Rpd3C(S) recruitment and deacetylation of nucleosomes in the wake of the elongating polymerase. The production of cryptic transcripts from the FLO8 and STE11 genes in set2Δ or eaf3Δ mutants led to the hypothesis that HDACs are co-transcriptionally recruited to actively transcribing genes to essentially re-set the chromatin behind the elongating polymerase.1,8 Although our recent results confirmed the cryptic transcript production and the increase in H4 acetylation at FLO8 and STE11 in set2Δ strains, we did not observe any reduction in Rpd3C(S) occupancy in the coding regions of these genes.14 This result suggested that while the H3K36me2 mark was indeed important for the deacetylation function of the Rpd3C(S) complex, it was not absolutely required for recruitment of the complex to chromatin.
The importance of H3K36 methylation for Rpd3C(S) binding to nucleosomes was confirmed by our co-immunoprecipitation (Co-IP) experiments in that the interaction of the Rpd3C(S)-specific subunit Rco1 with H3 was lost in set2Δ cells.14 Such a result, however, did not explain the efficient recruitment of Rpd3C(S) that we observed through chromatin immunoprecipitation (ChIP) in the absence of Set2. Together, these results indicated that while H3K36me2/3 was required for Rpd3C(S) binding to nucleosomes, some other factor was responsible for its recruitment to chromatin. Because various factors involved in elongation or mRNA processing, including Set2, were shown to be recruited to chromatin through elongating Pol II,4,5,20–23 we sought to determine whether Rpd3C(S) physically interacted with Pol II, phosphorylated at the C-terminal Domain (CTD). Co-IP experiments revealed that Rco1 co-immunoprecipitated with Ser5 phosphorylated (Ser5P) but not hypophosphorylated Pol II, and that the interaction required Kin28, a Ser5 and Ser7 Pol II CTD kinase.14 Furthermore, in vitro peptide binding assays showed that reconstituted Rpd3C(S) bound strongly to peptides phosphorylated at Ser5, and to di-phosphorylated peptides (Ser2P and Ser5P) even more so, when compared to unmodified peptides or peptides with Ser2 or 5 mutated to aspartic acid.14 Consistent with these observations, Rco1 occupancies, measured by ChIP, at the ARG1 ORF were significantly reduced in both kin28-ts and kin28-as (sensitive to the ATP-analog NMPP1) mutants. Furthermore, the physical interaction of Rpd3C(S) with both H3 and Ser5P was lost in the kin28-ts mutant, while the interaction with H3 but not Ser5P was affected in set2Δ mutants.14 Overall, these results suggest that Phospho-Pol II is important for the initial recruitment of Rpd3C(S) to coding sequences, and that the H3K36me2/3 modification is important for the binding of Rpd3C(S) to nucleosomes and for its histone deacetylase activity. These results are additionally supported by a recent genome-wide study showing that Rpd3C(S) occupancy was affected at the ORFs of only one third of genes in set2Δ or H3K36A mutants, while the occupancy was virtually lost genome-wide in the kin28-as mutant strain.15
Hos2 of the Set3C complex also physically interacts with Pol II phosphorylated at Ser5, but not with hypo-phosphorylated Pol II,14 suggesting that a similar mechanism is responsible for the co-transcriptional recruitment of Hos2 to coding regions.5 Hos2-Set3C was shown to be recruited to ORFs by the recognition of Set1-mediated H3K4me2 by the PHD domain in Set3.24 Hos2 occupancy at the ARG1 ORF, however, was not affected in a bre1Δ mutant, defective for H3K4 di and tri methylation, suggesting that H3K4me2 is not required for Hos2 recruitment at least to this gene.14 Consistent with our coIP data, Hos2 occupancy was reduced in the kin28-ts mutant, suggesting that similar to Rpd3C(S), Phospho-Pol II is important for the recruitment of Hos2 to coding regions and that H3K4me2 is important for its subsequent binding to nucleosomes. 14 In further support of this hypothesis, the physical interaction of Hos2 with H3 but not with Ser5P was affected in bre1Δ cells.14 Overall, these results provide evidence for a two-step model in which HDACs are recruited to coding regions through an interaction with elongating Pol II, and that particular histone modifications, in this case H3K36me2/3 and H3K4me2, are subsequently important for nucleosome binding and deacetylation function. It is possible that the modifications directly recruit HDACs at certain subsets of genes, and that Phospho-Pol II is required for recruitment to others, as suggested by the genome-wide study of Rpd3C(S) occupancy mentioned above.15 Such an observation would explain the discrepancy between our results in bre1Δ mutant cells and those of an earlier study.24
Histone Acetylation is Inversely Correlated with Histone Occupancy in Coding Regions
HAT complexes were initially observed to be targeted to promoters, and acetylation in coding regions was thus thought to be a result of untargeted global activity. 25 Recently, multiple studies have shown that both HATs and HDAs are co-transcriptionally recruited to coding regions.1,5,8,14,24 The activities of these factors in coding regions have additionally been linked to the facilitation of transcription elongation. For example, acetylation by SAGA (Gcn5) and NuA4 (Esa1) in the GAL1 ORF mediates co-transcriptional histone eviction and Pol II processivity through coding regions.4,5 Furthermore, we recently showed that multiple HDACs are recruited to the coding region of ARG1 upon induction.14 Double and triple mutants for Rpd3, Hda1 and Hos2 exhibited an increase in acetylation and a decrease in histone occupancy, suggesting that the function of these HDACs is to minimize co-transcriptional histone eviction.14
Chromatin Remodeling Complexes Potentially Function Coordinately with HATs and HDACs During Transcription Elongation
As mentioned above, histone acetylation is inversely correlated with histone occupancy in coding regions.5,14 The association of HATs and HDACs with elongating Pol II thus provides a potential explanation for how histone occupancy and consequently transcription elongation are regulated at actively transcribed genes. HAT complexes are recruited through an interaction with elongating Pol II to acetylate nucleosomes, most likely in front of the polymerase.4,5,21 These acetylated nucleosomes are then somehow targeted for eviction to allow the polymerase to pass through.5,14 Nucleosomes are then immediately reassembled behind the polymerase to reduce the possibility of untargeted transcription from cryptic promoters or damage to accessible DNA. This rapid reassembly is evident from the unaltered histone acetylation and occupancy observed in coding regions in wild-type cells.14 Presumably, the HDACs co-transcriptionally recruited along with HAT complexes allow for rapid and efficient deacetylation of nucleosomes that are reassembled behind the elongating polymerase.1,8 As described above, both the HATs and HDACs required for establishing the acetylation patterns that facilitate histone eviction and reassembly are recruited through an interaction with the elongating polymerase. Similarly, the factors responsible for establishing the methylation patterns required for the nucleosome binding and histone deacetylase function of the HDACs are recruited through elongating Pol II. It follows then, that the factors that evict and reassemble the histones may interact with elongating Pol II as well. In this way, the local concentration of factors necessary for modulating histone occupancy and thus elongation would be increased at the coding regions of actively transcribed genes.
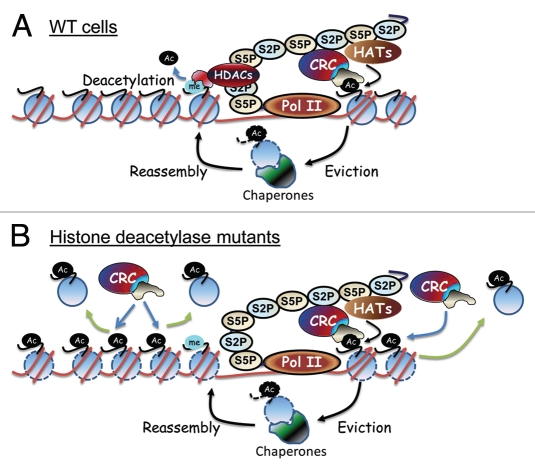
In support of this hypothesis, the SWI/SNF chromatin remodeling complex has been shown to travel with elongating Pol II,26 and the RSC chromatin remodeling complex interacts with the Rpb5 subunit of RNA Polymerases I, II and III.27 Additionally, histone chaperones such as Spt6, Asf1 and FACT interact with elongating Pol II.28,29 The SWI/SNF family chromatin remodeling complexes represent candidate effector molecules that could recognize the acetylation patterns generated by the co-transcriptionally recruited HATs and HDACs to evict ORF nucleosomes. Both the SWI/SNF and RSC complexes possess bromodomains which are capable of recognizing and binding acetylated histones.30,31 The Swi2/Snf2 bromodomain has been shown to be important for SWI/SNF binding to nucleosomes in vivo.31 This bromodomain has further been shown to be important for the removal of SAGA acetylated promoter nucleosomes by SWI/SNF in vitro.32 RSC has been shown to mediate Pol II elongation through nucleosomes in vitro, and prior acetylation by SAGA and NuA4 enhanced this function of RSC.33 Additionally, acetylation by NuA4 was shown to promote both RSC and SWI/SNF recruitment to the GAL1 coding region in vivo.4 Based on these studies, we propose a model in which HAT complexes are recruited to coding regions upon induction (Fig. 1A). Chromatin remodeling complexes such as SWI/SNF or RSC, which interact with the elongating polymerase, are able to recognize acetylation patterns through bromodomain-containing subunits to bind and evict histones. The evicted histones are likely transferred by RSC or SWI/SNF to histone chaperones such as Asf1 or FACT which can then aid in reassembly. In fact, both RSC and SWI/SNF interact genetically with Asf1 and Spt16.34,35 The HDACs that are recruited to coding regions during activation can then rapidly deacetylate the reassembled histones to prevent them from immediate re-eviction. In this way, the coding regions of actively transcribed genes are protected from both cryptic transcription and damage to accessible DNA.
Figure 1.
(A) Histone acetyltransferase (HAT) complexes associate with elongating polymerase and acetylate nucleosomes in front of Pol II. Bromodomain-containing remodeling complexes (CRC) that interact with Pol II are able to recognize, bind and evict acetylated histones. The evicted histones, which presumably are hyperacetylated, are reassembled behind elongating Pol II with the aid of histone chaperones such as Asf1 or FACT. The reassembled histones are then rapidly deacetylated by the histone deacetylase complexes (HDACs) associated with Pol II. (B) The hyperacetylated reassembled nucleosomes are potential targets for non-Pol II associated bromodomain-containing complexes. If HDACs are not present to rapidly deacetylate reassembled histones, the histones may be recognized by free bromodomain-containing remodelers and evicted.
We speculate that the decrease in histone occupancy in coding regions that we observe in HDAC mutants stems from eviction of the hyperacetylated nucleosomes reassembled behind the elongating polymerase. The acetylation levels or patterns of these nucleosomes likely render them accessible to untargeted activity of the bromodomain-containing chromatin remodeling complexes normally recruited by Pol II to evict histones directly in front of the polymerase (Fig. 1B). Similarly, it is possible that hyperacetylated nucleosomes far in front of the elongating polymerase are targeted by free remodelers and evicted as well, resulting in the overall lower level of histones that we observe in coding regions in HDAC mutants.
Conclusions
Our results indicate that the Pol II CTD in S. cerevisiae is involved in recruiting both HATs and HDACs to modulate histone occupancy in the coding regions of actively transcribed genes. These results, together with evidence that the Pol II CTD recruits additional chromatin modifying complexes, histone chaperones and elongation factors, suggest that Phospho-Pol II is integral in coordinating the activities of the many factors required for regulating histone dynamics and consequently transcription elongation at actively transcribing genes.
References
- 1.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 2.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 3.Williams SK, Tyler JK. Transcriptional regulation by chromatin disassembly and reassembly. Curr Opin Genet Dev. 2007;17:88–93. doi: 10.1016/j.gde.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Ginsburg DS, Govind CK, Hinnebusch AG. NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5. Mol Cell Biol. 2009;29:6473–6487. doi: 10.1128/MCB.01033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG. Gcn5 promotes acetylation, eviction and methylation of nucleosomes in transcribed coding regions. Mol Cell. 2007;25:31–42. doi: 10.1016/j.molcel.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 6.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 7.Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 9.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 10.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 11.Bottomley MJ. Structures of protein domains that create or recognize histone modifications. EMBO Rep. 2004;5:464–469. doi: 10.1038/sj.embor.7400146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karam CS, Kellner WA, Takenaka N, Clemmons AW, Corces VG. 14-3-3 mediates histone cross-talk during transcription elongation in Drosophila. PLoS Genet. 2010;6:1000975. doi: 10.1371/journal.pgen.1000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, et al. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 14.Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, et al. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drouin S, Laramee L, Jacques PE, Forest A, Bergeron M, Robert F. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genet. 2010;6:1001173. doi: 10.1371/journal.pgen.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 17.Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- 18.Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, et al. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu H, Hu C, Wong CM, Hinnebusch AG. The Spt4p subunit of yeast DSIF stimulates association of the Paf1 complex with elongating RNA polymerase II. Mol Cell Biol. 2006;26:3135–3148. doi: 10.1128/MCB.26.8.3135-3148.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wittschieben BO, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, et al. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- 22.Qiu H, Hu C, Hinnebusch AG. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol Cell. 2009;33:752–762. doi: 10.1016/j.molcel.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 24.Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol. 2003;4:276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- 26.Schwabish MA, Struhl K. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol Cell Biol. 2007;27:6987–6995. doi: 10.1128/MCB.00717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soutourina J, Bordas-Le Floch V, Gendrel G, Flores A, Ducrot C, Dumay-Odelot H, et al. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol Cell Biol. 2006;26:4920–4933. doi: 10.1128/MCB.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Mason PB, Struhl K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol. 2003;23:8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cairns BR, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg RD, Winston F. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH and bromodomains. Mol Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- 31.Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, et al. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- 32.Chandy M, Gutierrez JL, Prochasson P, Workman JL. SWI/SNF displaces SAGA-acetylated nucleosomes. Eukaryot Cell. 2006;5:1738–1747. doi: 10.1128/EC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol Cell. 2006;24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 35.Malone EA, Clark CD, Chiang A, Winston F. Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5710–5717. doi: 10.1128/mcb.11.11.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]