Abstract
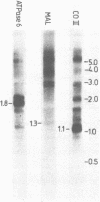
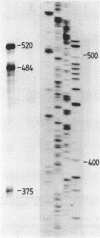
Neurospora mitochondrial DNA is transcribed into long molecules containing the information of several genes. Processing leads to formation of functionally active RNAs. It has been shown previously that when tRNA sequences are present in these transcripts excision of mRNAs occurs at the acceptor stem of these tRNA sequences. We have investigated the processing of precursor RNAs transcribed from a region of the mitochondrial genome devoid of tRNA genes. This region comprises the genes encoding subunit 6 of the mitochondrial ATPase, subunit 2 of cytochrome aa3 and a mitochondrial ATPase proteolipid-like gene. We have proved that a common precursor of the putative mRNAs of these genes exists and we have determined the positions of the 5' and 3' ends of processing intermediates and of the mature mRNAs. We will discuss possible processing routes and secondary structures that substitute for tRNA sequences as processing sites.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Apirion D. RNA processing in a unicellular microorganism: implications for eukaryotic cells. Prog Nucleic Acid Res Mol Biol. 1983;30:1–40. doi: 10.1016/s0079-6603(08)60682-0. [DOI] [PubMed] [Google Scholar]
- Breitenberger C. A., Browning K. S., Alzner-DeWeerd B., RajBhandary U. L. RNA processing in Neurospora crassa mitochondria: use of transfer RNA sequences as signals. EMBO J. 1985 Jan;4(1):185–195. doi: 10.1002/j.1460-2075.1985.tb02335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G., Helmer Citterich M., Nelson M. A., Werner S., Macino G. RNA processing in Neurospora crassa mitochondria: transfer RNAs punctuate a large precursor transcript. EMBO J. 1985 Jan;4(1):197–204. doi: 10.1002/j.1460-2075.1985.tb02336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G., Werner S. The mitochondrial URF1 gene in Neurospora crassa has an intron that contains a novel type of URF. J Mol Biol. 1985 Nov 20;186(2):231–242. doi: 10.1016/0022-2836(85)90100-7. [DOI] [PubMed] [Google Scholar]
- Collins R. A., Lambowitz A. M. RNA splicing in Neurospora mitochondria. Defective splicing of mitochondrial mRNA precursors in the nuclear mutant cyt18-1. J Mol Biol. 1985 Aug 5;184(3):413–428. doi: 10.1016/0022-2836(85)90291-8. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Garriga G., Bertrand H., Lambowitz A. M. RNA splicing in Neurospora mitochondria: nuclear mutants defective in both splicing and 3' end synthesis of the large rRNA. Cell. 1984 Mar;36(3):623–634. doi: 10.1016/0092-8674(84)90342-8. [DOI] [PubMed] [Google Scholar]
- Green M. R., Grimm M. F., Goewert R. R., Collins R. A., Cole M. D., Lambowitz A. M., Heckman J. E., Yin S., RajBhandary U. L. Transcripts and processing patterns for the ribosomal RNA and transfer RNA region of Neurospora crassa mitochondrial DNA. J Biol Chem. 1981 Feb 25;256(4):2027–2034. [PubMed] [Google Scholar]
- Green M. R., Grimm M. F., Goewert R. R., Collins R. A., Cole M. D., Lambowitz A. M., Heckman J. E., Yin S., RajBhandary U. L. Transcripts and processing patterns for the ribosomal RNA and transfer RNA region of Neurospora crassa mitochondrial DNA. J Biol Chem. 1981 Feb 25;256(4):2027–2034. [PubMed] [Google Scholar]
- Hu N., Messing J. The making of strand-specific M13 probes. Gene. 1982 Mar;17(3):271–277. doi: 10.1016/0378-1119(82)90143-3. [DOI] [PubMed] [Google Scholar]
- Jacobson A. B., Good L., Simonetti J., Zuker M. Some simple computational methods to improve the folding of large RNAs. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):45–52. doi: 10.1093/nar/12.1part1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macino G., Morelli G. Cytochrome oxidase subunit 2 gene in Neurospora crassa mitochondria. J Biol Chem. 1983 Nov 10;258(21):13230–13235. [PubMed] [Google Scholar]
- Meinkoth J., Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984 May 1;138(2):267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- Morelli G., Macino G. Two intervening sequences in the ATPase subunit 6 gene of Neurospora crassa. A short intron (93 base-pairs) and a long intron that is stable after excision. J Mol Biol. 1984 Sep 25;178(3):491–507. doi: 10.1016/0022-2836(84)90235-3. [DOI] [PubMed] [Google Scholar]
- de Vries H., Haima P., Brinker M., de Jonge J. C. The Neurospora mitochondrial genome: the region coding for the polycistronic cytochrome oxidase subunit I transcript is preceded by a transfer RNA gene. FEBS Lett. 1985 Jan 7;179(2):337–342. doi: 10.1016/0014-5793(85)80547-0. [DOI] [PubMed] [Google Scholar]
- van den Boogaart P., Samallo J., Agsteribbe E. Similar genes for a mitochondrial ATPase subunit in the nuclear and mitochondrial genomes of Neurospora crassa. Nature. 1982 Jul 8;298(5870):187–189. doi: 10.1038/298187a0. [DOI] [PubMed] [Google Scholar]
- van den Boogaart P., van Dijk S., Agsteribbe E. The mitochondrially made subunit 2 of Neurospora crassa cytochrome aa3 is synthesized as a precursor protein. FEBS Lett. 1982 Oct 4;147(1):97–100. doi: 10.1016/0014-5793(82)81019-3. [DOI] [PubMed] [Google Scholar]