Abstract
There is a lack of diagnostic tests for leptospirosis in technology-restricted settings. We developed loop-mediated isothermal amplification (LAMP) specific for the 16S ribosomal RNA gene (rrs) of pathogenic and intermediate group Leptospira species. The lower limit of detection was 10 genomic equivalents/reaction, and analytical specificity was high; we observed positive reactions for pathogenic/intermediate groups and negative reactions for non-pathogenic Leptospira species and other bacterial species. We evaluated this assay in Thailand by using a case–control study of 133 patients with laboratory-proven leptospirosis and 133 patients with other febrile illnesses. Using admission blood, we found that the rrs LAMP showed positive results in 58 of 133 cases (diagnostic sensitivity = 43.6, 95% confidence interval [CI] = 35.0–52.5) and in 22 of 133 controls (diagnostic specificity = 83.5, 95% CI = 76.0–89.3). Sensitivity was high for 39 patients who were culture positive for Leptospira spp. (84.6, 95% CI = 69.5–94.1). The rrs LAMP can provide an admission diagnosis in approximately half of patients with leptospirosis, but its clinical utility is reduced by a lower specificity.
Introduction
Leptospirosis is an acute febrile illness caused by pathogenic species belonging to the genus Leptospira. This zoonotic disease has a worldwide distribution, but has the greatest impact on health in developing countries where it is often grossly under-recognized. Clinical features are similar to a range of other infectious diseases that occur in the same geographic regions, including scrub typhus, dengue, and malaria.1 Accurate laboratory diagnosis of leptospirosis is important for correct patient management, but there is a paucity of diagnostic tests that provide timely information around the time of admission. Culture can be performed during the first week of symptoms but is rarely undertaken because it requires considerable expertise, is time-consuming, and may not provide a positive result for weeks or months.
The microscopic agglutination test (MAT) is commonly used as a serologic diagnostic gold standard. This test relies on detecting an increase in antibody titer between serum samples obtained at least two weeks apart and provides a retrospective diagnosis that is useful for epidemiologic purposes but does not benefit individual patient management. Conventional and real-time polymerase chain reactions have been described for detection of pathogenic Leptospira in blood at the time of patient presentation,2–5 and are useful for investigation of patients presenting within the first week of illness (during the leptospiremic phase of infection) in settings that have the necessary equipment and trained laboratory personnel. However, these assays are not in routine use in many countries where leptospirosis is highly endemic because the equipment required is costly, and performing the assay to the required standard demands considerable expertise and training.
Loop-mediated isothermal amplification (LAMP) is an alternative method of rapid DNA amplification. The final products are stem-loop DNAs with several inverted repeats of the target and cauliflower-like structures with multiple loops formed by annealing between alternately inverted repeats of the target in the same strand.6 The reaction results in the accumulation of 109 copies of target and simply requires a laboratory water bath or heating block to maintain a constant temperature of 60–65°C, making it particularly suited to lower-technology settings. The LAMP has been developed for detection of a range of pathogenic bacteria and viruses,7–9 including Leptospira species.10 The published Leptospira assay targets lipL41, a gene encoding the outer membrane protein LipL41, which is expressed by pathogenic Leptospira.11 The lower limit of detection was reported to be approximately 100 genomic equivalents (GEs) per reaction, and the assay was specific for Leptospira in a laboratory evaluation. The assay detected Leptospira in seven mouse kidney samples obtained from animals captured during environmental surveillance,10 but has not undergone clinical evaluation to date.
Leptospira genomes contain two 16S ribosomal RNA genes for which there are multiple sequences of variable length in public databases, phylogenetic analysis of which has identified three clades within the genus composed of the pathogenic species, non-pathogenic species, and a clade with intermediate 16S ribosomal RNA gene sequence relatedness.12 The latter group is considered to represent opportunistic/intermediate pathogens, although there are an increasing number of reports associating them with human leptospirosis.13–15 The objective of this study was to use the phylogenetic distinctions inherent in the 16S ribosomal RNA gene sequence to develop an alternative LAMP assay (rrs LAMP) that can detect Leptospira spp. in the pathogenic and intermediate groups, and to undertake a clinical case–control evaluation of the diagnostic accuracy of rrs and lipL41 LAMP in rural Thailand.
Materials and Methods
Laboratory strains.
The rrs LAMP was initially developed by using genomic DNA extracted from L. interrogans serovar Autumnalis strain L0551 isolated in 2001 from a patient in northeastern Thailand, and then further evaluated by using an additional 23 Leptospira isolates shown in Table 1. The specificity of the assay for Leptospira spp. was evaluated by using one clinical isolate of each of the following bacterial species: Staphylococcus aureus, Enterococcus sp., Escherichia coli, Salmonella enterica serovar Typhi, Klebsiella pneumoniae, Pseudomonas aeruginosa, Burkholderia pseudomallei, Orientia tsutsugamushi strain Kato, and Rickettsia typhi. Genomic DNA was extracted from laboratory cultures by using the Wizard® Genomic DNA Extraction Kit (Promega, Madison, WI) with the addition of 5 μL of lysostaphin (10 mg/mL) during the extraction of S. aureus DNA.
Table 1.
Leptospira spp. used in the study, Thailand*
| Species | Serogroup | Serovar | Strain |
|---|---|---|---|
| L. interrogans | Autumnalis | Autumnalis | L0551 |
| L. interrogans | Autumnalis | Autumnalis | Akiyami A |
| L. interrogans | Bataviae | Bataviae | Swart |
| L. interrogans | Djasiman | Djasiman | Djasiman |
| L. interrogans | Hebdomadis | Hebdomadis | Hebdomadis |
| L. interrogans | Icterohaemorrhagiae | Lai | Lai |
| L. kirshneri | Grippotyphosa | Grippotyphosa | Moskva |
| L. kirschneri | Cynopteri | Cynopteri | 3522C |
| L. borgpetersenii | Ballum | Ballum | Mus 127 |
| L. borgpetersenii | Javanica | Javanica | Veldrat Batavia 46 |
| L. santarosai | Autumnalis | Alice | Alice |
| L. alexanderi | Manhao | Manhao3 | L60 |
| L. noguchii | Autumnalis | Fortbragg | Fort Bragg |
| L. weilii | Celledoni | Celledoni | Celledoni |
| L. weilli | Sarmin | Sarmin | Sarmin |
| L. alstonii | Ranarum | Pingchang | 80-412 |
| L. inadai | Lyme | Lyme | 10 |
| L. biflexa | Semaranga | Patoc I | Patoc I |
| L. wolffii | Khorat | Khorat | H2 |
| L. meyeri | Ranarum | Ranarum | Iowa City Frog |
| L. meyeri | Semaranga | Semaranga | Veldrat Semaranga 173 |
| L. wolbachii | Codice | Codice | CDC |
| L. terpstrae | Icterohaemorrhagiae | Hualin | LT11-33 |
| L. yanagawae | Semaranga | Saopaulo | Sao Paulo |
Isolates were obtained from the World Health Organization/Food and Agriculture Organization of the United Nations/World Organisation for Animal Health Collaborating Centre for Reference and Research on Leptospirosis, Brisbane, Queensland Australia; the Bureau of Emerging Infection Disease, Ministry of Public Health, Bangkok, Thailand; the American Type Culture Collection, Manassas, Virigina; the Royal Tropical Institute (KIT), Amsterdam, The Netherlands; and Dr. Thareerat Kalambaheti (Mahidol University, Bangkok, Thailand).
Primer design.
A total of 39 sequences for the full-length 16S ribosomal RNA gene of Leptospira spp. were downloaded from GenBank, of which 23 sequences were from 8 pathogenic Leptospira spp., 6 sequences were from 3 intermediate group Leptospira spp., and 10 sequences were from 6 non-pathogenic Leptospira spp. Accession numbers are pathogenic species Leptospira interrogans (n = 10, accession nos. AY631894, AY996790, AY996791, AY996792, AY996893, AY996794, AY996796, AY996797, AY9967698, AY99680), L. borgpetersenii (n = 2, accession nos. AY631884, AY887899), L. kirschneri (n = 3, accession nos. AY631895, AY996801, AY996802), L. alexanderi (n = 3, accession nos. AY631880, AY996803, AY996804), L. santarosai (n = 2, accession nos. AY631883, AY996805), L. noguchii (n = 1, accession no. AY631886), L. alstonii (n = 1, accession no. AY631881), L. weilii (n = 1, accession no. AY631877); intermediate species L. inadai (n = 3, accession nos. AY631887, AY631891, AY631896), L. broomii (n = 1, accession no. AY796065), L. fainei (n = 2, accession nos. AY631885, AY996789); and non-pathogenic species L. biflexa (n = 2, accession nos. AY631876, AY631893), L. meyeri (n = 3, accession nos. AY631878, AY631889, AY631892), L. wolbachii (n = 2, accession nos. AY631879, AY631890), L. vanthielii (n = 1, accession no. AY631897), L. terpstrae (n = 1, accession no. AY631888), and L. yanagawae (n = 1, accession no. AY631882). The partial 16S ribosomal RNA sequence of three recently described Leptospira spp. were also included: pathogenic L. kmetyi (n = 1, accession no. AB279549) and intermediate group L. licerasiae (n = 9, accession nos. EF612280-8) and L. wolffii (n = 1, accession no. EF025496).
All sequences were aligned and compared by using Clustal X version 1.8316 and Genedoc (http://www.nrbsc.org/gfx/genedoc/index.html).17 A set of five primers consisting of two outer primers (F3 and B3), two inner primers (forward inner primer FIP and backward inner primer BIP), and one loop primer (loop B (LBP)) were designed by using PrimerExplorer version 4 (http://primerexplorer.jp/e/). The target sequences were regions that were specific to the 16S ribosomal RNA gene sequence of all pathogenic and intermediate but not non-pathogenic Leptospira spp., as shown in Figure 1A. Primers are shown in Figure 1B.
Figure 1.
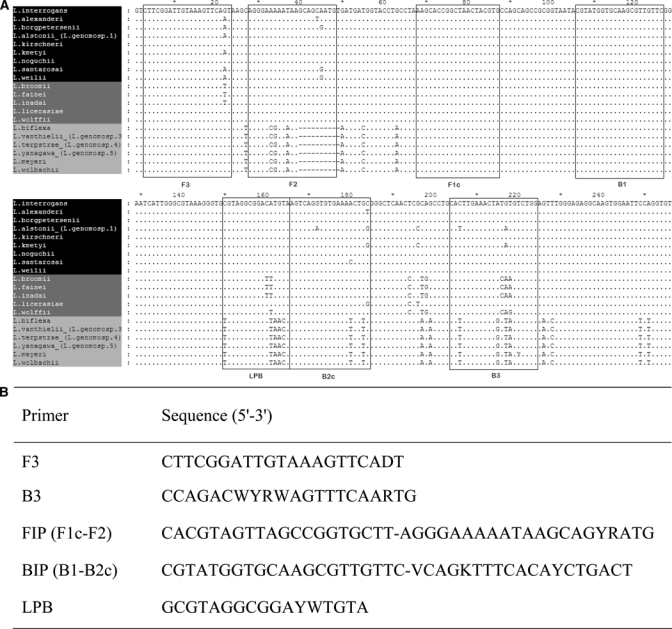
A, Sequence alignment of a region of the 16S ribosomal RNA gene (rrs) of 20 Leptospira spp. to demonstrate the regions selected for primer design. The objective was to design primers that annealed to the target gene of Leptospira spp. that belonged to either the pathogenic (white letters in black box) or intermediate group (white letters in gray box), but that failed to anneal to the target gene of Leptospira spp. in the non-pathogenic group (black letters in gray box). F1c, F2, F3, B1, B2c, B3, and LPB refer to the names and locations of target sequences for the primers used. B, Sequences of rrs loop-mediated isothermal amplification (LAMP) primers. Primer FIP consisted of the F1complementary sequence (20 nucleotides) and the F2 direct sequence (21 nucleotides). Primer BIP consisted of the B1 direct sequence (21 nucleotides) and the B2 complementary sequence (19 nucleotides).
LAMP assays.
The lipL41 LAMP was performed by using primers and conditions described.10 The rrs LAMP was carried out in a reaction mixture of 25 μL containing 5 pmol of each outer primer (F3, B3), 40 pmol of each inner primer (FIP, BIP), 20 pmol of loop primer (LPB), 1× ThermoPol reaction buffer, 0.8 M Betaine (Sigma, St. Louis, MO), 1.4 mM dNTP, 6 mM MgSO4, 8 units of Bst DNA polymerase, large fragment (New England Biolabs, Ipswich, MA), and 1 μL (approximately 20–100 ng) of genomic DNA extracted from laboratory cultures of Leptospira spp. The reaction mixture was incubated at 63°C for 2 hours and then heated at 80°C for 5 minutes to terminate the reaction. The presence of product was defined for both LAMP assays on the basis of detecting a white precipitate with the naked eye after centrifugation at 13,000 × g for 2 minutes. All results were confirmed by examining 3 μL of amplified products by using 2% agarose gel electrophoresis and staining with ethidium bromide. Positive and negative controls were genomic DNA from L. interrogans serovar Lai and reaction mixture without DNA, respectively. The analytical sensitivity of rrs LAMP was determined by using a 10-fold serial dilution (ranging from 2 × 106 to 0.02 GEs) of L. interrogans serovar Lai genomic DNA. The analytical specificity of this assay was determined by using DNA from non-pathogenic Leptospira spp. and a range of other bacterial species.
Clinical validation.
Patients with (cases) or without leptospirosis (controls) were selected from a prospective cohort of 418 patients with an acute febrile illness who came to the Udon Thani Regional Hospital in northeastern Thailand during January 10, 2001–June 16, 2002.18 Briefly, patients were recruited into the study during twice a day ward rounds. Inclusion criteria were patients who were ≥ 15 years of age with fever (> 37.8°C) of unknown cause who agreed to participate and to attend out-patient follow-up for a convalescent-phase serum sample. The exclusion criterion was the presence of a definable infection at the time of admission.
Blood was obtained on admission from all patients for Leptospira culture, serologic testing, and molecular diagnostics, and a second (convalescent-phase) sample was obtained for serologic testing approximately two weeks later. Leptospira culture was performed as described,19 and isolates sent to the World Health Organization/Food and Agriculture Organization of the United Nations/World Organisation for Animal Health Collaborating Centre for Reference and Research on Leptospirosis Collaborating Center for Reference and Research on Leptospirosis (Brisbane, Queensland, Australia) for serovar identification by using the cross-agglutinin absorption test.20 The MAT was performed by the World Health Organization/Food and Agriculture Organization of the United Nations/World Organisation for Animal Health Collaborating Centre for Reference and Research on Leptospirosis as described20 by using a live panel of antigens representing ubiquitous and locally prevalent serovars. A diagnosis of leptospirosis was based on isolation of Leptospira from blood and/or a positive MAT result, which was defined as a four-fold increase in titer between acute-phase and convalescence-phase samples or a single titer of ≥ 1:400. Patients who did not meet these criteria were defined as not having current or recent leptospirosis.
All patients diagnosed as having leptospirosis during the prospective cohort study (n = 133) were selected as cases. Laboratory confirmation was made on the basis of culture-positive and MAT-positive results in 21 (16%) patients, culture- positive results and MAT-negative results in 18 (13%) patients, and culture-negative results and MAT-positive results in 94 (71%) patients. A positive MAT result was based on a four-fold increasing titer in 97 cases and a single titer of ≥ 1:400 in 27 cases. Controls (n = 133) were randomly selected from those patients who did not meet the diagnostic criteria for leptospirosis. Patients in this group had a convalescent-phase serum sample obtained a median of 17 days (range = 10–43 days, interquartile range [IQR] = 13–21 days) after the onset of symptoms.
A database was created in which cases and controls were entered, randomized, and blinded to the technician prior to performing the two LAMP assays. DNA was extracted from a 5-mL blood sample (plus EDTA) obtained from cases and controls on admission and stored at –80°C prior to use. Extraction was performed by using the Nucleon BACC 3 Kit B (GE Healthcare Biosciences, Piscataway, NJ), and the extract was suspended in 1 mL of Tris-EDTA buffer. The LAMP assays were described as above except that 5 μL of DNA extracted from whole blood was used per reaction. All patient samples were evaluated in duplicate for each of the two LAMP assays. A positive result for either one or both samples was interpreted as positive.
The study protocol was reviewed and approved by the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand.
Statistical analysis.
Statistical analyses were performed by using STATA/SE version 10.0 (StataCorp., College Station, TX). Diagnostic sensitivity (DSe) and specificity (DSp) of each PCR assay was defined against the combined result for culture and MAT (a positive result for either or both being interpreted as diagnostic for leptospirosis), and expressed as a proportion with exact 95% confidence intervals (CIs). Comparison of DSe and DSp for the two LAMP assays was performed by using the McNemar test. The DSe of each assay was re-evaluated in the subset of patients who were culture positive.
Results
Analytical sensitivity and specificity.
Primers were designed that were predicted to anneal to a region of rrs of Leptospira spp. belonging to pathogenic and intermediate groups but not to the non-pathogenic group. The analytical specificity of these primers was assessed using genomic DNA extracted from 15 Leptospira spp., including representatives of each of the three groups (Figure 2) demonstrates confirmation of the expected annealing pattern. Specificity was further evaluated by testing a range of bacterial species that commonly cause febrile illness in our patient population. No product was observed using genomic DNA from one isolate of each of the following bacteria: S. aureus, Enterococcus spp., E. coli, S. enterica serovar Typhi, K. pneumoniae, P. aeruginosa, B. pseudomallei, O. tsutsugamushi strain Kato, and R. typhi. The analytical sensitivity (lower limit of detection) determined by using genomic DNA extracted from pure laboratory culture of L. interrogans serovar Lai was equivalent to10 GEs/reaction.
Figure 2.
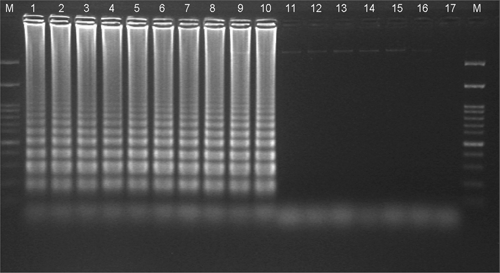
16S ribosomal RNA gene loop-mediated isothermal amplification products separated by 2% agarose gel electrophoresis. Lanes 1–8, pathogenic Leptospira spp. (L. interrogans, L. kirshneri, L. noguchii, L. alexanderi, L. weilli, L. borgpetersenii, L. santarosai, and L. alstonii, respectively); lanes 9 and 10, intermediate group Leptospira spp. (L. inadai and L. wolffii, respectively). No products were observed for non-pathogenic species (lanes 11–16, L. biflexa, L. meyeri, L. wolbachii, L. terpstrae, L. yanagawae, and Turneriella parva, respectively). Lane M, 100-basepair DNA marker; lane 17, negative (no template) control.
Diagnostic sensitivity and specificity of rrs LAMP.
In total, 133 patients with laboratory confirmed leptospirosis (cases) and 133 patients who did not have leptospirosis (controls) were evaluated. Patient recruitment and diagnostic testing are summarized in Figure 3. Of 133 patients, 18 (13%) were culture positive and MAT negative, 94 patients (71%) were MAT positive and culture negative, and 21 (16%) were MAT positive and culture positive. The median (IQR, range) age was 35 years (26–46, 15–74 years) for cases and 42 years (29–54, 15–79 years) for controls (P = 0.01). The median duration of illness prior to admission was 4 days (IQR = 2–5 days, range = 1–12 days) for cases and 6 days (IQR = 3–9 days, range = 0–33 days) for controls (P < 0.001). Most (120 of 133, 90.2%) of the case-patients came to the hospital with ≤ 7 days of the onset of symptoms. Fourteen case-patients (11%) had received antimicrobial drug therapy that would be predicted to be bacteriocidal for Leptospira prior to blood sample collection. The discharge diagnoses of controls were as follows: scrub typhus (n = 54), other bacterial septicemia (n = 8), E. coli (n = 2), K. pneumoniae (n = 2), Acinetobacter baumanii (n = 1), Corynebacterium jeikeium (n = 1), Enterococcus sp. (n = 1), Streptococcus pneumoniae (n = 1), dengue fever (n = 5), murine typhus (n = 4), melioidosis (n = 2), human immunodeficiency virus–related infections (n = 2), other diagnoses (n = 7), and unknown diagnosis (n = 51). Five cases (4%) and four controls (3%) died during hospital admission.
Figure 3.
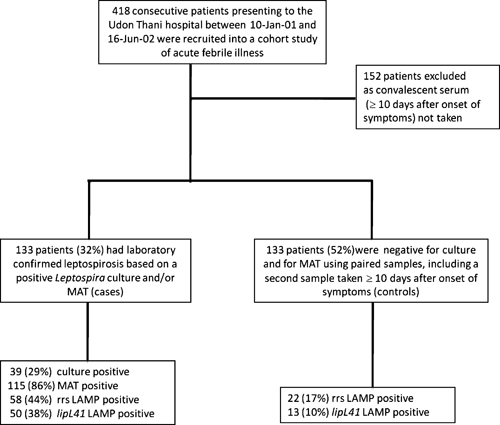
Patient recruitment and result of laboratory tests for leptospirosis.
The rrs LAMP result was positive in 58 of 133 patients (DSe = 43.6, 95% CI = 35.0–52.5), and 22 of 133 controls (DSp = 83.5, 95% CI = 76.0–89.3). The diagnoses for the 22 positive controls were scrub typhus (8), dengue (2), enterococcal septicemia (1), murine typhus (1), tuberculosis (1), and unknown (9). The duration of symptoms prior to presentation was significantly different between rrs LAMP-positive cases and rrs LAMP-negative cases (median = 3 days, IQR = 2–4 days, range = 1–8 days versus median 4 days, IQR = 3–6 days, range = 1–12 days, respectively; P < 0.001).
Clinical evaluation of lipL41 LAMP and comparison with rrs LAMP.
The lipL41 LAMP result was positive in 50 of 133 cases, (DSe = 37.6, 95% CI = 29.3–46.4), and 13 of 133 controls (DSp = 90.2, 95% CI = 83.9–94.7). The diagnoses for these 13 positive controls were scrub typhus (4), dengue (2), eosinophilic meningitis (cause unknown) (1), murine typhus (1), and unknown (5). The differences in DSe and DSp values between the rrs and lipL41 LAMP assays did not reach statistical significance (P = 0.13 and 0.06, respectively).
Concordance between the two assays was as follows. For cases, 43 were positive by both assays, 15 were positive by rrs, and 7 were positive by lipL41. For controls, 8 were positive by both assays, 14 were positive by rrs, and 5 were positive by lipL41.
Sensitivity of LAMP assays in patients with leptospiremia.
Sensitivity was reanalyzed for 39 patients who were culture positive for Leptospira spp. The rrs LAMP result was positive in 33 patients (sensitivity = 84.6, 95% CI = 69.5–94.1), and the lipL41 LAMP result was positive in 29 patients (sensitivity = 74.4, 95% CI = 57.9–87.0) (P = 0.29). Six culture-positive patients were negative by both assays, and all culture-positive and LAMP-negative patients were infected with L. interrogans serovar Autumnalis, the dominant cause of leptospirosis in Thailand during the study period.21
Discussion
The ease with which LAMP can be performed to detect pathogens in clinical samples has led to its development for major infectious diseases in the developing world, including tuberculosis and malaria.22,23 Most of the published literature on LAMP describes assay development and analytical sensitivity and specificity, but relatively few studies have reported clinical evaluation of DSe and DSp. We developed an rrs LAMP assay for the diagnosis of leptospirosis and demonstrated 100% analytical sensitivity and specificity, and proceeded to a clinical evaluation by using samples obtained from a cohort of consecutive patients with an acute febrile illness who came to a hospital in northeastern Thailand. Having identified that the frequency of laboratory proven leptospirosis for this group was 18%, we elected to use a case–control design on the basis of cost. The DSe of rrs LAMP was disappointingly low (43.6%). The sensitivity of rrs LAMP for a subset of 39 patients who were culture positive for Leptospira spp. was considerably higher (84.6%). However, this observation has little practical utility because it is not possible to predict those patients with leptospiremia on the basis of clinical features alone.
A LAMP assay specific for lipL41 has been developed previously for the diagnosis of leptospirosis and was included in our clinical evaluation. Possible reasons for the lower diagnostic sensitivity of lipL41 LAMP compared with rrs LAMP include the difference in lower limit of detection for the two assays (10 GE/reaction and 100GE/reaction for rrs and lipL41, respectively), or infection with intermediate group Leptospira spp., which would not be detected by lipL41. However, the latter possibility is not the case because all 39 isolates from patients who were culture positive have been identified and none belonged to the intermediate group.
There are several possible explanations for the low DSe of both LAMP assays. A wide range of oral antimicrobial drugs are available over the counter in Thailand, and it is common for persons to self-medicate in the period leading up to hospital admission. Many of the available antimicrobial drugs would be predicted to be effective against Leptospira spp., which may lead to a false-negative LAMP result although not necessarily a negative MAT result. We were aware that 14 case-patients had received an antimicrobial drug at the time of admission (of which 4 case-patients had positive results for both assays), but use of a drug may have been more common. The LAMP assays may have had false-negative results because the number of organisms in the sample was below the lower limit of detection for the assay, or because of failure of primer annealing, although we consider this unlikely for rrs, given the conserved nature of the target. Although further refinements of the LAMP assay may result in an improvement in the lower limit of detection and diagnostic sensitivity, we are pessimistic that molecular tests will prove highly sensitive in unselected patients in settings where antibiotic use is uncontrolled.
The DSp of both LAMP assays were lower than anticipated, and it is possible that some of the LAMP-positive controls showed false-negative results by culture and MAT. The sensitivity of MAT is reported to be greater than 90%,24 and the finding in our study that 18 cases were culture positive but MAT negative highlights the imperfect sensitivity of MAT in our setting. The observation that 8 controls (with a diagnosis of scrub typhus [3], dengue [2], and unknown [3]) were positive by rss and lipL41 assays is a further hint that these patients may have been leptospiremic. However, in the absence of definitive evidence to support true infection rather than another explanation such as laboratory contamination, we must assume that these cases had false-positive results rather than true positive results.
In conclusion, we have demonstrated that the rss LAMP can identify approximately half of patients with leptospirosis at the time of presentation in northeastern Thailand, but this finding is tempered by an imperfect specificity. We are unable to recommend use of LAMP in routine clinical practice until the results of additional clinical evaluations become available.
ACKNOWLEDGMENTS
We thank our colleagues at Udon Thani Hospital for their support, Professor Yupin Suputtamongkol for performing the scrub typhus immunofluorscent antibody assay, and the staff of the World Health Organization/Food and Agriculture Organization of the United Nations/World Organisation for Animal Health Collaborating Center for Reference and Research on Leptospirois, Queensland Health Forensic and Scientific Services for providing serologic testing and isolate identification.
Footnotes
Financial support: This study was supported by the Thailand Research Fund and the Wellcome Trust.
Authors' addresses: Piengchan Sonthayanon, Department of Molecular Tropical Medicine and Genetics, Mahidol-Oxford Tropical Medicine Research Unit, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand, E-mail: piengchan@tropmedres.ac. Wirongrong Chierakul, Department of Clinical Tropical Medicine, and Mahidol-Oxford Tropical Medicine Research Unit, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand, E-mail: kae@tropmedres.ac. Vanaporn Wuthiekanun, Janjira Thaipadungpanit, Siriphan Boonsilp, and Premjit Amornchai, Mahidol-Oxford Tropical Medicine Research Unit, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand, E-mails: lek@tropmedres.ac, janjira@tropmedres.ac, siriphan@tropmedres.ac, and kung@tropmedres.ac. Thareerat Kalambaheti, Department of Microbiology and Immunology, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand, E-mail: tmtlk@mahidol.ac.th. Lee D. Smythe, World Health Organization/Food and Agriculture Organization of the United Nations/World Organisation for Animal Health Collaborating Centre for Reference and Research on Leptospirosis, Western Pacific Region, Communicable Disease Unit, Queensland Health Forensic and Scientific Services, Brisbane, Queensland, Australia, E-mail: Lee_Smythe@health.gld.gov.au. Direk Limmathurotsakul, Department of Tropical Hygiene and Mahidol-Oxford Tropical Medicine Research Unit, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand, E-mail: direk@tropmedres.ac. Nicholas P. Day, Mahidol-Oxford Tropical Medicine Research Unit, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand and Center for Clinical Vaccinology and Tropical Medicine, Nuffield Department of Clinical Medicine, University of Oxford, Oxford, United Kingdom, E-mail: nickd@tropmedres.ac. Sharon J. Peacock, Department of Microbiology and Immunology, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand and Department of Medicine, University of Cambridge, Addenbrooke's Hospital, Cambridge, United Kingdom, E-mail: sharon@tropmedres.ac.
Reprint requests: Piengchan Sonthayanon, Department of Molecular Tropical Medicine and Genetics and Mahidol-Oxford Tropical Medicine Research Unit, Faculty of Tropical Medicine, Mahidol University, 420/6 Rajvithi Road, Bangkok, 10400, Thailand, E-mail: piengchan@tropmedres.ac.
References
- 1.Suttinont C, Losuwanaluk K, Niwatayakul K, Hoontrakul S, Intaranongpai W, Silpasakorn S, Suwancharoen D, Panlar P, Saisongkorh W, Rolain JM, Raoult D, Suputtamongkol Y. Causes of acute, undifferentiated, febrile illness in rural Thailand: results of a prospective observational study. Ann Trop Med Parasitol. 2006;100:363–370. doi: 10.1179/136485906X112158. [DOI] [PubMed] [Google Scholar]
- 2.Merien F, Baranton G, Perolat P. Comparison of polymerase chain reaction with microagglutination test and culture for diagnosis of leptospirosis. J Infect Dis. 1995;172:281–285. doi: 10.1093/infdis/172.1.281. [DOI] [PubMed] [Google Scholar]
- 3.Smythe LD, Smith IL, Smith GA, Dohnt MF, Symonds ML, Barnett LJ, McKay DB. A quantitative PCR (TaqMan) assay for pathogenic Leptospira spp. BMC Infect Dis. 2002;2:13. doi: 10.1186/1471-2334-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palaniappan RU, Chang YF, Chang CF, Pan MJ, Yang CW, Harpending P, McDonough SP, Dubovi E, Divers T, Qu J, Roe B. Evaluation of lig-based conventional and real time PCR for the detection of pathogenic leptospires. Mol Cell Probes. 2005;19:111–117. doi: 10.1016/j.mcp.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Kositanont U, Rugsasuk S, Leelaporn A, Phulsuksombati D, Tantitanawat S, Naigowit P. Detection and differentiation between pathogenic and saprophytic Leptospira spp. by multiplex polymerase chain reaction. Diagn Microbiol Infect Dis. 2007;57:117–122. doi: 10.1016/j.diagmicrobio.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kubo T, Agoh M, Mai LQ, Fukushima K, Nishimura H, Yamaguchi A, Hirano M, Yoshikawa A, Hasebe F, Kohno S, Morita K. Development of a reverse transcription-loop-mediated isothermal amplification assay for detection of pandemic (H1N1), 2009, virus as a novel molecular method for diagnosis of pandemic influenza in resource-limited settings. J Clin Microbiol. 2009;48:728–735. doi: 10.1128/JCM.01481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chantratita N, Meumann E, Thanwisai A, Limmathurotsakul D, Wuthiekanun V, Wannapasni S, Tumapa S, Day NP, Peacock SJ. Loop-mediated isothermal amplification method targeting the TTS1 gene cluster for detection of Burkholderia pseudomallei and diagnosis of melioidosis. J Clin Microbiol. 2008;46:568–573. doi: 10.1128/JCM.01817-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nemoto J, Sugawara C, Akahane K, Hashimoto K, Kojima T, Ikedo M, Konuma H, Hara-Kudo Y. Rapid and specific detection of the thermostable direct hemolysin gene in Vibrio parahaemolyticus by loop-mediated isothermal amplification. J Food Prot. 2009;72:748–754. doi: 10.4315/0362-028x-72.4.748. [DOI] [PubMed] [Google Scholar]
- 10.Lin X, Chen Y, Lu Y, Yan J, Yan J. Application of a loop-mediated isothermal amplification method for the detection of pathogenic Leptospira. Diagn Microbiol Infect Dis. 2009;63:237–242. doi: 10.1016/j.diagmicrobio.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Shang ES, Summers TA, Haake DA. Molecular cloning and sequence analysis of the gene encoding LipL41, a surface-exposed lipoprotein of pathogenic Leptospira species. Infect Immun. 1996;64:2322–2330. doi: 10.1128/iai.64.6.2322-2330.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morey RE, Galloway RL, Bragg SL, Steigerwalt AG, Mayer LW, Levett PN. Species-specific identification of Leptospiraceae by 16S rRNA gene sequencing. J Clin Microbiol. 2006;44:3510–3516. doi: 10.1128/JCM.00670-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slack AT, Kalambaheti T, Symonds ML, Dohnt MF, Galloway RL, Steigerwalt AG, Chaicumpa W, Bunyaraksyotin G, Craig S, Harrower BJ, Smythe LD. Leptospira wolffii sp. nov., isolated from a human with suspected leptospirosis in Thailand. Int J Syst Evol Microbiol. 2008;58:2305–2308. doi: 10.1099/ijs.0.64947-0. [DOI] [PubMed] [Google Scholar]
- 14.Levett PN, Morey RE, Galloway RL, Steigerwalt AG. Leptospira broomii sp. nov., isolated from humans with leptospirosis. Int J Syst Evol Microbiol. 2006;56:671–673. doi: 10.1099/ijs.0.63783-0. [DOI] [PubMed] [Google Scholar]
- 15.Arzouni JP, Parola P, La Scola B, Postic D, Brouqui P, Raoult D. Human infection caused by Leptospira fainei. Emerg Infect Dis. 2002;8:865–868. doi: 10.3201/eid0808.010445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholas KB, Nicholas HB, Deerfield DWII. GeneDoc: Analysis and Visualization of Genetic Variation. EMBNEW. NEWS. 1997;4:14. [Google Scholar]
- 18.Sonthayanon P, Chierakul W, Wuthiekanun V, Blacksell SD, Pimda K, Suputtamongkol Y, Pukrittayakamee S, White NJ, Day NP, Peacock SJ. Rapid diagnosis of scrub typhus in rural Thailand using polymerase chain reaction. Am J Trop Med Hyg. 2006;75:1099–1102. [PubMed] [Google Scholar]
- 19.Wuthiekanun V, Chierakul W, Limmathurotsakul D, Smythe LD, Symonds ML, Dohnt MF, Slack AT, Limpaiboon R, Suputtamongkol Y, White NJ, Day NP, Peacock SJ. Optimization of culture of Leptospira from humans with leptospirosis. J Clin Microbiol. 2007;45:1363–1365. doi: 10.1128/JCM.02430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stallman ND. International Committee on Systematic Bacteriology Subcommittee on the Taxonomy of Leptospira: Minutes of the Meeting, 6 to 10 August 1982, Boston, Massachusetts. Int J Syst Bacteriol. 1984;34:258–259. [Google Scholar]
- 21.Thaipadungpanit J, Wuthiekanun V, Chierakul W, Smythe LD, Petkanchanapong W, Limpaiboon R, Apiwatanaporn A, Slack AT, Suputtamongkol Y, White NJ, Feil EJ, Day NP, Peacock SJ. A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand. PLoS Negl Trop Dis. 2007;1:e56. doi: 10.1371/journal.pntd.0000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han E-T, Watanabe R, Sattabongkot J, Khuntirat B, Sirichaisinthop J, Iriko H, Jin L, Takeo S, Tsuboi T. Detection of four Plasmodium species by genus- and species-specific loop-mediated isothermal amplification for clinical diagnosis. J Clin Microbiol. 2007;45:2521–2528. doi: 10.1128/JCM.02117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwamoto T, Sonobe T, Hayashi K. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J Clin Microbiol. 2003;41:2616–2622. doi: 10.1128/JCM.41.6.2616-2622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dassanayake D, Wimalaratna H, Agampodi S, Liyanapathirana V, Piyarathna T, Goonapienuwala B. Evaluation of surveillance case definition in the diagnosis of leptospirosis, using the microscopic agglutination test: a validation study. BMC Infect Dis. 2009;9:48. doi: 10.1186/1471-2334-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]


