Abstract
Recent genome-wide association studies (GWASs) have identified a locus on chromosome 1p13 as strongly associated with both serum low-density lipoprotein cholesterol (LDL-C) and myocardial infarction (MI) in humans. Here we show through a series of studies in human cohorts and human-derived hepatocytes that a common noncoding polymorphism at the 1p13 locus, rs12740374, creates a C/EBP transcription factor binding site and alters the hepatic expression of the SORT1 gene. With siRNA knockdown and viral overexpression in mouse liver, we demonstrate that Sort1 alters plasma LDL-C and very low-density lipoprotein (VLDL) particle levels by modulating hepatic VLDL secretion. Thus, we provide functional evidence for a novel regulatory pathway for lipoprotein metabolism and suggest that modulation of this pathway may alter risk for MI in humans. We also demonstrate that common noncoding DNA variants identified by GWASs can directly contribute to clinical phenotypes.
MI is the leading cause of death in the developed world. LDL-C is a causal risk factor for the disease, as demonstrated by (1) the increased and early burden of MI in individuals with the Mendelian disorder of familial hypercholesterolemia1 and (2) the success of LDL-C-lowering medications in reducing the incidence of MI in clinical trials in many populations2. Despite aggressive use of statin drugs, many individuals do not achieve the LDL-C levels recommended by clinical guidelines3. There remains a need for additional methods of reducing LDL-C.
GWASs for serum lipoprotein traits have identified a number of common single nucleotide polymorphism (SNP) variants that are strongly associated with serum LDL-C4-10. Many of these SNPs are in or near genes known to cause Mendelian dyslipidemias (LDLR, APOB, and PCSK9) or established molecular targets for LDL-C-lowering therapies (HMGCR). However, several of the LDL-C loci contain genes not previously implicated in lipoprotein metabolism. Of the newly mapped loci, the novel SNPs most strongly associated with LDL-C all lie on chromosome 1p13; indeed, in a meta-analysis of ~100,000 individuals (reported in this issue of Nature10) this locus has the strongest association with LDL-C of any locus in the genome (P = 1 × 10−170). The same 1p13 SNPs have also been independently linked to coronary artery disease and MI in GWASs10-12. Individuals of European descent who are homozygous for the major alleles of these SNPs have up to 16 mg/dl higher LDL-C as well as ~40% increased risk of MI11, 12 when compared with minor allele homozygotes. Thus, the same genetic locus is linked to both a recognized intermediate phenotype as well as a hard clinical disease endpoint.
As compelling as these associations are, they do not explain how human genetic variation at the 1p13 locus confers change in serum LDL-C and thereby alters risk of MI. We therefore sought to identify (1) the causal DNA variant in the 1p13 locus, (2) the gene regulated by the locus, (3) the mechanism by which the DNA variant affects the gene, and (4) the mechanism by which the gene influences lipoprotein metabolism.
1p13 SNPs Associated with LDL Particles
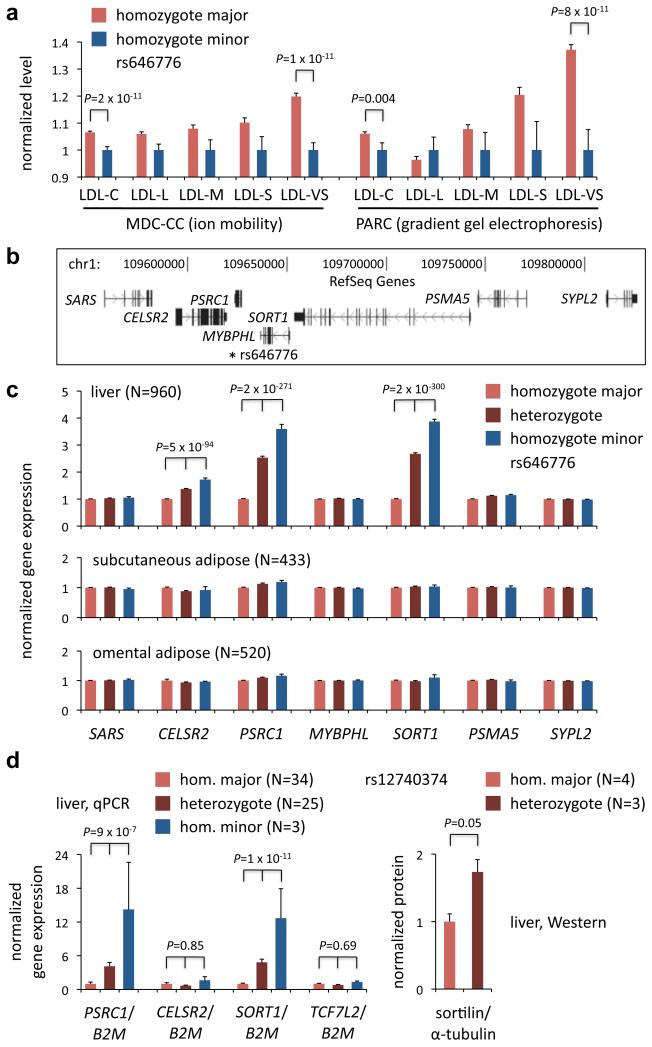
LDL-C comprises a variety of lipoprotein particles that range in size and density, and it has been hypothesized that smaller LDL particles are more atherogenic than larger LDL particles13. To determine whether the 1p13 locus selectively affects certain LDL subclasses, we used different methodologies—ion mobility and gradient gel electrophoresis—to measure lipoprotein subclasses in two different cohorts—the Malmö Diet and Cancer Study – Cardiovascular Cohort (MDC-CC)14 and the Pharmacogenomics and Risk of Cardiovascular Disease (PARC) study15. We found that an index SNP in the 1p13 locus, rs646776, was most highly associated with changes in the very small LDL (LDL-VS) lipoprotein subclass (20% increase in major allele homozygotes vs. minor allele homozygotes with P = 1.1 × 10−11 in MDC-CC; 37% increase with P = 8.0 × 10−11 in PARC); progressively smaller changes were seen with larger LDL subclasses (Fig.1a; Supplementary Fig. 1a, b).
Figure 1. The human chromosome 1p13 locus is preferentially associated with very small LDL and liver gene expression.
a, Mean serum lipid and lipoprotein particle levels in homozygotes for the minor haplotype of the 1p13 locus (minor allele of rs646776) vs. homozygotes for the major haplotype (major allele of rs646776), normalized to the mean level in minor haplotype homozygotes, in the MDC-CC cohort (measured by ion mobility) and the PARC cohort (measured by gradient gel electrophoresis). LDL-L = large LDL; LDL-M = medium LDL; LDL-S = small LDL; LDL-VS = very small LDL. b, Relative gene positions in and around the 1p13 locus; * indicates position of rs646776. c, Mean expression of local genes in homozygotes for the major 1p13 haplotype (major allele of rs646776) vs. heterozygotes vs. homozygotes for the minor 1p13 haplotype (minor allele of rs646776), normalized to the mean level in major haplotype homozygotes, in samples of human liver, human subcutaneous adipose, and human omental adipose. d, Mean expression of PSRC1, CELSR2, SORT1, and TCF7L2 (negative control) mRNA, standardized to B2M expression, and sortilin protein, standardized to α-tubulin, in samples of human liver from homozygotes for the major 1p13 haplotype (major allele of rs12740374) vs. heterozygotes vs. homozygotes for the minor 1p13 haplotype (minor allele of rs12740374) if available, normalized to the mean level in major haplotype homozygotes. P values derived from linear regression analyses or unpaired t test. Error bars show s.e.m.
1p13 SNPs and Liver-Specific Expression
The SNPs in the 1p13 locus previously reported to be most highly associated with LDL-C—rs646776, rs599839, rs12740374, and rs629301—lie in a noncoding DNA region between two genes, CELSR2 and PSRC1, whose functions are unknown (Fig. 1b, 2a)4-10. As noncoding DNA variants may alter gene expression, we previously used expression quantitative trait locus (eQTL) analyses to explore whether 1p13 SNPs are cis-acting regulators of nearby genes in human liver4, 7. We have now extended these studies by measuring expression of genes in or near the 1p13 locus in three types of human tissue samples: liver (960 samples), subcutaneous fat (433 samples), and omental fat (520 samples).
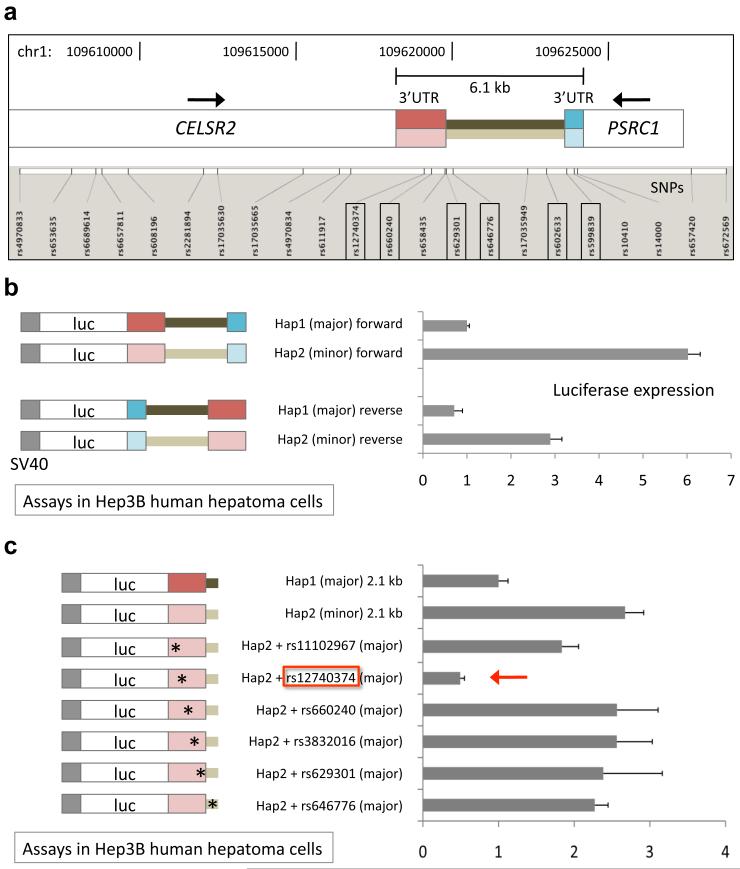
Figure 2. rs12740374 is responsible for haplotype-specific difference in transcriptional activity.
a, Map of 1p13 SNPs genotyped in ~20,000 individuals of European descent relative to CELSR2 and PSRC1 genes. The six SNPs with strongest association with LDL-C (indicated with boxes), comprising a single haplotype, define the 6.1 kb region between the stop codons of the two genes. b, Firefly luciferase expression from constructs transfected into Hep3B human hepatoma cells. Both the major (darker colors) and minor (lighter colors) haplotypes of the 6.1 kb region were subcloned in forward and reverse orientations into a basal firefly luciferase construct with the SV40 promoter. Shown are ratios of firefly luciferase expression to Renilla luciferase expression (expressed from cotransfected plasmid), measured 48 hours after transfection, normalized to the mean ratio from the major haplotype, forward orientation construct. Error bars show s.e.m., N = 2. c, Both the major and minor haplotypes of a minimal 2.1 kb region were subcloned into the basal construct. Single nucleotide alterations were introduced individually into the minor haplotype, changing minor alleles of SNPs into major alleles. Shown are ratios of firefly luciferase expression to Renilla luciferase expression normalized to the mean ratio from the major haplotype construct. Error bars show s.e.m., N = 4.
In liver, presence of the minor allele of rs646776 was highly associated with elevated transcript levels of three genes: CELSR2, PSRC1, and SORT1 (Fig. 1c). SORT1 displayed the largest expression change. We replicated these liver eQTL findings in an independent cohort of 62 human liver samples, from which rs12740374 (the putative causal 1p13 SNP, see below) was directly genotyped and SORT1, PSRC1, and CELSR2 expression were individually measured. Minor allele homozygotes displayed more than 12-fold higher SORT1 and PSRC1 expression than major allele homozygotes, with no significant change for CELSR2 (Fig. 1d). Immunoblot analysis of liver lysates demonstrated a significant increase in abundance of the SORT1 protein product (sortilin) in heterozygotes compared to major allele homozygotes (Fig. 1d, Supplementary Fig. 1c).
Notably, none of the gene expression changes in liver were seen in the two adipose tissue types (Fig. 1c), and minimal changes were reported in lymphocytes16, suggesting that the regulatory mechanism underlying the allele-specific gene expression is liver-specific.
A Causal 1p13 Noncoding DNA Variant
We performed fine mapping of the 1p13 locus to define the minimal DNA region responsible for the LDL-C association. Because rs646776, rs599839, rs12740374, and rs629301 lie between CELSR2 and PSRC1, we used data from a recent GWAS of ~20,000 individuals of European descent7 to perform association analyses with LDL-C on these and other SNPs spanning the two genes. Out of 18 other SNPs, we identified two SNPs with P values comparable to rs646776, rs599839, rs12740374, and rs629301 (P values ranging from 1.8 × 10−42 to 8.3 × 10−41) and no SNPs with lower P values (Supplementary Fig. 2a). Together these six best SNPs cluster in a noncoding DNA region that is 6.1 kb in size, spanning the 3′ untranslated region (3′UTR) of CELSR2, the intergenic region, and the PSRC1 3′UTR oriented in the opposite direction (Fig. 2a). The six SNPs are in high linkage disequilibrium (LD) and comprise two predominant haplotypes in HapMap Europeans (CEU), with the “major” haplotype present on 68% of chromosomes 1 and the “minor” haplotype on 29% (Supplementary Fig. 3).
We identified two human bacterial artificial chromosomes (BACs) harboring the major and minor haplotypes of the 6.1 kb region. We sequenced the region on each of the BACs in full and identified 16 polymorphisms (Supplementary Fig. 2b). From each BAC, the region spanning precisely between the stop codon of CELSR2 and the stop codon of PSRC1 was subcloned into firefly luciferase expression constructs just distal to the stop codon of the luciferase gene in either the “forward” (CELSR2) or “reverse” (PSRC1) orientation. Upon transfection of the constructs into Hep3B cultured human hepatocellular carcinoma cells, we found that in both orientations, the minor haplotype produced significantly greater luciferase expression than the major haplotype, consistent with the human liver eQTL analyses (Fig. 2b; compare to Fig. 1c). After localizing the haplotype-specific effect to the proximal 2.1 kb of the region (Supplementary Fig. 4), we tested an array of constructs in which single polymorphisms in the minor haplotype were switched to major alleles. We identified the SNP rs12740374 as being sufficient to confer the haplotype-specific effect (Fig. 2c, 3c).
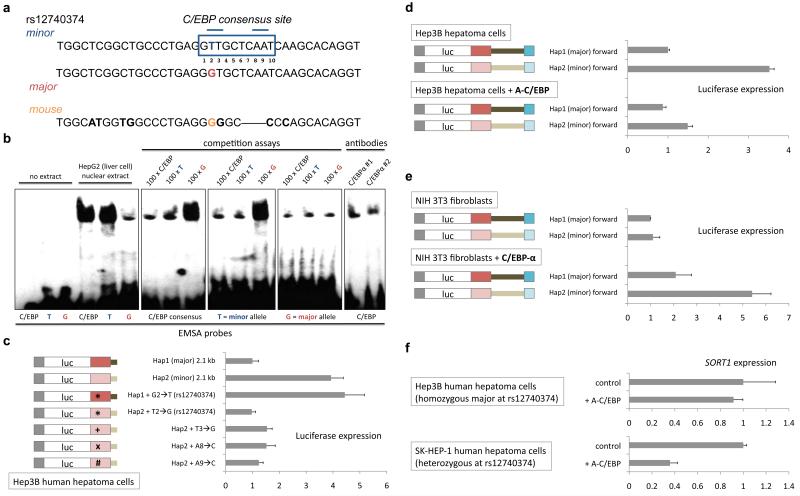
Figure 3. rs12740374 alters a C/EBP transcription factor binding site.
a, The human DNA sequence surrounding rs12740374, major and minor alleles, and orthologous DNA sequence in mouse. The major allele of rs12740374 disrupts one of two core elements (position 2-3, 8-9) in the predicted consensus binding site on which a C/EBP dimer binds21. b, Electrophoretic mobility shift assays (EMSA) with labeled probes matching the C/EBP consensus binding site18, the rs12740374 minor allele (T) sequence, and the rs12740374 major allele (G) sequence. Competition assays were performed with 100-fold excess of cold probe. Either of two C/EBPα antibodies was used to compete for binding and/or shift the protein-DNA complex. c, Relative firefly luciferase expression from constructs with haplotypes of 2.1 kb region transfected into Hep3B cells. Single nucleotide alterations were introduced into constructs as indicated, altering rs12740374 and the three other core recognition nucleotides in the predicted C/EBP binding site. d, e, Relative firefly luciferase expression from constructs with haplotypes of 6.1 kb region transfected into (d) Hep3B human hepatoma cells with or without concomitant transduction with A-C/EBP (dominant negative C/EBP) cDNA via lentivirus and (e) NIH 3T3 fibroblasts with or without concomitant transduction with C/EBP-α cDNA via lentivirus. f, Relative SORT1 expression, determined as a ratio with B2M expression by qRT-PCR, in Hep3B cells [homozygous major (GG) at rs12740374] or SK-HEP-1 human hepatoma cells [heterozygous (GT) at rs12740374] with or without concomitant transduction with A-C/EBP cDNA via lentivirus. Error bars show s.e.m., N = 3 for each experiment.
We genotyped rs12740374 and 15 other SNPs in or near the 6.1 kb noncoding region in ~9,000 African Americans. Whereas six SNPs have indistinguishable evidence for association with LDL-C in Europeans, we found that, in African Americans, rs12740374 alone had the strongest evidence for association (P = 2.3 × 10−20 for rs12740374 vs. 9.2 × 10−15 at the next best 1p13 SNP) (Supplementary Fig. 2a). This is consistent with rs12740374 being in high LD with nearby SNPs in HapMap Europeans (CEU) but not so in HapMap Africans (YRI) (Supplementary Fig. 3).
We observed that rs12740374 alters a predicted binding site for CCAAT/enhancer binding protein (C/EBP) transcription factors, with the minor allele creating the site and the major allele disrupting it; the binding site is not present in the orthologous DNA region in mice (Fig. 3a). C/EBPα is a liver-enriched transcriptional factor that regulates the expression of numerous hepatic genes involved in a variety of metabolic processes17. We tested binding of the rs12740374 minor and major allele sequences by C/EBP with electrophoretic mobility shift assays and found the minor allele sequence to be shifted as much as a classic C/EBP binding sequence18, with minimal shifting of the major allele sequence; addition of either of two C/EBPα antibodies impaired the binding (Fig. 3b).
We tested luciferase constructs in Hep3B cells expressing a dominant negative C/EBP protein (A-C/EBP)19, 20 and found significantly reduced differences in haplotype-specific expression (Fig. 3d, Supplementary Fig. 5b). We also tested luciferase constructs in NIH 3T3 cultured mouse fibroblast cells (Fig. 3e) and found no haplotype-specific expression difference, consistent with liver specificity. Addition of C/EBPα to the 3T3 cells restored the haplotype-specific effect (Fig. 3e). We altered other nucleotides besides rs12740374 in the consensus binding site predicted to be critical for C/EBP protein-DNA interactions21 and found that they were needed for transcriptional activation by the minor haplotype (Fig. 3c). Furthermore, we determined that C/EBPα binds to the site of rs12740374 in homozygous minor allele cells by chromatin immunoprecipitation (Supplementary Fig. 5c).
We tested whether C/EBP proteins can influence SORT1 expression via rs12740374. When we added A-C/EBP to Hep3B cells that are homozygous for the major allele, there was no difference in SORT1 expression (Fig. 3f). In contrast, when we added A-C/EBP to SK-HEP-1 cultured human hepatoma cells that are heterozygous (one minor allele), we observed a three-fold reduction in SORT1 expression (Fig. 3f). When we added C/EBPα to human embryonic stem (ES) cells that are homozygous for the minor allele (HUES-1), there was no change in SORT1 expression, presumably because ES cells do not harbour cofactors needed for transcriptional activation (Supplementary Fig. 5d). When we differentiated HUES-1 cells into endoderm, the first step towards hepatocyte differentiation22, addition of C/EBPα resulted in significantly increased SORT1 expression; in contrast, human ES cells homozygous for the major allele (HUES-9), when differentiated into endoderm, showed no expression difference (Supplementary Fig. 5d).
Together, these findings indicate that rs12740374 is the causal variant responsible for the liver-specific association between the 1p13 locus and gene expression and, by extension, the associations with LDL-C and MI risk.
Sort1 in Mouse Liver Alters Plasma Lipids
Of the genes differentially expressed in human liver by 1p13 genotype, the SORT1 gene showed the largest difference (Fig. 1c). SORT1 encodes the sortilin protein23, also known as neurotensin receptor 3, a protein that functions as a multiligand sorting receptor. Sortilin localizes to various intracellular compartments including the Golgi apparatus and has roles in both endocytosis and intracellular trafficking of other proteins24. To model the functional effects of altered SORT1 expression on lipids and lipoproteins, we performed knockdown and overexpression studies of Sort1 in the livers of mice. Importantly, we chose approaches to specifically alter gene expression in liver, because variation at the 1p13 locus results in a large SORT1 expression change in liver but no change in adipose tissues (Fig. 1c). Since sortilin is known to be highly expressed and have important physiological roles in adipocytes and neurons25, 26, we felt it was most appropriate to restrict knockdown and overexpression to liver to model the effects of the 1p13 locus on phenotype. Because wild-type mice have very low levels of plasma LDL-C compared to humans, we used “humanized” mice of various genetic backgrounds for our studies (Supplementary Fig. 6).
Adeno-associated virus serotype 8 (AAV8) has been demonstrated to appropriately target genes for specific expression in liver27, 28. AAV8 vector encoding the murine Sort1 gene driven by a liver-specific promoter (thyroglobulin) was delivered to mouse liver via intraperitoneal injection. A null AAV8 vector was used as a control. The Sort1 AAV resulted in increased sortilin levels in liver with no change in adipose (Supplementary Fig. 7a). Use of these viral vectors did not result in elevated alanine aminotransferase (ALT) levels (Supplementary Fig. 7b).
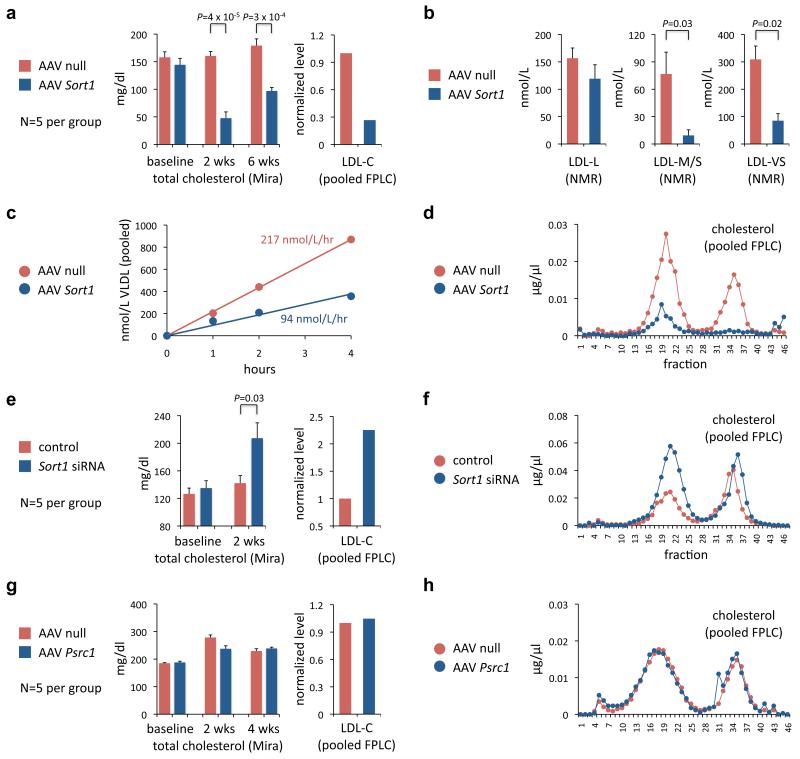
When compared with mice injected with null virus, Sort1-overexpressing Apobec1−/−; APOB Tg mice showed a marked decrease in total plasma cholesterol (70% reduction at two weeks, 46% reduction at six weeks) and LDL-C (73% reduction at two weeks) (Fig. 4a, d); consistent results were seen in three other mouse backgrounds (Supplementary Fig. 6, 7c–f). At six weeks the mice displayed a 73% reduction in very small LDL particles and an 88% reduction in medium small LDL particles (Fig. 4b), resulting in increased LDL peak particle size (22.0 nm vs. 20.9 nm, P = 0.05). These gain-of-function studies in mice were concordant with the genetic findings in human cohorts, in whom the 1p13 minor haplotype was associated with increased liver SORT1 expression as well as decreased LDL-C and, especially, very small LDL particles (Fig. 1a, c).
Figure 4. Overexpression or knockdown of Sort1 in mouse liver alters plasma lipids and lipoproteins.
Adeno-associated virus 8 (AAV8) vectors either containing no gene, murine Sort1 cDNA, or murine Psrc1 cDNA were administered via intraperitoneal injection; phosphate-buffered saline or siRNA duplex targeting firefly luciferase or mouse Sort1 and prepared in lipidoid formulation was administered weekly via tail vein injection at 2.0 mg/kg. Plasma samples were collected prior to injection and at various time points after injection, and were subjected: individually to analytical chemistry (Mira autoanalyzer) to measure total cholesterol (a, e, g); as pooled samples to FPLC (d, f, h), from which fractions 10 to 26 were used to calculate LDL-C levels (a, e, g); individually to NMR to measure LDL particle concentrations (b). P values calculated with unpaired t test, shown if P < 0.05. Error bars show s.e.m. a-d, Apobec1−/−; APOB Tg mice (five mice per group). b, NMR measurements at six weeks. LDL-L = large LDL; LDL-M/S = medium small LDL; LDL-VS = very small LDL. c, The mice were intraperitoneally injected with Pluronic F-127 detergent to block VLDL triglyceride lipolysis and permit assessment of the rate of VLDL secretion. Plasma samples were collected at baseline, one hour, two hours, and four hours after injection. VLDL particle concentrations were measured from pooled samples with NMR. e, f, Apobec1−/−; APOB Tg mice (five mice per group). g, h, Ldlr−/− mice (five mice per group).
To study hepatic VLDL secretion, we administered Pluronic F-127 detergent to the AAV-injected mice and measured lipoproteins at serial time points. We found a 57% decrease in the rate of VLDL secretion (Fig. 4c) and a similar decrease in the rate of triglyceride secretion (data not shown) in Sort1-overexpressing mice.
Chemically synthesized small interfering RNA (siRNA)-mediated knockdown of apoB or Pcsk9 in liver has been successful in determining the effects of these genes on plasma lipid levels29, 30. We used a similar approach to reduce Sort1 expression in mouse liver. We identified siRNA duplexes that effected >90% knockdown of Sort1 expression in cells (Supplementary Fig. 8a). We selected one chemically modified duplex with a low half-maximal inhibitory concentration (IC50) that did not induce cytokines in a human peripheral blood mononuclear cell assay (Supplementary Fig. 8b; data not shown) for large-scale preparation in a lipidoid formulation optimized for liver-specific delivery31 and injection into mouse tail veins. As a negative control in some experiments, we used a chemically modified, non-immunostimulatory siRNA duplex specific for the firefly luciferase gene. Sort1 siRNA achieved 70%-80% reduction in Sort1 expression in liver, confirmed to be due to siRNA-mediated cleavage, as well as reduced sortilin levels in liver with no change in adipose (Supplementary Fig. 9a–c).
Sort1 knockdown in Apobec1−/−; APOB Tg mice resulted in a 46% increase in total cholesterol compared to control mice at two weeks, with a more than two-fold increase in LDL-C (Fig. 4e, f). Consistent results were seen in two other mouse backgrounds, as well as a significant increase in the plasma VLDL level (Supplementary Fig. 6, 9e–g). We also compared plasma lipid levels in Sort1 knockout mice32 and wild-type mice and observed significantly higher total cholesterol and LDL-C levels in the knockout mice (Supplementary Fig. 9h), consistent with the results of liver-specific knockdown.
To confirm that the altered plasma VLDL levels in the overexpression and knockdown mice were due specifically to altered VLDL secretion from hepatocytes, we performed labelling experiments using primary hepatocytes isolated from these mice. With Sort1 knockdown, we observed a significant increase in labelled apoB-100 secretion; with Sort1 overexpression, there was decreased apoB-100 secretion (Supplementary Fig. 10).
Besides SORT1, PSRC1 displayed the greatest differential expression in human liver by 1p13 genotype (Fig. 1c). We used an AAV8 vector encoding the murine Psrc1 gene for mouse liver overexpression and did not observe any significant changes in total cholesterol or LDL-C levels (Fig. 4g, h).
A Novel Lipoprotein Regulatory Pathway
Through a series of studies in human cohorts, mice, and hepatocytes, we provide evidence that a single noncoding DNA variant at the chromosome 1p13 locus, rs12740374, influences LDL-C and MI risk via liver-specific transcriptional regulation of the SORT1 gene by C/EBP transcription factors. The clinical importance of this novel pathway is defined by the ~40% difference in MI risk between alternative 1p13 homozygotes, an effect comparable to those of common variants of LDLR and PCSK9 and larger than the effects of common variants in HMGCR (the target of statin drugs)11, 12. As the 1p13 minor allele frequency is about 30% in Europeans and is also common in other ethnicities including African Americans, Hispanics, Asian Indians, and Chinese8, 33, this locus is an important global genetic determinant of MI risk. We note that among lipid-regulating genes related to MI, SORT1 is unique for having been identified by GWAS mapping of common DNA variants, rather than by discovery of rare gene variants underlying Mendelian disorders.
In conclusion, our results nominate SORT1 as the causal gene at the 1p13 locus for LDL-C and MI and the sortilin pathway as a promising new target for therapeutic intervention in the reduction of LDL-C and prevention of MI. They also provide insights into mechanisms by which common noncoding genetic variants can lead to clinical phenotypes, rather than simply being markers for disease.
Methods Summary
The full Methods provides information about all experimental procedures: (1) description of association analyses in the population cohort; (2) description of genotype-expression analyses in human liver, subcutaneous adipose, and omental adipose samples; (3) details for generation of luciferase expression constructs; (4) details for conducting luciferase expression assays; (5) details for conducting SORT1 expression assays; (6) details for performing electrophoretic mobility shift assays; (7) details for performing chromatin immunoprecipitation assays; (8) description of siRNA screening and validation; (9) details for performing gene knockdown studies in mouse liver; (10) details for performing gene overexpression studies in mouse liver; (11) details for measuring lipids and lipoproteins by analytic chemistry, FPLC, and NMR; (12) details for performing VLDL secretion studies; and (13) details for performing hepatocyte apoB studies.
Supplementary Material
Acknowledgments
We thank David Altshuler, Edward Fisher, and John Maraganore for advice and guidance, and Akin Akinc, Jeffrey Billheimer, Robert Brown, Raymond Camahort, Deborah Cromley, Edwige Eduoard, Ilia Fuki, Casey Geaney, Greg Hinkle, Indu Kohaar, Satya Kuchimanchi, William Lagor, Frank Lau, David Lum, Martin Maier, Dawn Marchadier, Rachel Meyers, John Millar, Stuart Milstein, David Nguyen, Denise Perez, Derek Peters, Valeska Redon, Alessandra Rigamonti, Robert Schinzel, Mao-Sen Sun, Sue-Anne Toh, Aisha Wilson, and Katie Wojnoonski for assistance and suggestions. We acknowledge the National Heart, Lung, and Blood Institute (NHLBI) Gene Therapy Resource Program for providing support for viral vector production as well as the Vector Core laboratory of the University of Pennsylvania for producing the vectors. We acknowledge the members of the NHLBI Candidate Gene Association Resource (CARe) lipids working group for the contribution of association data in African Americans. This work was supported in part by a T32 grant in Cell and Molecular Training for Cardiovascular Biology from the United States National Institutes of Health (NIH), K99-HL098364 from the NIH, and the Clinician Scientist Program of the Harvard Stem Cell Institute (K.M.); a Medical Scientist Training Program grant from the NIH (A.S.); the intramural research program of the Division of Cancer Epidemiology & Genetics, National Cancer Institute, NIH (M.P.-O.); the Swedish Medical Research Council, Heart-Lung Foundation, and Påhlsson Foundation (M.O.-M., O.M.); U01-HL069757 from the NIH and research support from Quest Diagnostics, Inc. (R.M.K.); RC2-HL101864 from the NIH (S.K.); and P01-HL059407 and RC2-HL101864 from the NIH and a “Freedom to Discover” Unrestricted Cardiovascular Research Grant from Bristol-Myers Squibb (D.J.R.).
References
- 1.Rader DJ, et al. Monogenic hypercholesterolemia: new insights in pathogenesis and treatment. J. Clin. Invest. 2003;111:1795–1803. doi: 10.1172/JCI18925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown MS, Goldstein JL. Heart attacks: gone with the century? Science. 1996;272:629. doi: 10.1126/science.272.5262.629. [DOI] [PubMed] [Google Scholar]
- 3.Waters DD, et al. Lipid treatment assessment project 2: a multinational survey to evaluate the proportion of patients achieving low-density lipoprotein cholesterol goals. Circulation. 2009;120:28–34. doi: 10.1161/CIRCULATIONAHA.108.838466. [DOI] [PubMed] [Google Scholar]
- 4.Kathiresan S, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willer CJ, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallace C, et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am. J. Hum. Genet. 2008;82:139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kathiresan S, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aulchenko YS, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabatti C, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat. Genet. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teslovich TM, et al. Biological, clinical, and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samani NJ, et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myocardial Infarction Genetics Consortium et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berneis KK, Krauss RM. Metabolic origins and clinical significance of LDL heterogeneity. J. Lipid Res. 2002;43:1363–1379. doi: 10.1194/jlr.r200004-jlr200. [DOI] [PubMed] [Google Scholar]
- 14.Musunuru K, et al. Ion mobility analysis of lipoprotein subfractions identifies three independent axes of cardiovascular risk. Arterioscler. Thromb. Vasc. Biol. 2009;29:1975–1980. doi: 10.1161/ATVBAHA.109.190405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siri-Tarino PW, Williams PT, Fernstrom HS, Rawlings RS, Krauss RM. Reversal of small, dense LDL subclass phenotype by normalization of adiposity. Obesity (Silver Spring) 2009;17:1768–1775. doi: 10.1038/oby.2009.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linsel-Nitschke P, et al. Genetic variation at chromosome 1p13.3 affects sortilin mRNA expression, cellular LDL-uptake and serum LDL levels which translates to the risk of coronary artery disease. Atherosclerosis. 2010;208:183–189. doi: 10.1016/j.atherosclerosis.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 17.Darlington GJ, Wang N, Hanson RW. C/EBP alpha: a critical regulator of genes governing integrative metabolic processes. Curr. Opin. Genet. Dev. 1995;5:565–570. doi: 10.1016/0959-437x(95)80024-7. [DOI] [PubMed] [Google Scholar]
- 18.Osada S, Yamamoto H, Nishihara T, Imagawa M. DNA binding specificity of the CCAAT/enhancer-binding protein transcription factor family. J. Biol. Chem. 1996;271:3891–3896. doi: 10.1074/jbc.271.7.3891. [DOI] [PubMed] [Google Scholar]
- 19.Olive M, Williams SC, Dezan C, Johnson PF, Vinson C. Design of a C/EBP-specific, dominant-negative bZIP protein with both inhibitory and gain-of-function properties. J. Biol. Chem. 1996;271:2040–2047. doi: 10.1074/jbc.271.4.2040. [DOI] [PubMed] [Google Scholar]
- 20.Ahn S, et al. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol. Cell. Biol. 1998;18:967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller M, Shuman JD, Sebastian T, Dauter Z, Johnson PF. Structural basis for DNA recognition by the basic region leucine zipper transcription factor CCAAT/enhancer-binding protein alpha. J. Biol. Chem. 2003;278:15178–15184. doi: 10.1074/jbc.M300417200. [DOI] [PubMed] [Google Scholar]
- 22.Si-Tayeb K, et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersen CM, et al. Molecular identification of a novel candidate sorting receptor purified from human brain by receptor-associated protein affinity chromatography. J. Biol. Chem. 1997;272:3599–3605. doi: 10.1074/jbc.272.6.3599. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen MS, et al. The sortilin cytoplasmic tail conveys Golgi-endosome transport and binds the VHS domain of the GGA2 sorting protein. EMBO J. 2001;20:2180–2190. doi: 10.1093/emboj/20.9.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nykjaer A, et al. Sortilin is essential for proNGF-induced neuronal cell death. Nature. 2004;427:843–848. doi: 10.1038/nature02319. [DOI] [PubMed] [Google Scholar]
- 26.Shi J, Kandror KV. Sortilin is essential and sufficient for the formation of Glut4 storage vesicles in 3T3-L1 adipocytes. Dev. Cell. 2005;9:99–108. doi: 10.1016/j.devcel.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Kitajima K, et al. Complete prevention of atherosclerosis in apoE-deficient mice by hepatic human apoE gene transfer with adeno-associated virus serotypes 7 and 8. Arterioscler. Thromb. Vasc. Biol. 2006;26:1852–1857. doi: 10.1161/01.ATV.0000231520.26490.54. [DOI] [PubMed] [Google Scholar]
- 28.Tanigawa H, et al. Expression of cholesteryl ester transfer protein in mice promotes macrophage reverse cholesterol transport. Circulation. 2007;116:1267–1273. doi: 10.1161/CIRCULATIONAHA.107.704254. [DOI] [PubMed] [Google Scholar]
- 29.Soutschek J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 30.Frank-Kamenetsky M, et al. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc. Natl. Acad. Sci. U. S. A. 2008;105:11915–11920. doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akinc A, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng J, Racicott J, Morales CR. The inactivation of the sortilin gene leads to a partial disruption of prosaposin trafficking to the lysosomes. Exp. Cell Res. 2009;315:3112–3124. doi: 10.1016/j.yexcr.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 33.Keebler ME, et al. Association of blood lipids with common DNA sequence variants at 19 genetic loci in the multiethnic United States National Health and Nutrition Examination Survey III. Circ. Cardiovasc. Genet. 2009;2:238–243. doi: 10.1161/CIRCGENETICS.108.829473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.