Abstract
The binding site for allosteric inhibitor (amino acid) is highly conserved between human liver pyruvate kinase (hL-PYK) and the rabbit muscle isozyme (rM1-PYK). To detail similarities/differences in the allosteric function of these two homologs, we quantified the binding of 45 amino acid analogues to hL-PYK and their allosteric impact on affinity for the substrate, phosphoenolpyruvate (PEP). This complements a similar study previously completed for rM1-PYK. In hL-PYK, the minimum chemical requirements for effector binding are the same as those identified for rM1-PYK (i.e. the L-2-aminopropanaldehyde substructure of the effector is primarily responsible for binding). However different regions of the effector determine the magnitude of the allosteric response in hL-PYK vs. rM1-PYK. This finding is inconsistent with the idea that allosteric pathways are conserved between homologs of a protein family.
All pyruvate kinase isozymes catalyze the conversion of phosphoenolpyruvate and ADP to pyruvate and ATP as the last step in glycolysis. However, various isozymes, including mammalian enzymes, differ in regulatory properties. The pyruvate kinase isozyme from human liver (hL-PYK) has decreased affinity for PEP when allosterically inhibited by Ala. In contrast, Ala binding to the isozyme from rabbit muscle (rM1-PYK) elicits minimal change in the affinity for PEP (1). Instead, rM1-PYK has decreased affinity for PEP when allosterically inhibited by Phe. Despite their different functional outcomes, Ala and Phe competitively bind to rM1-PYK, indicating that they bind to the same site on the protein. Thus, the differential effects of Ala on PEP binding by rM1-PYK and hL-PYK provide a means for investigating how allosteric function can vary between homologous proteins.
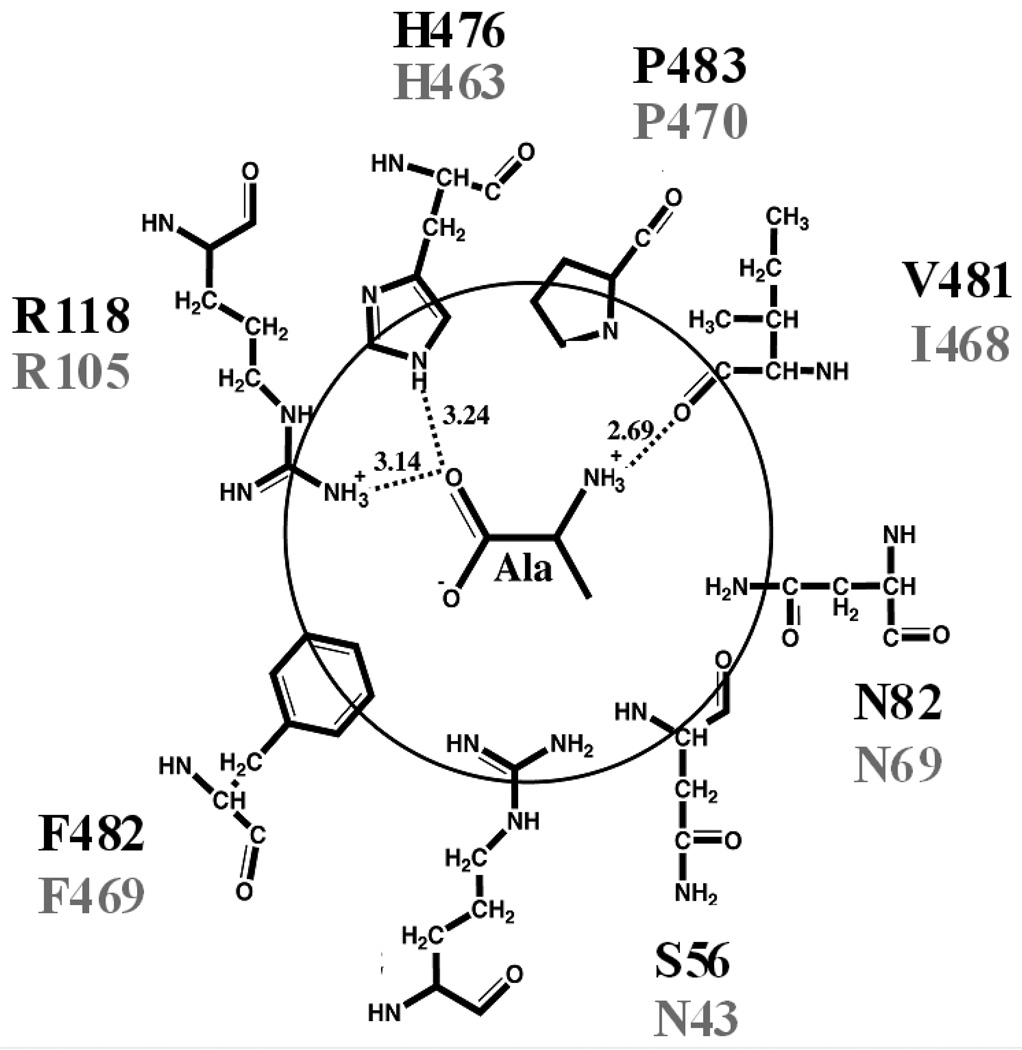
Previously, we identified the amino acid binding site of rM1-PYK by co-crystallization with Ala (Figure 1; PDB 2G50) (1). Ala binding in the equivalent site of hL-PYK has been confirmed by mutagenesis (e.g. H476L as shown in Supporting Information). Although, hL-PYK and rM1-PYK share 67% identity and 81% similarity overall, all of the residue side-chains in the amino acid binding site are completely conserved in these two proteins. The two non-conserved protein residues in this binding site have their backbone atoms exposed to the effector.
Figure 1.
Schematic representation of the amino acid binding site of rM1-PYK with bound Ala (1). The residues of hL-PYK and rM1-PYK are in black and gray, respectively. Note that the rotomeric position of H476 is not firmly established (9).
We report here how 45 different amino acid analogues bind to and allosterically regulate hL-PYK. This analogue series parallels that previously used in the study of amino acid inhibition of rM1-PYK (1). Using the analogue series, we distinguished the chemical moieties of the effector required for binding to hL-PYK and those that determine the magnitude of the allosteric response. A comparison of these properties in the hL-PYK and rM1-PYK (1) systems supports that effector binding, but not allosteric function, are conserved between the two homologues.
Materials and Methods
Materials
The potassium salts of ADP and PEP were purchased from Chem-Impex International, Inc. NADH was from Sigma. L-lactic dehydrogenase (Type III bovine heart) was purchased from Calzyme Laboratories, Inc. Other buffer components were from Fisher Scientific and Sigma. The pLC11 plasmid encoding hL-PYK was obtained as a gift from Drs. Andrea Mattevi and Giovanna Valentini (2).
Since the low solubility and low affinity for some analogues (listed by supplier) may have prevented an observed allosteric effect, the highest concentrations used to determine allosteric responses are shown in brackets, and that used in competitive binding (when completed) are in italicized brackets. The L-forms of Ala [559 mM], Arg [559 mM], Asn [50.3 mM], Asp [6.50 mM], Cys [83.80 mM], Gln [119 mM], Gly [977.5 mM], Glu [12.3 mM], His [81 mM], Ile [61.4 mM], Leu [56.4], Lys [559 mM], Met [167 mM], Phe [83.8 mM], Pro [559 mM], Ser [559 mM], Thr [265 mM], Trp [22.3 mM], Tyr [1.7 mM], and Val [111.7 mM] were purchased from Fisher Scientific. Ethanolamine [81 mM], ethylamine [40 mM] [13 mM] and isopropylamine [80 mM] [30 mM] were purchased from Sigma. (S)-(+)-2-Phenylglycine [8.4 mM], L-homophenylalanine HCl [1 mM], 2-aminoisobutyric acid [559 mM], butylamine [42 mM] [13 mM], D/L-2-aminocaprylic acid [0.15 mM], 4-nitro-L-phenylalanine [3.8 mM], and Omethyl-L-tyrosine [18.8 mM] were from by Aldrich. L-Homoserine [95 mM], L-(+)-2,3-diaminopropionic acid [56 mM], L-(+)-2-aminobutyric acid [559 mM], propionic acid [20.1 mM] [7.7 mM], and L-alanine methyl ester HCl [100 mM] [29 mM], L-norvaline [69.3 mM], L-norleucine [0.44 mM], N-methyl-L-alanine [559 mM], D-alanine [559 mM], 3-cyclohexyl-L-alanine [12.7 mM], D/L-2-aminoheptanoic acid [10.5 mM], and L-alaninol [160 mM] [40 mM] were obtained from Fluka. N-Formyl-L-alanine [15.3 mM] [10 mM], and N-acetyl-L-alanine [20.5 mM] [10 mM] were purchased from MP Biomedicals. D-phenylalanine [67 mM] and (S)-(+)-2-amino-2-methyl-3-phenylpropanoic acid [38.50 mM] were purchased from Acros Organics.
Mutagenesis and Protein Expression and Purification
Mutagenesis of the hL-PYK gene to create the H476L gene was with Quikchange (Stratagene). Wild type and H476L were expressed in the FF50 strain of Escherichia coli (3). Cell lysis, ammonium sulfate fractionation and DEAE-cellulose column purification were carried out as previously reported (3). Purified proteins were used in all studies with amino acid analogs.
Kinetic Assays
Activity measurements were carried out at 30 °C using a lactate dehydrogenase coupled assay (4). Reactions were in 350 µL bicine buffer containing 50 mM bicine/KOH, 5 mM MgCl2, 0.1 mM EDTA, 0.18 mM NADH, 19.6 U/mL lactate dehydrogenase and 5 mM ADP at pH 7.5. PEP and effector ligand concentrations were varied as indicated. PEP and effector ligand stock solutions were adjusted to pH 7.5 with KOH before addition, and dilutions were in KCl to maintain constant total K+ concentration of 150 mM in all assays (4). The enzymatic reaction was initiated with PEP and monitored at 340 nm over time. Data were collected in a 96-well plate using a Molecular Devices Spectramax Plus384 spectrometer. Initial rates were collected from the linear portion of the progress curve.
Data Analysis
Throughout this work, “K-type” heterotropic allostery is defined to occur “when one ligand binds to a protein differently in the absence, versus the presence, of a second ligand,” given that the two ligands bind at different locations on the protein (5). As previously discussed (5–7), this linked-equilibrium view of allostery defines allosteric coupling (Qax) as a ratio of binding constants:
| (1) |
where Kia = the dissociation constant for the first ligand, A, binding to the protein in the absence of the second ligand, X; Kia/x = the dissociation constant for A binding to the protein with X prebound, Kix = the dissociation constant for X binding to the protein in the absence of A; and Kix/a = the dissociation constant for molecule X binding to the protein with A pre-bound. Qax = 1 means no allosteric response, Qax > 1 defines allosteric activation, and Qax < 1 defines allosteric inhibition. Since Qax is a ratio, the magnitude of this allosteric coupling is independent of the magnitude of any one ligand dissociation constant. Therefore, individual atom-atom interactions between atoms from protein residues and atoms from chemical moieties of the ligand may contribute uniquely to ligand binding vs. allostery.
Data fitting was with the nonlinear least-squares analysis of Kaleidagraph (Synergy) software. Fits of PEP titrations of initial rates (ν) used to obtain Kapp-PEP are as previously described (3, 4). Although the potassium concentration used throughout ligand concentration range was kept constant, a control experiment was included to ensure that other counter ions and/or non-specific effects of the various Ala analogues were not contributing to the observed regulation. For this control, the impact of the analogues on the affinity of H476L for PEP was monitored. H476L completely removes an Ala elicited response but maintains an affinity for PEP similar to that of wild type hL-PYK (see supporting information). Responses of wild type protein to analogues were corrected by dividing the Kapp-PEP vs. analogue response for wild type by the Kapp-PEP vs. analogue response for H476L (i.e. subtraction of the respective free energies).
The magnitude of Qax is measured by plotting the Kapp-PEP values as a function of effector concentration and fit to equation 2 (7):
| (2) |
where Ka = Kapp-PEP when [Effector] = 0. In several cases, it was not possible to obtain formation of the ternary substrate-enzyme-effector complex (i.e. formation of the upper plateau when plotting Kapp-PEP as a function of effector analogue concentration) within the working concentration range of effector analogues. When sufficient data cannot be collected to define the upper plateau, Johnson and Reinhart (8) demonstrated that eq 2 simplifies to:
| (3) |
This simplified equation is equivalent to competitive binding between A and X. Therefore, when the upper plateau could not be obtained, data were fit to eq3 as a means of evaluating effector analogue binding. Data for effector analogues fit in this manner can be used to evaluate which region of the effector contribute to effector binding, but give no insights into the region of the effector that contribute to eliciting the allosteric response. To highlight this distinction, parameters obtained from fits to eq2 and 3 are segregated in Table 1.
Table 1.
Fit Parameters for the Allosteric Response caused by Different Amino Acid Analogues
| Fit Parameters from Equation 2 | Fit Parameters from Equation 3 | |||
|---|---|---|---|---|
| Allosteric Response | ||||
| Commercial Name | Commercial Name | |||
| Kix (mM) | Qax | Kix (mM) | ||
| L-Ala | 0.33±0.01 | 0.100±0.001 | ||
| L-Phe | 3.9±0.3 | 0.39±0.01 | ||
| Carboxyl modifications | ||||
| L-alanine methyl esterb | 1.90±0.05 | 0.120±0.002 | Ethanolamineb | 50±20 |
| Ethylamineb,c | 40±10 | |||
| Isopropylamineb,c | 25±2 | |||
| L-alaninolb,c | 25±1 | |||
| Butylamineb,c | —a | |||
| Amino modifications | ||||
| N-methyl-L-alab | 1.1±0.2 | 0.16±0.01 | Propionic Acidb,c | —a |
| L-Pro | 1.40±0.04 | 0.063±0.001 | N-formyl-L-alab,c | —a |
| N-acetyl-L-alab,c | —a | |||
| Chiral carbon modifications | ||||
| D-alanineb | 10±2 | 0.37±0.02 | D-phenylalanineb | —a |
| 2-aminoisobutyric acidb | 11±2 | 0.16±0.01 |
S(+)-2-amino-2-methyl-3- phenyl-propionic acidb |
—a |
| Side Chain modifications | ||||
| Glyb | 23±2 | 0.26±0.01 | L-homoserine | 10±4 |
| L-Cys | 0.101±0.003 | 0.124±0.002 | L-Ilee | 31±8 |
| L-Ser | 4.6±0.4 | 0.24±0.01 | L-norvaline | 29±3 |
| L-(+)-2,3-diaminopropionic acid | 8±3 | 0.15±0.05 | L-Asn | —a |
| L-(+)-2-aminobutyric acid (2AB) | 0.43±0.02 | 0.05±0.01 | L-Asp | —a |
| L-Val | 7.6±0.4 | 0.090±0.004 | L-Leu | —a |
| L-Thr | 19.6±0.7 | 0.160±0.003 | L-Glu | —a |
| L-Met | 21±2 | 0.35±0.01 | L-norleucine | —a |
| O-methyl-L-tyr | 3.8±0.6 | 0.20±0.04 | 2-aminoheptanoic acid | —a |
| 2-aminocaprylic acid | —a | |||
| L-His | —a | |||
| L-Arg | —a | |||
| L-Lys | —a | |||
| 3-cyclohexyl-L-alanine | —a | |||
| (S)-(+)-2-phenylglycine | —a | |||
| 4-nitro-L-phenylalanine | —a | |||
| L-homophenylalanine | —a | |||
| L-Tyr | —a | |||
| L-Trp | —a | |||
Ka-PEP was not responsive to the amino acid analogue within the working concentration defined in Materials and Methods.
Corrected using H476L as described in supporting information
Binding tested by competition with Ala binding. When no Kix value is listed, no competitive binding was detected.
When an amino acid analogue failed to elicit an allosteric response, the ability of that compound to bind competitively with Ala to hL-PYK was used to test for binding. In this approach Kix-Ala was determined (as described above using fits to eq2) at varying concentrations of analogue (see supporting information). If competitive binding is observed, a Kix value for the analogue could be determined by fitting such data to the competitive equation, i.e. same form as eq3. However we did not identify any analogues that showed competitive binding with Ala that did not also elicit sufficient influence on Kapp-PEP to allow an evaluation of Kix via fitting the allosteric response to eq2 or 3. Our discussion will consider quantitative comparisons of fit parameters (Table 1).
Due to the relatively low affinity of hL-PYK for Ala, our studies used very high ligand concentrations. Two controls can be considered to gain confidence that the results represented herein are not due to non-specific effects. First, we can consider the response of wild type hL-PYK to very high concentrations of Ala (see supporting information). Once Ala concentrations are sufficiently high to saturate the effector binding site (i.e. the upper plateau is obtained at 25 mM), Kapp-PEP is not further responsive to very high (up to 500 mM) concentrations of Ala. Therefore, it does not appear that very high concentrations of Ala alter Kapp-PEP due to non-specific effects. It follows that when mutant proteins bind Ala with affinities lower than the wild type protein, Ala concentrations up to 100 mM can be used to evaluate binding and allosteric properties. Secondly, Ala analogues (and/or counter ions associated with those analogues), as opposed to Ala may promote non-specific binding effects. H476L binds PEP with a similar affinity as the wild type protein (see supporting information). However, the Kapp-PEP of H476L shows no response to Ala up to the 100 mM concentration range used. It follows that any response of this mutant protein to high concentrations of an Ala analogue must be due to non-specific effects. Therefore, allosteric responses of wild type protein to analogues smaller than Ala (i.e. compounds that represent a fractionation of the Ala molecule) were corrected by dividing the Kapp-PEP vs. analogue response for wild type by the Kapp-PEP vs. analogue response for H476L (i.e. subtraction of the respective free energies).
Results and Discussion
Minimum requirement for effector binding
The first question to be addressed with the analogue series is which chemical moieties of the effector are the minimum required for binding to hL-PYK. Since the amino and carboxyl groups of the effector were required for binding to rM1-PYK, modification of these regions of the effector were first considered. Complete removal of the carboxyl group of the effector (ethylamine vs. Ala) or replacement of this moiety with either a methyl (isopropylamine vs. Ala) or a methyl alcohol (alaninol vs. Ala) group reduces binding sufficiently to prevent formation of the upper plateau within the working analogue concentration range. However, L-alaninol and isopropylamine bind to hL-PYK with similar affinities, indicating that the hydroxyl oxygen contributes little to binding affinity. Therefore, we can speculate that the carbonyl oxygen must contribute to effector binding. Like rM1-PYK (1), hL-PYK is also regulated by L-alanine-methyl ester, indicating that the effector (and allosteric response) does not require a charge on the carboxylate group (i.e. a charge on the carboxylate group is not required for binding or allosteric functions). Overall, this data trend follows that observed for rM1-PYK, with the exception that there is less distinction between the binding affinities of hL-PYK for ethylamine vs. isopropylamine and alaninol.
Also similar to the finding in the rM1-PYK system (1), complete removal of the amino group (propionic acid) prevents binding of the effector to hL-PYK. Although the addition of larger chemical moieties at the amino position (N-formyl-L-ala and N-acetyl-L-ala) prevented an allosteric response, the addition of a methyl and a cyclic group lead to a minimal decrease both in binding affinity and allosteric inhibition (N-methyl-L-ala and Pro vs. Ala). Both of the respective analogues contain secondary rather than primary amines, indicating that the primary amine of standard amino acids is not required for binding or allosteric functions. Of these, the regulation by N-methyl-L-ala shows the most contrast with the results obtained for rM1-PYK; this analogue did not elicit an allosteric response in the muscle protein. However, since rM1-PYK is allosterically inhibited by Pro (1, 9), differences between the two pyruvate kinase isozymes appear limited to the ability to accommodate various chemical moieties attached to the amino group, rather than one isozyme selecting against any additions to the effector amino group. However, it appears that effector amino nitrogen is required for binding to hL-PYK, consistent both with results from effector analogue studies of rM1-PYK and the predicted contribution based on how the effector coordinates to the protein (Figure 1).
Since both the amino nitrogen and the carbonyl oxygen (above) are required for binding and removal of additional moieties (Ala vs. Gly, ethanolamine, ethylamine, and butylamine) greatly reduce affinity, the L-2-aminopropanaldehyde substructure appears to be the primary requirement for effector binding to hL-PYK. This same L-2-aminopropanaldehyde substructure was found to be required for effector binding to rM1-PYK.
Determinant of allostery
The second question to be addressed in this study is which region of the effector elicits the allosteric response. With regards to the modification introduced at the amino and carboxyl groups (discussed above), all effector analogues that bind to hL-PYK elicit an allosteric response. In addition, the effects of additions to the chiral carbon and additions of side chain atoms beyond the Cβ of Ala were considered. However due to the lack of a strong data trend, little can be concluded regarding the accommodation of additional chemical moieties at the chiral carbon of the effector. In contrast to the lack of response in rM1-PYK (1), hL-PYK shows inhibition by 2-aminoisobutyric acid and D-alanine. These analogues bind to hL-PYK with different affinities and elicit different magnitudes of allosteric coupling as compared to Ala (i.e. the L-form of alanine). The fact that these analogues bind and influence PEP affinity greatly differs from the lack of binding in the rM1-PYK isozyme. In contrast to the small Ala analogues, only the L-form of Phe (not D-phenylalanine) elicits an allosteric response. Replacement of the α-hydrogen of L-Phe with a methyl group (S(+)-2-amino-2-methyl-3-phenyl-propionic acid vs. L-Phe) also eliminates an allosteric response, which is in contrast to the minimal effect on coupling caused by the methyl group substitution to Ala (2-Aminoisobutyric acid vs. Ala). Therefore, although there is no obvious data trend for analogues with additions at the chiral carbon, the data consistently indicates that all effector analogues that bind to hL-PYK elicit an allosteric response.
Since Pro and Ala are the only two amino acid effectors that have been co-crystallized with any of the pyruvate kinase isozymes (1, 9), a full appreciation for how large effector side-chains interact with the binding site is currently lacking. Nonetheless, effector analogues can be used to describe the functional roles of the effector side-chain. Many types of chemical moieties can be added to the Cβ without preventing the allosteric responses (Ser, Cys, L-(+)-2,3-diaminopropionic acid, 2-aminobutyric acid, Pro, Val, Thr, Met, and homoserine). Like rM1-PYK, the effector site of hL-PYK is capable of accommodating amino acids much larger than Ala (e.g. O-methyl-L-tyr). These larger analogues elicit allostery, but there is a moderate trend indicating that binding affinity is reduced as the side-chain increases in hydrophobic bulk (O-methyl-L-tyr is the exception). Also, not all side-chains allow an allosteric response (Asp and Asn; No attempt was made to distinguish if these two ligands fail to bind or bind but fail to elicit a response). Ala, 2-aminobutyric acid, Cys, Pro, and Val elicit the largest magnitude of allosteric inhibition (i.e. smaller Qax values; Table 1); this maximum effect in hL-PYK is greatly reduced relative to the maximum inhibition in the rM1-PYK system (1). Of the analogues for which a full analysis of allosteric coupling was obtained, Phe elicits the smallest antagonism of PEP affinity (i.e. Qax value closest to 1). Therefore, the nature of the effector side-chain can modify the magnitude of the allosteric response, relative to Ala. However, the chemical moieties required to elicit an allosteric inhibition are included in Ala. Overall, it appears that the L-2-aminopropanaldehyde substructure of the effector that is required for binding is also the primary determinant of the allosteric regulation.
Summary
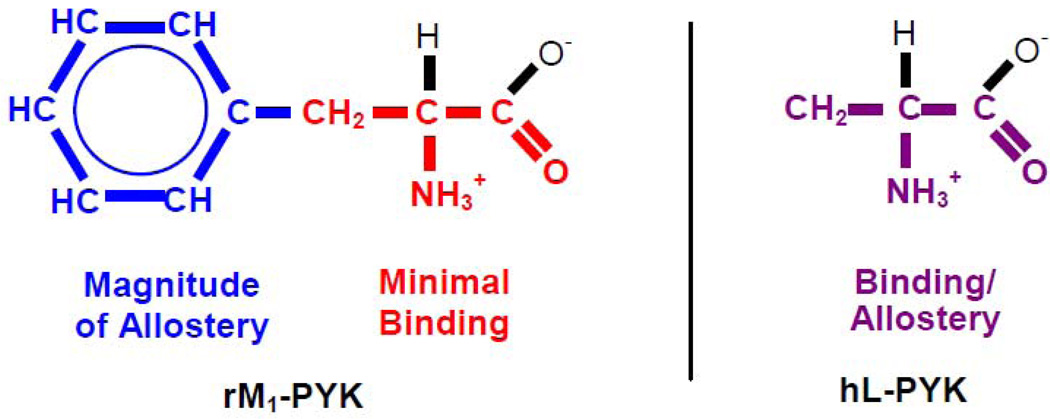
Several conclusions can be drawn from the use of amino acid analogues in this study. 1) Similar to rM1-PYK, the extent of allosteric regulation of hL-PYK is dependent on the effector chemistry (an inconsistency with those two-state models that assume all-or-none allostery). 2) The L-2-aminopropanaldehyde substructure of the amino acid is primarily responsible for effector binding to both rM1-PYK and hL-PYK (Figure 2). 3) Although the length of the hydrophobic side-chain determines the magnitude of the allosteric coupling in rM1-PYK, the primary allosteric determinants in the hL-PYK system are the amino and carboxyl groups of the effector. Consequently, there is a sharp contrast between the two isozyme systems: Although the determinants of effector affinity and the magnitude of the allosteric response were separate moieties of the effector in the rM1-PYK, this separation of function was not apparent for hL-PYK inhibition.
Figure 2.
Atoms of the effector required for binding and that determine the magnitude of the allosteric response. Although these two properties map to different regions of the effector for regulation of rM1-PYK (1), the same separation of function is not apparent in the regulation of hL-PYK.
The contrasting effector moieties that determine the magnitude of the allosteric coupling in hL-PYK vs. rM1-PYK has important implications for the common assumption of conserved allosteric function within protein families. Sequence based evolution/co-evolution of allostery within a family of homologues intrinsically assumes that this regulatory property is conserved within that family. There is growing concern that this assumption is not valid (5, 10–15). The observation that different regions of the amino acid inhibitor elicit the allosteric response in hL-PYK vs. rM1-PYK strengthens this concern.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Liskin Swint-Kruse for critically reading the manuscript and making helpful suggestions.
Abbreviations
- PYK
pyruvate kinase
- rM1-PYK
the pyruvate kinase isozyme found in rabbit brain and muscle
- hL-PYK
the pyruvate kinase isozyme expressed in human liver
- PEP
phosphoenolpyruvate
Footnotes
This work was supported by NIH grant DK78076.
Supporting Information
Supporting information includes a competitive binding example, a high ligand concentration control, and a table including all analogue structures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Williams R, Holyoak T, McDonald G, Gui C, Fenton AW. Differentiating a Ligand's Chemical Requirements for Allosteric Interactions from Those for Protein Binding. Phenylalanine Inhibition of Pyruvate Kinase(,) Biochemistry. 2006;45:5421–5429. doi: 10.1021/bi0524262. [DOI] [PubMed] [Google Scholar]
- 2.Valentini G, Chiarelli LR, Fortin R, Dolzan M, Galizzi A, Abraham DJ, Wang C, Bianchi P, Zanella A, Mattevi A. Structure and function of human erythrocyte pyruvate kinase. Molecular basis of nonspherocytic hemolytic anemia. J Biol Chem. 2002;277:23807–23814. doi: 10.1074/jbc.M202107200. [DOI] [PubMed] [Google Scholar]
- 3.Fenton AW, Hutchinson M. The pH dependence of the allosteric response of human liver pyruvate kinase to fructose-1,6-bisphosphate, ATP, and alanine. Arch Biochem Biophys. 2009;484:16–23. doi: 10.1016/j.abb.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fenton AW, Alontaga AY. Chapter 5: The Impact of Ions on Allosteric Functions in Human Liver Pyruvate Kinase. Methods Enzymol. 2009;466:83–107. doi: 10.1016/S0076-6879(09)66005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenton AW. Allostery: an illustrated definition for the 'second secret of life'. Trends Biochem Sci. 2008;33:420–425. doi: 10.1016/j.tibs.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber G. Ligand binding and internal equilibria in proteins. Biochemistry. 1972;11:864–878. doi: 10.1021/bi00755a028. [DOI] [PubMed] [Google Scholar]
- 7.Reinhart GD. Quantitative analysis and interpretation of allosteric behavior. Methods Enzymol. 2004;380:187–203. doi: 10.1016/S0076-6879(04)80009-0. [DOI] [PubMed] [Google Scholar]
- 8.Johnson JL, Reinhart GD. Failure of a two-state model to describe the influence of phospho(enol)pyruvate on phosphofructokinase from Escherichia coli. Biochemistry. 1997;36:12814–12822. doi: 10.1021/bi970942p. [DOI] [PubMed] [Google Scholar]
- 9.Fenton AW, Johnson TA, Holyoak T. The pyruvate kinase model system, a cautionary tale for the use of osmolyte perturbations to support conformational equilibria in allostery. Protein Sci. 2010;19:1796–1800. doi: 10.1002/pro.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pendergrass DC, Williams R, Blair JB, Fenton AW. Mining for allosteric information: Natural mutations and positional sequence conservation in pyruvate kinase. IUBMB Life. 2006;58:31–38. doi: 10.1080/15216540500531705. [DOI] [PubMed] [Google Scholar]
- 11.Jensen AA, Spalding TA. Allosteric modulation of G-protein coupled receptors. Eur J Pharm Sci. 2004;21:407–420. doi: 10.1016/j.ejps.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 12.May LT, Avlani VA, Sexton PM, Christopoulos A. Allosteric modulation of G protein-coupled receptors. Current pharmaceutical design. 2004;10:2003–2013. doi: 10.2174/1381612043384303. [DOI] [PubMed] [Google Scholar]
- 13.Hudson JW, Golding GB, Crerar MM. Evolution of allosteric control in glycogen phosphorylase. J Mol Biol. 1993;234:700–721. doi: 10.1006/jmbi.1993.1621. [DOI] [PubMed] [Google Scholar]
- 14.Royer WE, Jr, Knapp JE, Strand K, Heaslet HA. Cooperative hemoglobins: conserved fold, diverse quaternary assemblies and allosteric mechanisms. Trends Biochem Sci. 2001;26:297–304. doi: 10.1016/s0968-0004(01)01811-4. [DOI] [PubMed] [Google Scholar]
- 15.Royer WE, Jr, Zhu H, Gorr TA, Flores JF, Knapp JE. Allosteric hemoglobin assembly: diversity and similarity. J Biol Chem. 2005;280:27477–27480. doi: 10.1074/jbc.R500006200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.