Abstract
Orientia tsutsugamushi, an obligate intracellular Gram-negative bacterium, is the causative agent of scrub typhus, a vector-borne disease transmitted by infected chiggers (trombiculid mite larvae). In 2002, an outbreak of scrub typhus occurred among Royal Thai Army troops during the annual field training at a military base in Bothong district, Chonburi province, central Thailand. This report describes the outbreak investigation including its transmission cycle. Results showed that 33.9% of 174 trained troops had scrub typhus-like signs and symptoms and 9.8% of those were positive for O. tsutsugamushi-specific antibodies by indirect fluorescence antibody assay. One hundred thirty-five rodents were captured from this training area, 43% of them had antibodies against O. tsutsugamushi. Six new O. tsutsugamushi isolates were obtained from captured rodent tissues and successfully established in cell culture. Phylogenetic studies showed that these six isolates were either unique or related to a native genotype of previously described isolates from Thailand.
Introduction
Scrub typhus or tsutsugamushi disease is an acute, febrile, and potentially fatal disease, caused by infection with the obligate intracellular Gram-negative rod-shaped bacterium known as Orientia tsutsugamushi.1–3 Orientia tsutsugamushi is a member of the family Rickettsiaseae in the Proteobacteria α-subdivision based on 16s ribosomal RNA (rRNA) gene homology classification.2,4 The bacterium size is ∼0.5 μm wide and 1.2–3.0 μm long.2,5,6 The pathogen is transmitted to humans by the bite of infected larval stage trombiculid mites, Leptotrombidium spp., commonly called chiggers.3,6,7 This mite-borne disease distributes across the Asia-Pacific region and causes substantial morbidity in an area stretching from Pakistan8 in the west and Australia,9,10 Japan,11 Korea,12,13 and Thailand,14,15 in the east. This region commonly known as the tsutsugamushi triangle, hosts ∼1 billion people.16 Mortality rates for scrub typhus range from < 1% to 50% depending upon proper antibiotic treatment, health status of the patient, and virulence of the infected strain of O. tsutsugamushi encountered.17
Antigenic differences among isolates of O. tsutsugamishi were clearly demonstrated in 1962.18 Originally, there were three distinctive antigen prototype strains of O. tsutsugamishi described: Karp, Kato, and Gilliam.16,19 The antigenic variation of these prototype strains, subsequent strains, and isolates discovered depends on the diversity of the immuno-dominant 56-kDa type-specific antigen (TSA) located on the surface of O. tsutsugamushi.20 Recently, new O. tsutsugamushi isolates have been described in Thailand,21,22 Taiwan,23 and Malaysia,24 which implies diverse and antigenic distinct strains of O. tsutsugamushi are pervasive in the tsutsugamushi triangle region. Typing of new isolates can be accomplished by serotyping with immunofluorescence assays using strain- or type-specific monoclonal antibodies or hyperimmune sera, which recognizes 56-kDa TSA by genotyping with restriction fragment length polymorphism analysis or sequence analysis of the 56-kDa TSA gene.20,25 In addition, phylogenetic analysis based on DNA homologies has been used recently to further clarify the evolutionary relationship among individual isolates of O. tsutsugamushi.26,27
After the 2002 military annual field training in Chonburi province, central Thailand, a number of Royal Thai Army troops came down with scrub typhus, suggesting an outbreak. Here, we report the investigation of this outbreak including its transmission cycle. A total of 174 trained troops were interviewed, physically examined, and serially blood tested for evidence of scrub typhus infection. Rodents in training areas were periodically trapped from 2002 to 2008 and evaluated to determine whether they served as reservoir hosts of the scrub typhus agent. Chiggers as ectoparasites of trapped rodents were also collected and identified. As a result, six O. tsutsugamushi isolates were obtained from 10 species of captured rodents and successfully established in cell culture. Genetic characterization of these O. tsutsugamushi isolates showed that some of these were unique and may represent a native or local genotype specifically found in Thailand, because the 56-kDa TSA gene sequences ascertained did not cluster into a previous described genotype.
Materials and Methods
Outbreak investigation.
The outbreak contact population consisted of Royal Thai Army troops deployed to an army fort located in the Chonburi province, in the central region of Thailand about 80 km southeast of Bangkok. The deployed troops were not local residents, most originally came from different regions of Thailand. To investigate the scrub typhus outbreak, a total of 174 individuals were interviewed and physically examined by the Royal Thai Army outbreak response team. In addition, blood samples were serially collected and evaluated for evidence of O. tsutsugamushi infection by indirect fluorescence antibody (IFA) assay.
Rodent study sites.
Rodent study sites included six military training areas, where soldiers with scrub typhus had exercised during the military annual training for the year 2002. These included Ban Angkraphong, Ban Khlongpling, Ban Vangri, Ban Saunpa, Ban Thaprang, and Ban Thapsung villages (Figure 1) in Bothong district, Chonburi province.
Figure 1.
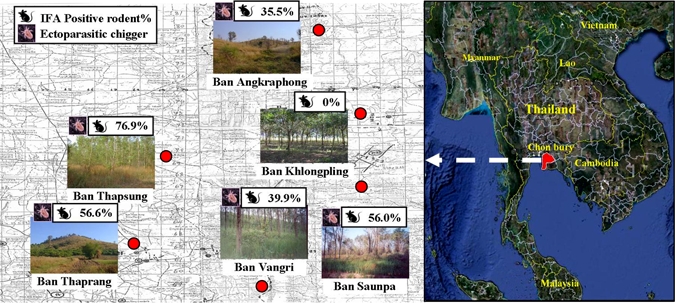
Rodent study sites included six military training areas: Ban Angkrapong, Ban Khlongpling, Ban Vangri, Ban Sounpa, Ban Thaprang, and Ban Thapsung villages in Bothong district, Chonburi province, central Thailand. Each area illustrated includes percent of seropositive captured rodents and the presence of scrub typhus vector, chiggers.
Rodent capture.
Rodents were captured using Sherman traps (3 in. × 3 in. × 10 in.) baited with banana, corn, and cucumber. At each study site, different terrains were chosen according to living habitats of rodents, which included palmaceous, para rubber, shrubby, and abandoned rice field terrains. In each trapped area, numerous trap locations were selected and one or more trap lines were set. Each trap line consisted of ∼20 traps. The traps were placed nearby the base of bushes, rodent burrows, or areas showing evidence of rodent activity and located ∼5–10 meters apart depending on ground surface. The traps were set during the early evening and were collected before 7:30 am of the following morning.
Rodent processing.
Captured rodents were euthanized using a CO2 chamber and prepared as voucher specimens. Information of each rodent such as gender, weight, and body length was recorded for species classification. Ectoparasites such as chiggers were observed and collected for further identification (Figure 2). Blood samples were collected by cardiac puncture, aliquoted into cryotubes containing EDTA, and subsequently stored in liquid nitrogen. Liver and spleen tissues were dissected and collected into cryotubes (Figure 2). All tissue and chigger samples were preserved in liquid nitrogen. The samples were then transported to the Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand (AFRIMS) for detection and isolation of O. tsutsugamushi.
Figure 2.
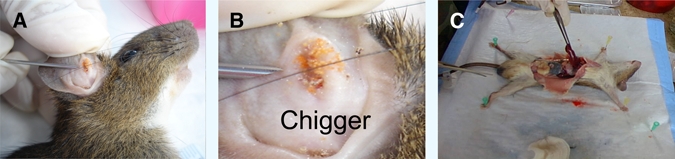
Rodent processing. (A, B) collection of ectoparasites; and (C) dissection of liver and spleen tissue.
Orientia tsutsugamushi-specific antibody (IgM and IgG) detection using IFA assay.
Trained soldiers and captured rodents were tested for evidence of O. tsutsugamushi infection using IFA according to methods described by Bozeman and Elisberg28 and Robinson and others29 with some modification. Briefly, antigens used were pooled from semi-purified O. tsutsugamushi Karp, Kato, and Gilliam strains infected mouse fibroblast cell culture. Fluorescein isothiocyanate (FITC) conjugated to rabbit anti-human immunoglobulins: immunoglobulin M (IgM) and IgG or rabbit anti-rat immunoglobulins: IgG (Sigma Chemicals, St. Louis, MO) were used to detect O. tsutsugamushi-specific antibodies in human and rodent samples, respectively. Serum samples were primarily screened against the pooled antigen at a 1:50 dilution. If a sample showed reactivity, the antibody titer was determined with serial dilutions of 1:100, 1:200, 1:400, 1:800, 1:1600, 1:3200, 1:6400, and 1:12800. Positive and negative control sera for human and rodent samples consisted of O. tsutsugamushi-infected human pooled serum and pooled normal human serum, and in-house produced rat hyperimmune serum and normal rat serum, respectively. The cut-off values for interpreting infection in humans were 1:50 and 1:400 for IgM and IgG, respectively, whereas the cut-off value for IgG in rodent sera was 1:50.
Isolation of O. tsutsugamushi using animal inoculation and mouse fibroblast cell (L-929) culture.
Orientia tsutsugamushi was isolated using animal inoculation as previously described.30 Approximately 0.4 mL of homogenized liver and spleen tissues were injected into the peritoneum of Swiss outbred ICR mice (Charles River Laboratories International, Inc., Wilmington, MA). Inoculated mice were observed daily for up to 14 days for signs and symptoms of illness (i.e., high temperature, rough coat, inactivity, and lack of appetite). Sick animals were euthanized and peritoneal scrapings were examined for the presence of Orientia by Giemsa stain and IFA. Sub-passages were carried out using the triturated liver and spleen tissues of the first inoculum until Orientia were identified.
Mouse fibroblast (L-929) cell culture was used to obtain O. tsutsugamushi isolates. The L-929 cells were previously propagated as described.31 Two days before inoculation, confluent L-929 cells were harvested by trypsinization, resuspended in M199 medium (Grand Island Biological Company [GIBCO], Grand Island, NY), and subsequently exposed to 3,000-rad irradiation in 60Co gamma irradiator (Atomic Energy of Canada, Ltd., Ottawa, Canada). Irradiated L-929 cells were dispensed into three 50-cm2 flasks to develop a monolayer of cells. Homogenized liver and spleen tissues containing O. tsutsugamushi in brain heart infusion (BHI) broth were inoculated into confluent monolayers of irradiated L-929 cells. Inoculated cells were incubated at room temperature for 1 hour on a shaker and then the BHI supernatant was discarded and replaced with fresh culture medium before incubation. Infected L-929 cell cultures were observed for cytopathic effect daily. Control irradiated L-929 cells were used in every batch of O. tsutsugamushi isolation attempts. Ten days after inoculation, the cultures were harvested by gently shaking 5-mm sterile glass beads over the cells. The cell suspensions were pelleted by centrifugation at 8,000 rpm for 30 minutes at 4°C. The pellets were then resuspended in sucrose renografin magnesium chloride (SRM) solution containing 1% renografin 76 (Bracco Diagnosis Inc., Princeton, NJ) and aliquoted for seed storage. To ensure stability of isolates, O. tsutsugamushi-infected cells were repeatedly passaged onto fresh irradiated L-929 cells until heavy infection was observed, typically three passages. Giemsa staining and IFA using O. tsutsugamushi hyperimmune serum and polymerase chain reaction (PCR) amplification of 56-kDa genus-specific gene were performed to identify O. tsutsugamushi in cell culture.
Detection of O. tsutsugamushi infection in cell culture using IFA.
Orientia tsutsugamushi-infected cell cultures were identified by antigen-specific IFA according to microimmunofluorescence methods as previously described.28,29 Briefly, infected L929 cells of each new isolate were spotted in duplicate on 12-well slides and allowed to air dry for 30 min, and then fixed with cold acetone for 10 min. Pooled Karp-, Kato-, and Gilliam-infected L929 cells and uninfected L-929 cells were used as positive and negative controls, respectively. Each spot of duplicated test cells was probed by either the in-house produced rat hyperimmune serum to pooled Karp, Kato, Gilliam strains of O. tsutsugamushi or the normal non-infected rat serum at a positive cut-off dilution of 1:400.
Animal use.
All animal handling and care was conducted according to the Guide for the Care and Use of Laboratory Animals.32 This study was approved by the Animal Research Committee of the Royal Thai Army Component, AFRIMS, Bangkok.
DNA extraction and PCR amplification.
Infected L-929 cells were pelleted by centrifugation at 1,000 rpm for 10 min and genomic DNA was extracted using DNeasy tissue kits (QIAGEN, Hilden, Germany) according to manufacturer's protocol. Each PCR reaction mixture was prepared with the following conditions: 1 × PCR buffer (Invitrogen, Carlsbad, CA), 1.5 mM MgCl2, 0.5 μM each forward and reverse primers, 0.2 mM dNTP, 2.5 U Taq DNA polymerase (Invitrogen) and 1–10 μL template DNA. The PCR profiles used were as follows: an initial denaturation step at 94°C for 3 min was followed by 35 cycles of 94°C for 1 min, 44.5°C (16s rRNA gene) or 53°C (56-kDa TSA gene) for 1 min, and 72°C for 1 min, and then a final incubation step of 72°C for 10 min. The primer sets for the 16s rRNA gene (EC16SF, 5′-GCT TAA CAC ATG CAA G; EC16SR, 5′-CCA TTG TAG CAC GCG T)33 and 56-kDa TSA gene (JG-OtF584, 5′-CAA TGT CTG CGT TGT CGT TGC (this study); RTS9, 5′-ACA GAT GCA CTA TTA GGC AA) yielded ∼1,000 bp and 900 bp amplicons, respectively. The PCR products were visualized using UV light to determine the presence and size of amplicons on ethidium bromide-stained 1% agarose gels after electrophoresis.
DNA sequencing and phylogenetic analysis.
The PCR products of expected amplicon size were purified using the High Pure PCR Template Preparation kit (Roche, Indianapolis, IN). The purified products were directly sequenced using the dye terminator method (BigDye Terminator sequencing kit, Applied Biosystems, Foster City, CA) by ABI PRISM 377 DNA sequencer (Applied Biosystem). Consensus sequences were constructed using sequencher 3.1 software (Gene Codes Corporation, Ann Arbor, MI) followed by removing unexpected gene regions and primer regions. The multi-partial DNA sequences were aligned with reference DNA sequences of each gene retrieved from GenBank (www.ncbi.nlm.nih.gov) using Lasergene DNAstar software (DNASTAR Inc., Madison, WI) according to ClustalW algorithm followed by manual modification based on amino acid translation. Nucleotide identity of each DNA sequence was calculated on the basis of pairwise comparison (Lasergene, DNAstar). Neighbor-joining (NJ), maximum parsimony (MP) and maximum likelihood (ML) methods were chosen to generate phylogenetic relationships using PAUP 4.0b10 software34 and MODELTEST 3.06.35,36 Heuristic searches where gaps were treated as missing data were chosen to generate MP trees. The Kimura two parameter distance model with consideration of the gamma shape parameter was used to generate NJ trees, whereas stepwise addition with consideration of the proportion of invariable sites and gamma-distributed rate heterogeneity across sites was performed for the ML tree. One thousand bootstrap replicates were applied to all three methods. The DNA sequences of 16s rRNA gene of Chlamydia psittaci and Chlamydia trachomatis were chosen to represent an outgroup for phylogenetic analysis. However, because of the lack of a suitable outgroup for the 56-kDa gene, all phylogenetic trees were treated with midpoint-rooting. All sequences were deposited in GenBank under accession nos. (16s rRNA gene: GU068050, GU068051, and GU068052 and for 56-kDa gene: GU068053, GU068054, GU068055, GU068056, GU068057, and GU068058).
Results and Discussion
Two weeks after the 2002 annual field training in Bothong district, Chonburi province, central Thailand, three trained soldiers were admitted to army hospitals because of high fever and severe headache. Diagnosis based on eschar presenting during physical examination and laboratory tests indicated scrub typhus. A number of trainees also reported sick with similar febrile illnesses at this time leading to an alert for a scrub typhus outbreak. The Royal Thai Army outbreak response team was subsequently sent to investigate this outbreak. A total of 174 troops were interviewed, physically examined, and blood tested for evidence of O. tsutsugamushi infection. The outbreak investigation revealed 59 of 174 training soldiers (33.9%) had febrile illnesses with scrub typhus-like signs and symptoms. Using IFA, 17 soldiers (9.8%) were seropositive to O. tsutsugamushi (Table 1). Among this seropositive group, 14 soldiers (82.4%, N = 17) were hospitalized with severe scrub typhus symptoms, whereas three soldiers (17.6%, N = 17) were asymptomatic (Table 1). The 45 seronegative soldiers might have been infected with other pathogens such as influenza virus, which show similar signs and symptoms of scrub typhus. A health education program was implemented to prevent O. tsutsugamushi infection during field exercises. To monitor for scrub typhus, blood tests after semi-annual training in August 2002 were serially followed up. No subsequent evidence of scrub typhus infection in training troops was found (Table 1).
Table 1.
Antibody titer, clinical signs, and symptoms of 17 soldiers with scrub typhus infection*
| Patient no. | Age (year) | IFA antibody positive titer to Orientia tsutsugamushi (IgM/IgG) | Clinical signs and symptoms | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Day 19 (19 Apr 02) | Day 96 (5 July 02) | Day 157 (4 Sep 02) | Day 198 (15 Oct 02) | Eschar | Fever | Chill | Headache | ||
| 1 | 21 | 12800/800 | 50/400 | 50/200 | 50/200 | − | + | + | + |
| 2 | 41 | ND | 50/800 | 50/200 | 50/200 | + | + | + | − |
| 3 | 43 | ND | 200/800 | 50/200 | 50/200 | + | + | + | + |
| 4 | 38 | ND | 100/400 | 50/200 | 50/200 | + | + | + | + |
| 5 | 42 | ND | 50/800 | 50/200 | 50/200 | − | + | + | + |
| 6 | 22 | ND | 50/800 | 50/200 | 50/200 | − | + | + | + |
| 7 | 43 | ND | 50/800 | 50/800 | ND | − | + | + | + |
| 8 | 23 | ND | 50/800 | 50/200 | 50/200 | − | + | + | − |
| 9 | 45 | ND | 200/800 | 50/200 | 50/100 | − | + | + | + |
| 10 | 22 | ND | 50/1600 | 50/800 | 50/400 | − | + | − | + |
| 11 | 21 | ND | 50/800 | ND | ND | − | − | − | − |
| 12 | 21 | ND | 50/800 | 50/800 | ND | − | + | + | + |
| 13 | 22 | ND | 100/800 | 50/800 | 50/200 | − | − | − | − |
| 14 | 23 | ND | 50/800 | ND | ND | − | + | + | + |
| 15 | 43 | ND | 50/400 | ND | ND | + | + | − | + |
| 16 | 22 | ND | 200/800 | ND | ND | − | − | − | − |
| 17 | 23 | ND | 200/800 | ND | ND | + | + | + | + |
IFA = indirect fluorescence antibody; ND = not determined.
To investigate how these trained troops contacted scrub typhus, a survey program for reservoir hosts and vectors of scrub typhus was conducted in these training areas. Rodents were periodically trapped from 2002 to 2008 and evaluated for evidence of O. tsutsugamushi infection. One hundred thirty-five wild rodents were captured from different terrains in the soldier training areas covering six villages (Figure 1). The terrain included palmaceous, para rubber, grassy, and rice fields, and shrubby areas (Figure 1). The captured rodents were identified and subsequently classified into 10 species including: Bandicota indica (N = 89), Bandicota savilei (N = 2), Menestes berdmorei (N = 6), Mus caroli (N = 3), Mus musculus (N = 3), Rattus exulands (N = 9), Rattus losea (N = 1), Rattus norvegicus (N = 2), Rattus rattus (N = 16), and Tupaia glis (N = 4) (Table 2). Information regarding gender and weight of captured rodents are presented in Table 2. The rodent blood samples were evaluated for evidence of O. tsutsugamushi infection using IFA except M. berdmorei and T. glis samples, which were excluded because of unavailability of specific secondary antibody. The IFA tests revealed positive results in 43% (58/125) of captured wild rodents including five of eight species tested indicating a history of O. tsutsugamushi infection occurred in these rodent species (Table 2). Orientia tsutsugamushi-infected rodents were found in five out of six training sites (Figure 1). Different infection rates with O. tsutsugamushi were observed in different rodent species as shown in Table 2. There was no evidence of infection found in captured M. musculus, M. caroli, and R. norvegicus. For the other species tested, infection rates ranged from 11.1% to 100% (Table 2). Rattus losea had 100% infection (N = 1) but sample size of this species was limited, therefore, it may not represent the true infected population. Prevalence of R. rattus (N = 11) was comparatively higher than B. indica (N = 44) even though sample size of B. indica was five times greater. Prevalence of antibodies against O. tsutsugamushi within R. rattus population (68.8%, N = 16) indicated susceptibility of this host to infection with O. tsutsugamushi was higher than other species and may indicate that R. rattus is a better reservoir host. The mean rate of O. tsutsugamushi infection among all IFA tested rodents was 34.9 ± 13.4 (mean ± SE). This broad diversity of infected host species showed a non-species-specific requirement for transmission of O. tsutsugamushi in these military training areas.
Table 2.
Characterization of captured rodents and collected ectoparasitic chiggers from scrub typhus outbreak sites in Chonburi, Thailand: demonstrating by indirect fluorescence antibody (IFA) seropositive rates and successful establishment rate of Orientia tsutsugaamushi isolates from different captured rodent species*
| Rodents species | Number of captured rodents | Mean of weight (g) | Ectoparasitic chigger | IFA positive number N (%) | Isolate number (%) | |||
|---|---|---|---|---|---|---|---|---|
| Male | Female | Total | Number of infested rodent N (%) | Number of collected chiggers N | ||||
| Bandicota indica | 55 | 34 | 89 | 331.2 | 56 (63) | 1,504 | 44 (49.4) | 4 (9) |
| Rattus rattus | 6 | 10 | 16 | 133.7 | 12 (75) | 238 | 11 (68.8) | 1 (9) |
| Rattus exulands | 3 | 6 | 9 | 28.3 | 0 (0) | 0 | 1 (11.1) | 0 (0) |
| Mus musculus | 1 | 2 | 3 | 10.0 | 1 (33) | 20 | 0 (0) | 0 (0) |
| Mus caroli | 2 | 1 | 3 | 17.0 | 1 (33) | 10 | 0 (0) | 0 (0) |
| Bandicota savilei | 1 | 1 | 2 | 175.0 | 2 (100) | 30 | 1 (50) | 0 (0) |
| Rattus norvegicus | 1 | 1 | 2 | 382.5 | 2 (100) | 20 | 0 (0) | 0 (0) |
| Rattus losea | 1 | 0 | 1 | 90.0 | 1 (100) | 24 | 1 (100) | 0 (0) |
| Menetes berdmorei | 4 | 2 | 6 | 209.2 | 2 (33) | 213 | ND | 1 (17) |
| Tupaia glis | 3 | 1 | 4 | 145.0 | 2 (50) | 189 | ND | 0 (0) |
| Total | 77 | 58 | 135 | 259.2 | 79 (59) | 2,248 | 58 (43) | 6 (4) |
ND = not determined.
The presence of specific arthropod vectors for scrub typhus was also examined. A total of 2,248 chiggers were collected as ectoparasites of 79 (59%) rodents captured from five training sites and were subsequently identified (Figure 1 and Table 2). The well-defined chigger species as specific vectors for scrub typhus, Leptotrombidium deliense and Leptotrombidium akamushi were found 33.1% and 10.1% of collected ectoparasitic chiggers, respectively. The remaining collected chiggers were identified as Gahrliepia spp. and Aschoschoengastia spp. Furthermore, collected chiggers were found to infest almost all seropositive rodents (Table 2). Our findings clearly showed the existence of reservoir hosts and vectors for scrub typhus in these training areas. Soldiers training in these areas, therefore, are at risk for scrub typhus. To prevent and protect soldiers from encountering O. tsutsugamushi, the selection of training and camping locations is crucial. In addition, it is strongly recommended that soldiers avoid long periods of training activities such as resting or camping in these risky areas.
In an attempt to isolate O. tsutsugamushi, 135 liver and spleen tissue samples from captured rodents were injected into the peritoneum of outbred-ICR mice. A total of 6 (4%) liver and spleen samples caused mice to present with symptoms of disease: 4 samples from B. indica (CB35, 41, 49, and 62), 1 sample from R. Rattus (CB19), and 1 sample from M. berdmorei (CB52) (Tables 2 and 3). The livers and spleens from the sick mice were harvested, homogenized, and inoculated into L-929 cell cultures for further propagation of O. tsutsugamushi. Six isolates, CB19, CB35, CB41, CB49, CB52, and CB62 were successfully established in L-929 cell culture and stored as seeds at −70°C in SRM solution. Isolation of viable O. tsutsugamushi from captured rodents on which L. deliense chiggers infested (Table 3) confirmed that these training areas are indeed endemic areas of scrub typhus. In addition, we noted that the successful isolation of O. tsutsugamushi was related to IFA results. Of the six successful isolates, five were established from livers and spleens of IFA-positive rodents (9% of infected B. indica [N = 4], 9% of infected R. rattus [N = 1]). The sixth isolate came from an animal (M. berdmorei) in which IFA was not performed because of unavailability of secondary antibody (Table 3). Interestingly, no isolate was obtained from IFA-negative rodents. This implies that the potential successful establishment of O. tsutsugamushi isolates may depend on the presence of anti-O. tsutsugamushi antibodies as an indicator of infection. The presence of antibodies and infection is contrary to a study in Malang, Indonesia where typhus rickettsial DNA was detected only in vectors from seronegative small mammals but not from seropositive animals,37 but is consistent with findings with laboratory murine studies in which isolation of O. tsutsugamushi occurred persistently in antibody positive animals38 and a human case where Rickettsia honei DNA was detected in a patient with high titer of antibodies to spotted fever group rickettsiae.39 Moreover, the titer level of antibody against O. tsutsugamushi within reservoir hosts did not have any effect on the successful isolation of the pathogen (Table 3). Isolates, CB35, 49 and 62 came from IFA-positive animals with a low titer of 50, whereas isolates CB19 and CB41 came from animals with high titers of 200 and 400, respectively. Thus, the level of agent-specific antibody did not affect the establishment of isolate lines. Our study showed that serological information of in-reservoir hosts may be important in determining which particular rodents should be processed further for isolation of O. tsutsugamushi. The failure to isolate O. tsutsugamushi from some rodents could possibly be caused by many factors including but not limited to the individual's genetic background, type of O. tsutsugamushi encountered, or viability of the agent.
Table 3.
Description of Orientia tsutsugamushi isolates obtained from captured wild rodents in military training areas, Bothong district, Chonburi province, central Thailand*
| Orientia tsutsugamushi isolate | Source (rodent species) | Training area | Number of Leptotrombidium deliense infested | IFA titer (IgG) |
|---|---|---|---|---|
| CB19 | Rattus rattus | Ban Vangri | 13 | 200 |
| 13°11′12.8″N | ||||
| 101°32′36.8″W | ||||
| CB35 | Bandicota indica | Ban Thaprang | 17 | 50 |
| 13°13′56.6″N | ||||
| 101°27′23.3″W | ||||
| CB41 | Bandicota indica | Ban Thapsung | 33 | 400 |
| 13°14′08.2″N | ||||
| 101°27′33.2″W | ||||
| CB49 | Bandicota indica | Ban Thapsung | 10 | 50 |
| 13°14′08.2″N | ||||
| 101°27′33.2″W | ||||
| CB52 | Menetes berdmorei | Ban Thaprang | 35 | ND |
| 13°13′56.6″N | ||||
| 101°27′23.3″W | ||||
| CB62 | Bandicota indica | Ban Thaprang | 31 | 50 |
| 13°13′56.6″N | ||||
| 101°27′23.3″W |
ND = not determined
To date, all isolates have been continuously passaged and maintained in L-929 mouse fibroblast cells. The obtained isolates were identified morphologically and serologically by Giemsa staining and IFA with O. tsutsugamushi hyperimmune serum, respectively. Giemsa staining of isolates revealed small rod-shaped organisms both intracellular in the L-929 cells and extracellular (Figure 3A). These isolates also reacted with O. tsutsugamushi hyperimmune serum showing bright apple green stained small rod shaped organisms scatter massively in extracellular spaces and inside L-929 cells as shown in Figure 3B.
Figure 3.
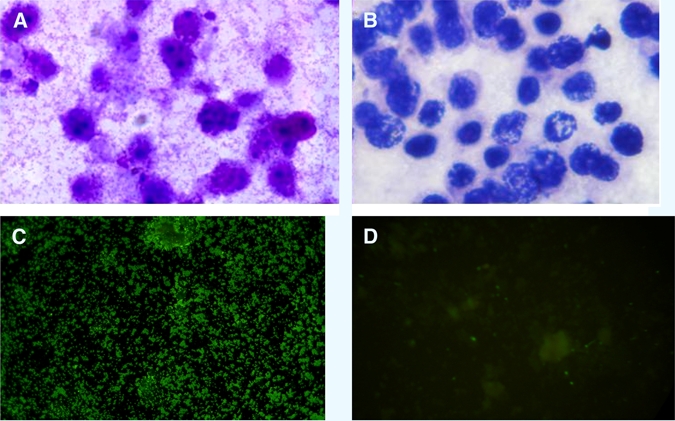
(A, B) Giemsa staining and (C, D) indirect fluorescence antibody assay (IFA) of CB19 isolate in L929 mouse fibroblast cell culture. A and C mouse fibroblast L929 cells infected with CB19 isolate and B and D uninfected mouse fibroblast cells.
The isolates were also genetically characterized. Fragments corresponding to universal 16s rRNA and Orientia genus-specific 56-kDa TSA genes of the six isolates were amplified and sequenced (Figure 4). The two gene sequences of the obtained six isolates were shown to be similar to 16s rRNA and 56-kDa TSA gene sequences of O. tsutsugamushi in GenBank using searching and comparing tools of BLASTN package of GenBank database (http://ncbi.nlm.nih.gov/blastn).40 These obtained six isolates were genetically verified and confirmed as O. tsutsugamushi isolates.
Figure 4.
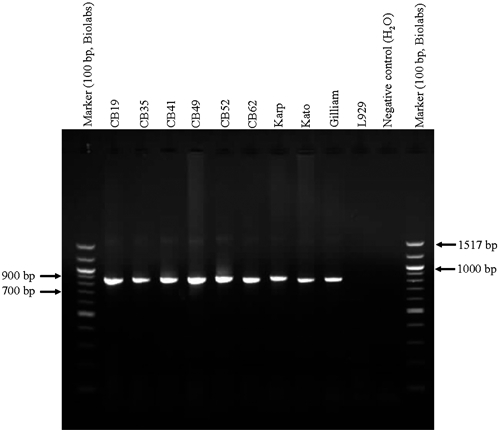
Fragments of Orientia tsutsugamushi 56-kDa type-specific antigen (TSA) gene (863 bp) amplified from L929 mouse fibroblast cells infected with six Thai isolate lines (CB19, CB35, CB41, CB49, CB52, CB62) obtained from wild rodents, Bothong district, Chonburi province, central Thailand. Control panel included L929 cells infected with Orientia tsutsugamushi, prototype strains, Karp, Kato, and Gilliam as positive control, negative controls included uninfected L929 and reagent. Marker used was 100 bp ladder (BioLabs, Ipswich, MA).
To resolve and characterize evolutionary relationships of these isolates, phylogenetic trees using three analyses: NJ, MP, and ML methods were performed for both 16s rRNA and 56-kDa TSA genes. The DNA sequences of three isolates for 16s rRNA and six isolates for 56-kDa TSA genes have been aligned and compared with reference sequences retrieved from GenBank (www.ncbi.nlm.nih.gov). Figure 5 illustrates three phylogenetic trees on the basis of the 16s rRNA gene generated by NJ, MP, and ML methods. Concordance was observed from the three tree topologies generated by these methods. Orientia tsutsugamushi showed monophyletic evolution and branched from the Rickettsia genus with a bootstrap value equal to 100%. Many previous studies have also shown that O. tsutsugamushi is closely related to but separate from Rickettsia spp.2
Figure 5.

Phylogenetic trees of Orientia tsutsugamushi isolates based on partial 16s rRNA gene sequences. The trees were rooted. Bootstrap values > 50% were labeled over branches (1,000 replicates). Thai O. tsutsugamushi isolates were shown in bold (accession nos.: CB49; GU068050, CB35; GU068051, and CB62; GU068052). (A) Neighbor-joining tree. The tree was generated by the Kimura two-parameter method with gamma correction (gamma shape parameter = 0.5371). (B) Maximum parsimony tree. The tree was generated using heuristic search with random stepwise addition (10 replicates). (C) Maximum-likelihood tree. The tree was generated by the stepwise addition method using general time-reversible method with consideration of gamma-distributed rate heterogeneity across sites (−lnL = 9952.68, gamma shape parameter = 0.5371).
The phylogenetic trees generated using the 16s rRNA gene did not show good resolution of O. tsutsugamushi phylogeny for characterizing evolutionary relationships between different O. tsutsugamushi strains or types caused by the low mutation rate of the 16s rRNA gene, therefore, the DNA variation within the gene could not resolve and characterize O. tsutsugamushi isolates variation.41 However, the hyper variable region of the 56-kDa TSA gene of O. tsutsugamushi evolves faster and carries a higher mutation rate and therefore was chosen and used for resolving and characterizing evolutionary relationships of each isolate. The alignment of the 56-kDa TSA gene from the isolates and reference strains showed that the isolates had comparatively distinct patterns in the variable regions implicating that the isolates would possibly represent an unrecognized genotype of O. tsutsugamushi. Figure 6 illustrates phylogenetic trees of the 56-kDa TSA gene generated by NJ, MP, and ML methods. There was no difference between node divergences of all tree topologies produced by the three methods except that an occurrence of node exchange was found within NJ tree, which Kato clade exchanged with TA clade (Figure 6A), whereas there was no exchange found in MP and ML trees (Figure 6B and C). Phylogenetic trees generated by the three methods showed similar tree topologies indicating a concordance of three analysis methods. The new Thai isolates could be classified into two genotypes and formed a polyphyletic evolution, which implies non-sharing ancestors. One genotype of the Thai isolates was found to cluster close to LA-1, which was first demonstrated in 1999 from an isolate of Leptotrombidium arenicola mite, a vector of scrub typhus.26 Although these isolates were closely related to LA-1 they were distinctly cluster apart from LA-1 (bootstrap value = 100; NJ, MP and ML) indicating a different set of isolates sharing the same genotype, therefore, we have grouped our isolates as LA like type. Another genotype of the new Thai isolates was observed and found to cluster far apart from the LA type. These Thai isolates were diverse from TA763 and shared the same ancestor with the TA genogroup of Thai O. tsutsugamushi.26 The isolates formed a distinctive assemblage within the TA group (Figure 6, bootstrap value = 100; NJ, MP and ML). Thus, this genotype probably represents a new genotype of O. tsutsugamushi within the TA group. The same concordance of non-reservoir species-specific association was found within this type as well as the LA type; therefore, O. tsutsugamushi does not appear to need specific reservoir species in their transmission cycle.
Figure 6.
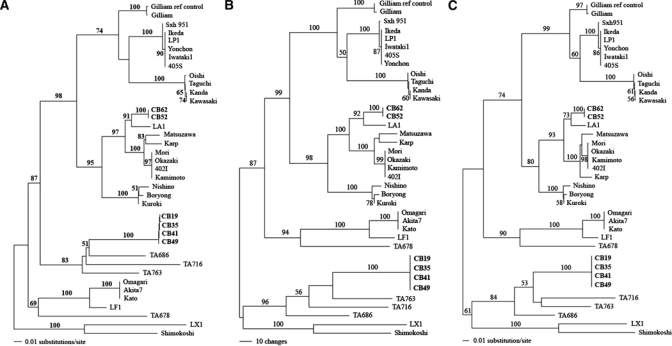
Phylogenetic trees of Orientia tsutsugamushi isolates based on partial 56-kDa type-specific antigen (TSA) gene. The trees were rooted. Bootstrap values > 50% were labeled over branches (1,000 replicates). Thai O. tsutsugamushi isolates were shown in bold (accession nos.: CB35; GU068053, CB52; GU068054, CB62; GU068055, CB49; GU068056, CB41; GU068057 and CB19; GU068058). (A) Neighbor-joining tree. The tree was generated by Kimura two-parameter method with gamma correction (gamma shape parameter = 1.0282). (B) Maximum parsimony tree. The tree was generated using heuristic search with random stepwise addition (10 replicates). (C) Maximum-likelihood tree. The tree was generated by the stepwise addition method using general time-reversible method with consideration of gamma-distributed rate heterogeneity across sites (−lnL = 6330.80, gamma shape parameter = 1.0282).
In this study, we have found that the new Thai isolates derived from rodent reservoir hosts were able to be classified into two distinct genotypes distantly related. One is a previously recognized genotype, LA, described by Enatsu and others,26 (bootstrap value = 100; NJ, MP and ML), whereas the other is an unrecognized genotype, which has not been described and clustered in a distinct clade. This unrecognized genotype shares the same ancestor as TA genotypes; TA763, TA716, and TA686, and formed a paraphyletic relationship indicating broad diversity of O. tsutsugamushi type in the Thai major group (TA group). This study has demonstrated the successful establishment of Thai isolate lines of two O. tsutsugamushi types, which will be useful in the development of type-specific scrub typhus vaccines16 and diagnosis kits in the future.
ACKNOWLEDGMENTS
We thank Atthaya Ruangpung for technical assistance. We also thank Fort Navamintracheenee Hospital for collecting data during the scrub typhus outbreak among Royal Thai Army troops during their annual field training in 2002.
Disclaimer: The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the U.S. Department of the Navy, or the Royal Thai Army. As an employee of the U.S. Government (ALR) this work was prepared as part of my official duties. Title 17 U.S.C. 105 provides that “Copyright protection under this title is not available for any work of the United States Government.” Title 17 U.S.C. 101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person's official duties.
Footnotes
Financial support: This work was supported by Thanphuying Viraya Chavakul Foundation for Medical Armed Forces Research Grant (2008), the Department of Biology, Faculty of Science and the Graduate school, Chiang Mai University Thailand, the Armed Forces Research Institute of Medical Science (AFRIMS) Royal Thai Army, Bangkok Thailand, and GEIS/AFHSC work unit number 847705.82000.25GB.A0074 for the financial support of this project.
Authors' addresses: Wuttikon Rodkvamtook, Toon Ruang-areerate, and Jariyanart Gaywee, Department of Epidemiology, Research Division Armed Forces Research Institute of Medical Science (AFRIMS), Royal Thai Army, Ratchathawee, Bangkok, Thailand, E-mails: wuttikornr@afrims.go.th, youangtr@yahoo.com, and jariyanartg@afrims.org. Allen L. Richards, Naval Medical Research Center, Viral and Rickettsial Diseases Department, Silver Spring, MD, E-mail: Allen.Richards@med.navy.mil. Pimmada Jeamwattanalert, Department of Enteric Diseases, Armed Forces Research Institute of Medical Sciences (AFRIMS), Ratchathawee, Bangkok, Thailand, E-mail: pimmadaj@afrims.org. Dharadhida Bodhidatta, Research Division, Armed Forces Research Institute of Medical Sciences (AFRIMS), Royal Thai Army, Ratchathawee, Bangkok, Thailand, E-mail: dbodhi@hotmail.com. Noppadon Sangjun, Department of Laboratory Animal, Analysis Division, Armed Forces Research Institute of Medical Sciences (AFRIMS), Royal Thai Army, Ratchathawee, Bangkok, Thailand, E-mail: noppadon625@yahoo.com. Anchana Prasartvit, Bureau of General Communicable Diseases, Department of Disease Control, Ministry of Public Health, Nonthaburi, Thailand, E-mail: A_Pasartvit@yahoo.com. Araya Jatisatienr and Chaiwat Jatisatienr, Department of Biology, Faculty of Science, Chiang Mai University, Muang District, Chiang Mai, Thailand, E-mails: Arayarj@yahoo.com and Jchaiwat@chiangmai.ac.th.
References
- 1.Philip CB. Tsutsugamushi disease in World War II. J Parasitol. 1948;34:169–191. [PubMed] [Google Scholar]
- 2.Ohashi N, Fukuhara M, Shimada M, Tamura A. Phylogenetic position of Rickettsia tsutsugamushi and the relationship among its antigenic variants by analyses of 16S rRNA gene sequences. FEMS Microbiol Lett. 1995;125:299–304. doi: 10.1111/j.1574-6968.1995.tb07372.x. [DOI] [PubMed] [Google Scholar]
- 3.Watt G, Parola P. Scrub typhus and tropical rickettsioses. Curr Opin Infect Dis. 2003;16:429–436. doi: 10.1097/00001432-200310000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Tamura A, Ohashi N, Urakami H, Miyamura S. Classification of Rickettsia tsutsugamushi in a new genus, Orientia gen. nov., as Orientia tsutsugamushi comb. nov. Int J Syst Bacteriol. 1995;45:589–591. doi: 10.1099/00207713-45-3-589. [DOI] [PubMed] [Google Scholar]
- 5.Tamura A, Urakami H, Ohashi N. A comparative view of Rickettsia tsutsugamushi and the other groups of rickettsiae. Eur J Epidemiol. 1991;7:259–269. doi: 10.1007/BF00145675. [DOI] [PubMed] [Google Scholar]
- 6.Kawamura AJ, Tanaka H, Tamura A, Kawamura AJ, Tanaka H, Tamura A. Tsutsugamushi Disease. Tokyo, Japan: University of Tokyo Press; 1995. [Google Scholar]
- 7.Seong SY, Choi MS, Kim IS. Orientia tsutsugamushi infection: overview and immune responses. Microbes Infect. 2001;3:11–21. doi: 10.1016/s1286-4579(00)01352-6. [DOI] [PubMed] [Google Scholar]
- 8.Shirai A, Wisseman CL., Jr Serologic classification of scrub typhus isolates from Pakistan. Am J Trop Med Hyg. 1975;24:145–153. doi: 10.4269/ajtmh.1975.24.145. [DOI] [PubMed] [Google Scholar]
- 9.Currie B. Medicine in tropical Australia. Med J Aust. 1993;158:612–615. doi: 10.5694/j.1326-5377.1993.tb137630.x. 609. [DOI] [PubMed] [Google Scholar]
- 10.Graves S, Wang L, Nack Z, Jones S. Rickettsia serosurvey in Kimberley, western Australia. Am J Trop Med Hyg. 1999;60:786–789. doi: 10.4269/ajtmh.1999.60.786. [DOI] [PubMed] [Google Scholar]
- 11.Ogawa M, Hagiwara T, Kishimoto T, Shiga S, Yoshida Y, Furuya Y, Kaiho I, Ito T, Nemoto H, Yamamoto N, Masukawa K. Scrub typhus in Japan: epidemiology and clinical features of cases reported in 1998. Am J Trop Med Hyg. 2002;67:162–165. doi: 10.4269/ajtmh.2002.67.162. [DOI] [PubMed] [Google Scholar]
- 12.Chong Y. Application of serologic diagnosis of tsutsugamushi disease (scrub typhus) in Korea where the disease was recently recognized to be endemic. Yonsei Med J. 1989;30:111–117. doi: 10.3349/ymj.1989.30.2.111. [DOI] [PubMed] [Google Scholar]
- 13.Yi KS, Chong Y, Covington SC, Donahue BJ, Rothen RL, Rodriguez J, Arthur JD. Scrub typhus in Korea: importance of early clinical diagnosis in this newly recognized endemic area. Mil Med. 1993;158:269–273. [PubMed] [Google Scholar]
- 14.Kollars TM, Jr, Bodhidatta D, Phulsuksombati D, Tippayachai B, Coleman RE. Short report: variation in the 56-kD type-specific antigen gene of Orientia tsutsugamushi isolated from patients in Thailand. Am J Trop Med Hyg. 2003;68:299–300. [PubMed] [Google Scholar]
- 15.Singhsilarak T, Leowattana W, Looareesuwan S, Wongchotigul V, Jiang J, Richards AL, Watt G. Short report: detection of Orientia tsutsugamushi in clinical samples by quantitative real-time polymerase chain reaction. Am J Trop Med Hyg. 2005;72:640–641. [PubMed] [Google Scholar]
- 16.Chattopadhyay S, Richards AL. Scrub typhus vaccines: past history and recent developments. Hum Vaccin. 2007;3:73–80. doi: 10.4161/hv.3.3.4009. [DOI] [PubMed] [Google Scholar]
- 17.Kelly DJ, Richards AL, Temenak J, Strickman D, Dasch GA. The past and present threat of rickettsial diseases to military medicine and international public health. Clin Infect Dis. 2002;34:S145–S169. doi: 10.1086/339908. [DOI] [PubMed] [Google Scholar]
- 18.Shishido A. Identification and serolgical classification of the causative agent of scrub typhus in Japan. Jpn J Sci Biol. 1962;15:308–321. [Google Scholar]
- 19.Eisemann CS, Osterman JV. Antigens of scrub typhus rickettsiae: separation by polyacrylamide gel electrophoresis and identification by enzyme-linked immunosorbent assay. Infect Immun. 1981;32:525–533. doi: 10.1128/iai.32.2.525-533.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura A, Ohashi N, Koyama Y, Fukuhara M, Kawamori F, Otsuru M, Wu PF, Lin SY. Characterization of Orientia tsutsugamushi isolated in Taiwan by immunofluorescence and restriction fragment length polymorphism analyses. FEMS Microbiol Lett. 1997;150:225–231. doi: 10.1016/s0378-1097(97)00119-5. [DOI] [PubMed] [Google Scholar]
- 21.Elisberg BL, Campbell JM, Bozeman FM. Antigenic diversity of Rickettsia tsutsugamushi: epidemiologic and ecologic significance. J Hyg Epidemiol Microbiol Immunol. 1968;12:18–25. [PubMed] [Google Scholar]
- 22.Tamura A, Takahashi K, Tsuruhara T, Urakami H, Miyamura S, Sekikawa H, Kenmotsu M, Shibata M, Abe S, Nezu H. Isolation of Rickettsia tsutsugamushi antigenically different from Kato, Karp, and Gilliam strains from patients. Microbiol Immunol. 1984;28:873–882. doi: 10.1111/j.1348-0421.1984.tb00743.x. [DOI] [PubMed] [Google Scholar]
- 23.Qiang Y, Tamura A, Urakami H, Makisaka Y, Koyama S, Fukuhara M, Kadosaka T. Phylogenetic characterization of Orientia tsutsugamushi isolated in Taiwan according to the sequence homologies of 56-kDa type-specific antigen genes. Microbiol Immunol. 2003;47:577–583. doi: 10.1111/j.1348-0421.2003.tb03420.x. [DOI] [PubMed] [Google Scholar]
- 24.Tay ST, Rohani YM, Ho TM, Shamala D. Sequence analysis of the hypervariable regions of the 56-kDa immunodominant protein genes of Orientia tsutsugamushi strains in Malaysia. Microbiol Immunol. 2005;49:67–71. doi: 10.1111/j.1348-0421.2005.tb03641.x. [DOI] [PubMed] [Google Scholar]
- 25.Ohashi N, Koyama Y, Urakami H, Fukuhara M, Tamura A, Kawamori F, Yamamoto S, Kasuya S, Yoshimura K. Demonstration of antigenic and genotypic variation in Orientia tsutsugamushi which were isolated in Japan, and their classification into type and subtype. Microbiol Immunol. 1996;40:627–638. doi: 10.1111/j.1348-0421.1996.tb01120.x. [DOI] [PubMed] [Google Scholar]
- 26.Enatsu T, Urakami H, Tamura A. Phylogenetic analysis of Orientia tsutsugamushi strains based on the sequence homologies of 56-kDa type-specific antigen genes. FEMS Microbiol Lett. 1999;180:163–169. doi: 10.1111/j.1574-6968.1999.tb08791.x. [DOI] [PubMed] [Google Scholar]
- 27.Kelly DJ, Fuerst PA, Ching WM, Richards AL. Scrub typhus: the geographical distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin Infect Dis. 2009;48:S203–S230. doi: 10.1086/596576. [DOI] [PubMed] [Google Scholar]
- 28.Bozeman FM, Elisberg BL. Serological diagnosis of scrub typhus by indirect immunofluorescence. Proc Soc Exp Biol Med. 1963;112:568–573. doi: 10.3181/00379727-112-28107. [DOI] [PubMed] [Google Scholar]
- 29.Robinson DM, Brown G, Gan E, Huxsoll DL. Adaptation of a microimmunofluorescence test to the study of human Rickettsia tsutsugamushi antibody. Am J Trop Med Hyg. 1976;25:900–905. doi: 10.4269/ajtmh.1976.25.900. [DOI] [PubMed] [Google Scholar]
- 30.Russell E, Monkanna T, Kenneth KJ, Strickman DJ, Frances SP, Tanskul P, Kollars TM, Inkaminlao JR, Watcharapichat P, Khlaimanee N, Phulsuksombati D, Sangjun N, Lerdthusnee K. Occurrence of Orientia tsutsugamushi in small mammals from Thailand. Am J Trop Med Hyg. 2003;69:519–524. [PubMed] [Google Scholar]
- 31.Lerdthusnee K, Khuntirat B, Leepitakrat W, Tanskul P, Monkanna T, Khlaimanee N, Inlao I, Kengluecha A, Mungviriya S, Chandranoi K, Krairojananan P, Bodhidatta D, Rodkwamthook W, Phulsuksombati D, Sangjun N, Watcharapichat P, Jones JW, Coleman RE. Scrub typhus vector competence of Leptotrombidium chiangraiensis chiggers and transmission efficacy and isolation of Orientia tsutsugamushi. Ann N Y Acad Sci. 2003;990:25–35. doi: 10.1111/j.1749-6632.2003.tb07333.x. [DOI] [PubMed] [Google Scholar]
- 32.Clark DC, Bladwin RL, Bayne KA, Brown MJ, Gebhart GF, Gonder JC, Gwathmey JK, Keeling ME, Kohn DF, Robb JW, Smith OA, Steggerda JA, Vandenbergh JG, White WJ, Williams-Blangero S, VandeBerg JL. Guide for the Care and Use of Laboratory Animals. Washington, DC: National Academy Press; 1996. [Google Scholar]
- 33.O'Neill SL, Giordano R, Colbert AM, Karr TL, Robertson HM. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc Natl Acad Sci USA. 1992;89:2699–2702. doi: 10.1073/pnas.89.7.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swofford DL. PAUP*. Phylogenetic analysis using pasimony (* and other methods) Sunderland, MA: Sinaneur Associates; 2002. Version 4.0 beta 10. [Google Scholar]
- 35.Posada D. Using MODELTEST and PAUP* to select a modezl of nucleotide substitution. Curr Protoc Bioinformatics. 2003 doi: 10.1002/0471250953.bi0605s00. Chapter 6: Unit 6.5. [DOI] [PubMed] [Google Scholar]
- 36.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 37.Richards AL, Soeatmandji DW, Widodo MA, Sardjono TW, Yanuwiadi B, Hernowati TE, Baskoro AD, Roebiyoso, Hakim L, Soendoro M, Rahardjo E, Putri MP, Saragih JM, Strickman D, Kelly DJ, Dasch GA, Olson JG, Church CJ, Corwin AL. Seroepidemiological evidence for murine and scrub typhus in Malang, Indonesia. Am J Trop Med Hyg. 1997;57:91–95. doi: 10.4269/ajtmh.1997.57.91. [DOI] [PubMed] [Google Scholar]
- 38.Frances SP, Watchrapichat P, Phunlsuksombati D. Development and persistence of antobodies to Orientia tsutsugamushi in the roof rat, Rattus rattus and laboratory mice following attachment of naturally infected Leptotrombidium deliense. Acta Trop. 2000;77:279–285. doi: 10.1016/s0001-706x(00)00144-3. [DOI] [PubMed] [Google Scholar]
- 39.Jiang J, Sangkasuwan V, Lerdthusnee K, Sukwit S, Chuenchitra T, Rozmajzl PJ, Eamsila C, Jones JW, Richards AL. Molecular evidence of human infection with TT-118, Rickettsia honei, from Thailand. Emerg Infect Dis. 2005;11:1475–1477. doi: 10.3201/eid1109.050011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamura A, Ohashi N, Urakami H, Takahashi K, Oyanagi M. Analysis of polypeptide composition and antigenic components of Rickettsia tsutsugamushi by polyacrylamide gel electrophoresis and immunoblotting. Infect Immun. 1985;48:671–675. doi: 10.1128/iai.48.3.671-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]


