Abstract
The Drosophila melanogaster fourth chromosome contains interspersed domains of active and repressive chromatin. We investigated a stock harboring a silenced transgene inserted into Dyrk3 and near Caps—two expressed genes on chromosome four. In an HP1a-deficient background, transgene expression was activated while, paradoxically, expression of Dyrk3 and Caps was reduced. We found that the promoters of Dyrk3 and Caps contained DNase I hypersensitive sites but also possessed methylated histone H3 and HP1a, marks of repressive chromatin. In HP1a-deficient flies, the Dyrk3 and Caps promoters displayed diminished accessibility to nuclease digestion, revealing a surprising role for HP1a in opening chromatin.
Key words: chromatin, Drosophila, gene regulation, heterochromatin, Heterochromatin Protein 1, histone methyltransferase
Eukaryotic genomes contain characteristic patterns of active and repressive chromatin, each harboring distinctly modified histones and associated factors.1 Histone N-terminal tails are methylated on various residues, which triggers the recruitment of partner proteins that affect chromatin structure. For example, centric heterochromatin, a form of repressive chromatin, is enriched for histone H3 that is di- and tri-methylated at lysine 9 (H3K9me2/3).2 H3K9me2/3 is a binding site for other factors associated with gene silencing, such as Heterochromatin Protein 1a (HP1a). Histone H3 can also be methylated on lysine 4 (H3K4me), a modification enriched in euchromatin. The H3K4me mark binds chromatin remodeling complexes (e.g., NURF) that activate gene expression.3
In Drosophila, when euchromatic genes are brought into juxtaposition with heterochromatin, they are silenced in a mosaic pattern; this phenomenon, known as position-effect variegation (PEV), implies that heterochromatin spreads into transposed euchromatin.4 PEV can be suppressed or enhanced by second-site mutations termed Su(var)s and E(var)s, respectively,4 which implies these mutations disrupt the formation and/or spread of heterochromatin.
Two Su(var) genes, Su(var)2-5 and Su(var)3-9, are of particular interest because they are part of a positive feedback loop predicted to propagate and maintain heterochromatin. Su(var)3-9 encodes a histone methyltransferase (HMT) that generates H3K9me.5,6 In turn, H3K9me2/3 is a binding site for HP1a [the protein product of Su(var)2-5]. HP1a is a conserved chromo domain protein7,8 that localizes to centric heterochromatin. HP1a is part of a protein family in which members posses both a chromodomain and chromoshadow domain, yet have diverse functions, including sex-specific roles.9 As loss of HP1a is lethal, none of the family members appear to have redundant function with HP1a. Within heterochromatin, HP1a is thought to recruit SU(VAR)3-9, which methylates adjacent histone H3 tails, generating additional HP1a binding sites and completing the positive feed-back loop.
Although euchromatic genes brought near heterochromatin are silenced, silencing can be relieved by Su(var) mutations and chromosomal translocations that return the gene to euchromatin.4 Conversely, genes that naturally reside in heterochromatin are not expressed well in Su(var) mutant backgrounds, or when placed near euchromatin by chromosomal translocations.10–13 The underlying cause of this reciprocal expression pattern is elusive; however, it may reflect an unexpected role for HP1a in gene expression.
The small fourth chromosome of Drosophila provides an attractive environment to study the role of HP1a in gene regulation because it uniquely contains interspersed domains of active and repressive chromatin.14 As with other Drosophila chromosomes, repressive domains on chromosome four are associated with H3K9me2/3; but, unique to chromosome four, this H3K9me2/3 is primarily generated by a different HMT—dSETDB1.15,16 While repressive domains on the fourth chromosome share some characteristics of centric heterochromatin, they differ in several respects.14 It is important to note that repressive domains can express resident genes. To take advantage of the unique situation of active and repressive domains, we examined the expression of a variegating hsp70-white P-element inserted in the fourth chromosome within the coding region of the expressed gene Dyrk3. The position of this insert provided an opportunity to better understand the mechanisms underlying differential control of gene expression by HP1a.
Here, we present data consistent with gene-specific roles for HP1a. This unique transgene insertion site revealed that HP1a-dependent gene expression and HP1a-dependent gene silencing can occur at nearly the same genomic location. Surprisingly, HP1a was required to establish or maintain an open chromatin structure at the promoter of Dyrk3, while, at the same time repressing expression of the hsp70-white reporter. These results suggest HP1a supports gene expression via a previously undiscovered mechanism that involves the generation of open chromatin.
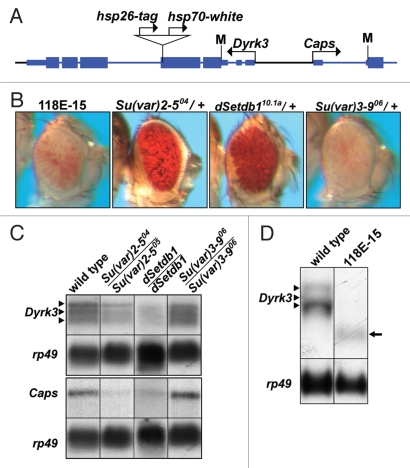
Previously we isolated stock 118E-15,17,18 in which an hsp70-white reporter gene inserted within the coding region of Dyrk3 (which encodes a serine/threonine kinase; Fig. 1A). PEV of the reporter was suppressed by a null allele of Su(var)2-5, demonstrating that the variegated silencing required HP1a (Fig. 1B).18 Mutations in the histone H3K9 HMT-encoding dSetdb1 also suppressed silencing of the reporter gene, whereas alleles of Su(var)3-9 did not, consistent with dSETDB1 functioning as the primary methyltransferase acting on the fourth chromosome.15,16,19
Figure 1.
An hsp70-white transgene inserted within Dyrk3 displays variegating expression. (A) Schematic representation of Dyrk3 and a portion of Caps (modified from the RefSeq track of the UCSC Genome Browser). Bent arrows represent the transcription start sites (+1). Coding sequences are displayed as thick, blue boxes; transcribed, but not translated sequences are shown as thin, blue boxes and introns shown as thin, blue lines. The translation start sites (M) and the location of the P-element insertion (triangle) are indicated. (B) Eye phenotypes of strains containing the hsp70-white transgene. Females of the P-element insertion stock, 118E-15, were crossed to males containing mutations in known modifiers of PEV. Photographs of eyes from the parental stock and the resulting heterozygous progeny are shown. (C) Northern analysis of Dyrk3 and Caps. RNA was isolated from third instar larvae of y,w67c23 (wild type) and Su(var)2-5, dSetdb1 or Su(var)3-9 mutants. Transcripts were detected by hybridization with an α-32P labeled fragment for Dyrk3 or Caps. Detection of rp49 was used for loading control. Three arrowheads indicate alternatively spliced Dyrk3 transcripts. Results shown are representative of three experiments. (D) Northern analysis of Dyrk3 in a wild-type background and stock 118E-15. Arrow heads indicate wild-type transcripts, arrow indicates the truncated transcripts observed in the P-element-containing 118E-15 stock.
PEV of the hsp70-white reporter gene suggested that the P-element had inserted into a region of repressive chromatin. Surprisingly, northern analyses revealed that Dyrk3 and Caps (a neighboring gene located ∼1.2 kb upstream of Dyrk3; Fig. 1C) were expressed. The P-element insertion did not prevent transcription initiation from the Dyrk3 promoter as evidenced by a shorter length message, consistent with transcriptional termination within the P-element (Fig. 1D). In larvae trans-heterozygous for null Su(var)2-5 or dSetdb1 alleles, expression levels of Dyrk3 and Caps were reduced by 50% or more (Fig. 1C). In contrast, a stock homozygous for a null allele of Su(var)3-9 expressed both transcripts similar to wild type. Despite a recently observed connection between HP1a and splicing factors,20 the mutant background did not appear to alter splicing of Dyrk3. Taken together, these results suggest that Dyrk3 and Caps reside in a repressive chromatin environment that requires dSETDB1 and HP1a for full transcriptional activity.
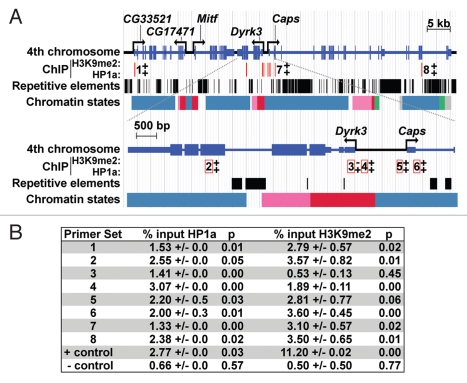
The region surrounding Dyrk3 and Caps is characteristic of much of the fourth chromosome: it is enriched for repetitive elements, but maintains a gene density similar to that of euchromatic regions (Fig. 2).14 The requirement of HP1a for two apparently opposite effects—active transcription of Dyrk3 and Caps, and PEV silencing of the reporter gene—led us to test whether HP1a is directly associated with the chromatin of this region. Approximately 70 kb of chromosome four, centered around Dyrk3 and Caps were analyzed via chromatin immunoprecipitation (ChIP). Third instar salivary gland nuclei were used for ChIP as available antibodies recognized multiple proteins in other tissues upon western analysis.21,22 Expression of Dyrk3 and Caps was readily detected by RT-PCR using RNA isolated from salivary glands and showed a 7- to 11-fold decrease in expression in the Su(var)2-5 mutant background (data not shown). Genomic fragments were immunoprecipitated using either anti-H3K9me2 or anti-HP1a antibodies, and amplified using eight unique primer sets (Fig. 2A and B). Antibodies recognizing H3K9me2 retrieved genomic fragments amplified by seven of eight primer sets, with a signal ranging from 1.89–3.60% of input (compared to 0.5% for the negative control; Fig. 2B). The exception was a region adjacent to the Dyrk3 transcription start site (TSS; +23 to +183), which gave values similar to the negative control (Fig. 2B). Surprisingly, all primer sets demonstrated that HP1a was associated with the target sequences; this included the primer set that was negative for histone H3K9me2. Values ranged from 1.33–3.07% of input (compared to 0.66% for the negative control; Fig. 2B). Data from the modENCODE project agreed with our findings, showing that Dyrk3 and Caps are expressed and are associated with HP1a and H3K9me2/3 in S2 cultured cells and during development, including the third instar larval stage (Fig. 2A; www.modencode.org). Histone H3K27me, a mark of developmental silencing via Polycomb was depleted throughout the region. modENCODE has annotated chromatin to include nine distinctive states.23 The TSS of Dyrk3 was designated as state 1, characterized by H3K4me2/3 in combination with H3K9ac; both are marks of active chromatin. The coding regions of both Dyrk3 and Caps possess regions in state 2, which are enriched in H3K36me3, a signature of transcription elongation. The entire genomic region, including some exons of Dyrk3 and Caps are designated as state 7, characterized by high levels of H3K9me2/3 similar to that present in centric heterochromatin and other regions of chromosome 4,23 (Fig. 2A). Taken together, these results suggest that HP1a associates with the genomic region encompassing Dyrk3 and Caps, even at sites where levels of histone H3K9me2 are undetectable and that HP1a promotes expression of Dyrk3 and Caps.
Figure 2.
The distribution of HP1a, histone H3K9me2, repetitive elements and chromatin states within the Dyrk3/Caps (A) genomic region. A diagram of the sub-telomeric region of the fourth chromosome is shown at the top; a magnification of Dyrk3 and the 5′ region of Caps is depicted below. Repetitive elements are represented by black boxes (taken from Repeat Masker track of the UCSD Genome Browser). Polyclonal antibodies to HP1a (Covance), H3K9me2 (Upstate) and GFP (Molecular probes) were used for ChIP from third instar salivary glands. The location of primer sets are indicated by red lines; numbers adjacent to (top) or within the open red boxes (bottom) correspond to the number of a primer set. +, a positive ChIP signal; −, a negative result. Primers for CG31999 at position 102B1 on chromosome 4 were used as a negative control. Primers corresponding to an hsp26 transgene integrated near the centric region of chromosome 4 were used as a positive control. This transgene is silenced in an HP1a-dependent manner.18 The chromatin states are described according to modENCODE:23 red represents enrichment of H3K4me3/2 and H3K9Ac (state 1) found at TSSs, pink represents enrichment for H3K36me3 (state 2) associated with transcription elongation; green represents enrichment for H4K16ac(state 5); blue represents enrichment for H3K9me2/3, which is found in heterochromatin (state 7); grey represents regions of very low transcriptional activity (state 9). (B) Data obtained from ChIP experiments; primer set numbers correspond to numbers in (A) and p denotes p values.
HP1a and H3K9me2/3 associate with sequences throughout the 70 kb encompassing Dyrk3 and Caps, suggesting that this region might be assembled into closed chromatin, a structure suggested by the PEV of the hsp70-white transgene.18 Paradoxically, this region actively expresses both Dyrk3 and Caps, suggesting it is in an open conformation. To explore this paradox, we used DNase I hypersensitivity assays to determine the chromatin conformation of the intergenic region between the Dyrk3 and the Caps TSS (Fig. 3). Consistent with active transcription, both the wild-type host stock and the P-element-containing 118E-15 stock showed three prominent DNase I hypersensitive sites, which corresponded approximately to positions +460, +160 and −240, relative to the Dyrk3 TSS (Fig. 3). Additionally, we detected a fourth hypersensitive site, at approximately position −130 relative to the Caps transcription start site (Fig. 3). Collectively, these data demonstrate that although HP1a associates with this region, the chromatin assumes an open conformation near the promoters of these two genes; in addition, insertion of the P-element had no effect on the open chromatin conformation of the endogenous genes.
Figure 3.
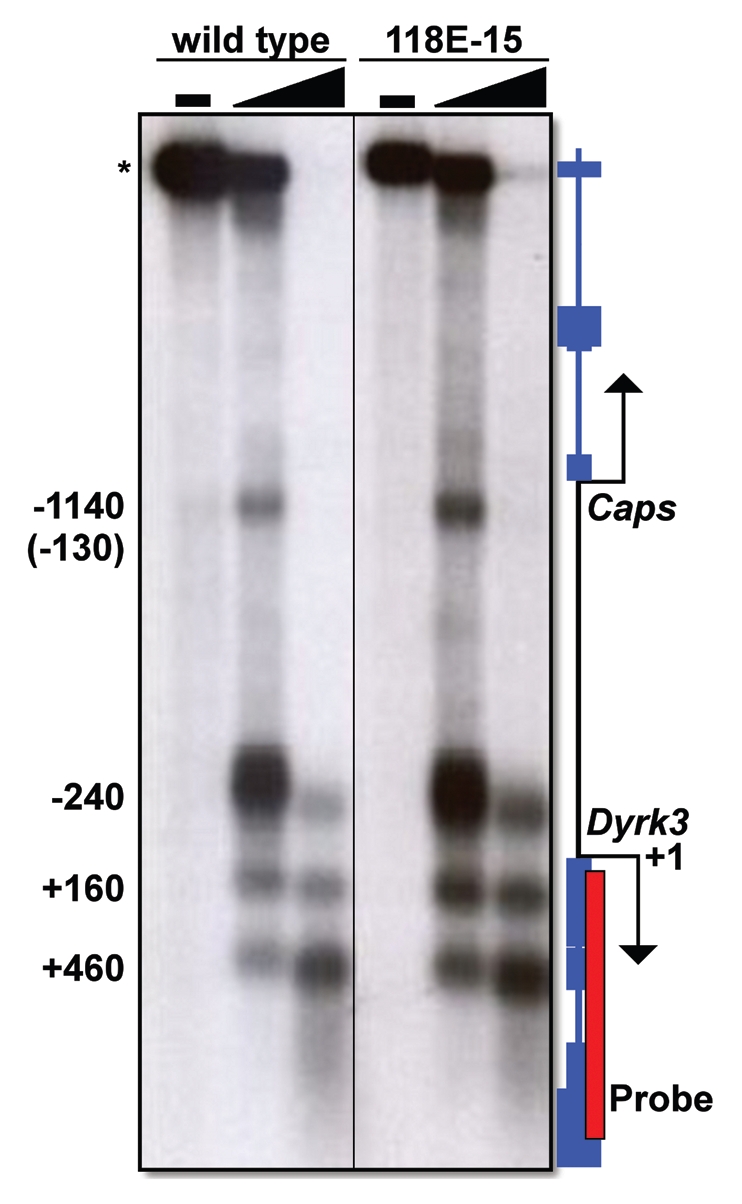
DNase I hypersensitivity analysis of the Dyrk3 and Caps promoter regions. Nuclei were isolated from larvae and treated with increasing amounts of DNase I. Genomic DNA was purified and digested to completion with NciI, which cleaves at sites flanking the region of interest. The DNA was separated by electrophoresis and transferred to a nylon membrane (Amersham). DNase I hypersensitive sites were detected following hybridization with α-32P labeled 842 bp fragment of Dyrk3 (red box). Numbers to the left of the radiograph denote the approximate locations of DNase I hypersensitive sites relative to the Dyrk3 transcription start site (+1).The * denotes the Ncil Fragment that is not cleaved by DNase 1. The number in parentheses represents the distance relative to the TSS of Caps.
In a Su(var)2-5 mutant background, Dyrk3 and Caps are downregulated. This suggests that, in the absence of HP1a, these genes assume a chromatin structure that is less transcriptionally active. To test this unexpected hypothesis, we performed restriction enzyme accessibility assays, which allow quantitative measurements of chromatin accessibility.17,18 In performing this assay, we took advantage of several AvaII restriction sites in the Dyrk3 promoter region. One site lies 110 bp upstream of the Dyrk3 transcription start site (approximately 270 bp and 130 bp from the DNase hypersensitive sites at +160 and −240, respectively); two more distal AvaII sites lie at −231 and −3,355 bp (Fig. 4). Nuclei from the wild-type host stock or Su(var)2-5 trans-heterozygotes were treated with an excess of AvaII, processed, and the cleavage products were resolved by electrophoresis and visualized by a radiolabeled probe (Fig. 4). In nuclei from the wild-type stocks, 61–65% of the promoter-containing fragment was cleaved at the proximal AvaII site (Fig. 4), confirming the promoter assembles into an open chromatin conformation.17,18 In a Su(var)2-5 trans-heterozygous null background, the same assay detected this site to be cleaved with only 36–37% efficiency (Fig. 4), showing that in the absence of HP1a this promoter region tends to adopt a relatively closed conformation. Surprisingly, these data indicate that HP1a allows the Dyrk3 promoter region to assume an open, transcriptionally active conformation.
Figure 4.
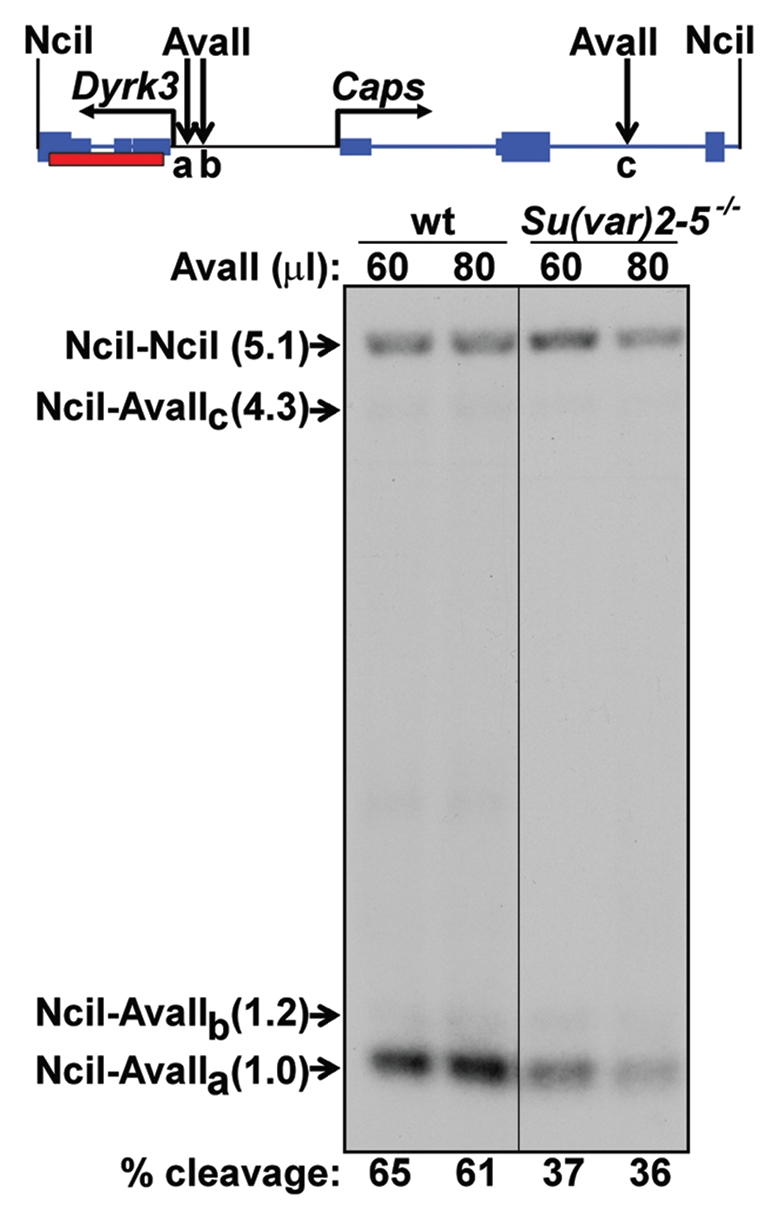
Restriction enzyme accessibility analysis of the Dyrk3 and Caps promoter regions. Top: schematic of the Dyrk3-Caps region flanked by two NciI sites, labels are the same as in Figure 3, with the addition of AvaII sites (down arrows). Bottom: Southern analysis of fragments resulting from nuclei isolated from wild type (wt) or trans-heterozygous [Su(var)2-5-/-] larvae treated with AvaII. Purified genomic DNA was cut with NciI. The arrow indicates the position of the NciI-NciI parental fragment. The positions of the two fragments resulting from cleavage at the distal AvaII sites (B and C) are indicated. NciI-AvaIIa indicates the position of the quantifiable fragment resulting from cleavage at the proximal AvaIIa site. The red box indicates a fragment of Dyrk3 used as a probe. The percent cleavage at the proximal AvaII site is reported at the bottom. Cleavage at the AvaIIa site was quantified using Labworks software (UVP BioImaging Systems) and is reported as the percentage of the total signal from all four bands.
Although heterochromatin contains proteins known to inhibit transcription from euchromatic genes, many transcriptionally competent genes are, in fact, embedded in heterochromatin.24 Here, we provide insights into a mechanism that might drive this transcription by showing that the promoter of a gene normally residing in a repressive chromatin environment is held in an open conformation only if it is packaged by chromatin containing HP1a. Moreover, across the length of the 70 kb of chromosome four containing Dyrk3 and Caps, we found HP1a associated with histone H3K9me2. These findings are consistent with recent reports that the epigenetic mark of histone H3K9me2 can be found associated with transcribed genes.21,25,26 Thus, the association of HP1a did not necessarily silence transcription, providing an explanation for the transcription of genes that naturally reside in heterochromatin.24 Most surprising, the Dyrk3 promoter requires bound HP1a to display an open chromatin configuration.
Several models have been put forth to describe the expression of heterochromatic genes.13 In D. melanogaster, studies comparing genes that reside in heterochromatin to their euchromatic orthologs in other Drosophila species found that the promoter structures are generally conserved.27 This strongly suggests unique promoter elements do not drive the contrasting expression patterns. Thus, in heterochromatin, distant enhancer elements might drive differential expression from otherwise unremarkable promoters. Although repetitive elements frequently interrupt heterochromatic genes and might displace enhancers, bound HP1a is predicted to dimerize at non-adjacent nucleosomes, allowing chromatin to loop, bringing distal enhancers in proximity with promoter elements.28 Such a model would explain why heterochromatic genes require HP1.
HP1a supports expression of selective euchromatic genes, including genes within heat-shock and developmentally induced puffs.20,21,29 In fact, HP1a interacts with, and stabilizes, transcripts from ∼100 highly transcribed genes.29 Recent studies into this effect uncovered that HP1a interacts with several heterogeneous nuclear ribonucleoproteins (hnRNPs).20 HP1 proteins have also recently been connected with transcription elongation; however, the major player seems to be one of the other family members, HP1c.30
Our data suggest an additional mechanism by which HP1a supports gene expression. We observed that the transgene inserted into Dyrk3 exhibited PEV, consistent with HP1a-dependent repressive chromatin in this region. Nevertheless, the open chromatin structure at the Dyrk3 and Caps promoter regions requires HP1a, suggesting that, at these promoters, HP1a promotes an open chromatin structure. One possible explanation of our data may be that HP1a bound near the Dyrk3 promoter recruits a chromatin-remodeling complex that locally opens the chromatin. Evidence for this comes from experiments in which overexpression of SNF2-type chromatin remodelers altered heterochromatic gene silencing.31,32
Acknowledgements
We would like to thank members of the Wallrath lab for comments on the manuscript, S. Mackey for assistance with photography, J. Danzer and J. Limas for technical assistance. This research was supported by a Ruth L. Kirschstein NRSA Postdoctoral Fellowship (GM085974) to M.W.V. and NIH Grant (GM61513) to L.L.W.
Authors' Contributions
D.E.C. performed gene expression/chromatin structure analysis and analyzed data; M.W.V. performed data analysis and prepared figures; L.L.W. designed experiments, analyzed the data and prepared the manuscript.
References
- 1.Mellor J, Dudek P, Clynes D. A glimpse into the epigenetic landscape of gene regulation. Curr Opin Genet Dev. 2008;18:116–122. doi: 10.1016/j.gde.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 4.Weiler KS, Wakimoto BT. Heterochromatin and gene expression in Drosophila. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- 5.Eskeland R, Czermin B, Boeke J, Bonaldi T, Regula JT, Imhof A. The N-terminus of Drosophila SU(VAR)3-9 mediates dimerization and regulates its methyltransferase activity. Biochemistry. 2004;43:3740–3749. doi: 10.1021/bi035964s. [DOI] [PubMed] [Google Scholar]
- 6.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 7.Eissenberg JC, James TC, Foster-Hartnett DM, Hartnett T, Ngan V, Elgin SC. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc Natl Acad Sci USA. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elgin SC, Grewal SI. Heterochromatin: silence is golden. Curr Biol. 2003;13:895–898. doi: 10.1016/j.cub.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Vermaak D, Henikoff S, Malik HS. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet. 2005;1:96–108. doi: 10.1371/journal.pgen.0010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu BY, Emtage PC, Duyf BJ, Hilliker AJ, Eissenberg JC. Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics. 2000;155:699–708. doi: 10.1093/genetics/155.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schulze SR, Sinclair DA, Fitzpatrick KA, Honda BM. A genetic and molecular characterization of two proximal heterochromatic genes on chromosome 3 of Drosophila melanogaster. Genetics. 2005;169:2165–2177. doi: 10.1534/genetics.103.023341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wakimoto BT, Hearn MG. The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. Genetics. 1990;125:141–154. doi: 10.1093/genetics/125.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasuhara JC, Wakimoto BT. Oxymoron no more: the expanding world of heterochromatic genes. Trends Genet. 2006;22:330–338. doi: 10.1016/j.tig.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 14.Riddle NC, Shaffer CD, Elgin SC. A lot about a little dot—lessons learned from Drosophila melanogaster chromosome 4. Biochem Cell Biol. 2009;87:229–241. doi: 10.1139/o08-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seum C, Reo E, Peng H, Rauscher FJ, 3rd, Spierer P, Bontron S. Drosophila SETDB1 is required for chromosome 4 silencing. PLoS Genet. 2007;3:76. doi: 10.1371/journal.pgen.0030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzeng TY, Lee CH, Chan LW, Shen CK. Epigenetic regulation of the Drosophila chromosome 4 by the histone H3K9 methyltransferase dSETDB1. Proc Natl Acad Sci USA. 2007;104:12691–12696. doi: 10.1073/pnas.0705534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cryderman DE, Cuaycong MH, Elgin SC, Wallrath LL. Characterization of sequences associated with position-effect variegation at pericentric sites in Drosophila heterochromatin. Chromosoma. 1998;107:277–285. doi: 10.1007/s004120050309. [DOI] [PubMed] [Google Scholar]
- 18.Wallrath LL, Elgin SC. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 1995;9:1263–1277. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- 19.Brower-Toland B, Riddle NC, Jiang H, Huisinga KL, Elgin SC. Multiple SET methyltransferases are required to maintain normal heterochromatin domains in the genome of Drosophila melanogaster. Genetics. 2009;181:1303–1319. doi: 10.1534/genetics.108.100271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piacentini L, Fanti L, Negri R, Del Vescovo V, Fatica A, Altieri F, et al. Heterochromatin protein 1 (HP1a) positively regulates euchromatic gene expression through RNA transcript association and interaction with hnRNPs in Drosophila. PLoS Genet. 2009;5:1000670. doi: 10.1371/journal.pgen.1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cryderman DE, Grade SK, Li Y, Fanti L, Pimpinelli S, Wallrath LL. Role of Drosophila HP1 in euchromatic gene expression. Dev Dyn. 2005;232:767–774. doi: 10.1002/dvdy.20310. [DOI] [PubMed] [Google Scholar]
- 22.Danzer JR, Wallrath LL. Mechanisms of HP1-mediated gene silencing in Drosophila. Development. 2004;131:3571–3580. doi: 10.1242/dev.01223. [DOI] [PubMed] [Google Scholar]
- 23.Kharchenko PV, Alekseyenko AA, Schwartz YB. Comprehensive analysis of the chromatin land scape in Drosophila melanogaster. Nature. 2010 doi: 10.1038/nature09725. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dimitri P, Caizzi R, Giordano E, Carmela Accardo M, Lattanzi G, Biamonti G. Constitutive heterochromatin: a surprising variety of expressed sequences. Chromosoma. 2009;118:419–435. doi: 10.1007/s00412-009-0211-y. [DOI] [PubMed] [Google Scholar]
- 25.de Wit E, Greil F, van Steensel B. High-resolution mapping reveals links of HP1 with active and inactive chromatin components. PLoS Genet. 2007;3:38. doi: 10.1371/journal.pgen.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yasuhara JC, Wakimoto BT. Molecular landscape of modified histones in Drosophila heterochromatic genes and euchromatin-heterochromatin transition zones. PLoS Genet. 2008;4:16. doi: 10.1371/journal.pgen.0040016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yasuhara JC, DeCrease CH, Wakimoto BT. Evolution of heterochromatic genes of Drosophila. Proc Natl Acad Sci USA. 2005;102:10958–10963. doi: 10.1073/pnas.0503424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Danzer JR, Alvarez P, Belmont AS, Wallrath LL. Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development. 2003;130:1817–1824. doi: 10.1242/dev.00405. [DOI] [PubMed] [Google Scholar]
- 29.Piacentini L, Fanti L, Berloco M, Perrini B, Pimpinelli S. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J Cell Biol. 2003;161:707–714. doi: 10.1083/jcb.200303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon SH, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. Heterochromatin protein 1 (HP1) connects the FACT histone chaperone complex to the phosphorylated CTD of RNA polymerase II. Genes Dev. 2010;24:2133–2145. doi: 10.1101/gad.1959110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneiderman JI, Sakai A, Goldstein S, Ahmad K. The XNP remodeler targets dynamic chromatin in Drosophila. Proc Natl Acad Sci USA. 2009;106:14472–14477. doi: 10.1073/pnas.0905816106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, et al. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]