Abstract
A novel cDNA clone encoding a cytochrome P450 gene has been isolated from the insecticide-susceptible strain of the red flour beetle, Tribolium castaneum (Herbst) (Coleoptera: Tenebrionidae). The nucleotide sequence of the clone, designated CYP345D3, was determined. The cDNA is 1554 bp in length and contains an open reading frame from base pairs 32 to 1513, encoding a protein of 493 amino acid residues and a predicted molecular weight of 57466 Daltons. The putative protein contains the classic heme-binding sequence motif FxxGxxxCxG (residues 430–439) conserved among all P450 enzymes as well as other characteristic motifs of the cytochrome P450s. Comparison of the deduced amino acid sequence with other CYP members shows that CYP345D3 shares 91% identity with the previously published sequence of CYP345D1 from the T. castaneum genome project and the nucleotide sequence identity between them is less than 80%. Phylogenetic analysis of amino acid sequences from members of various P450 families indicated close phylogenetic relationship of CYP345D3 with CYP6 of other insects than those from mammals and amore distant relationship to P450 from other families. CYP345D3 was submitted to GenBank, accession number EU008544.
Keywords: cytochrome P450, sequence analysis
Introduction
P450 enzymes (mixed function oxidases, cytochrome P450 monooxygenases), one of the most important enzyme systems involved in insecticide detoxification or activation, are a complex family of heme containing enzymes found in most organisms. To date, various kinds of P450 enzymes have been reported in animals, microorganisms, and plants, and classified into more than 36 families (Nelson et al. 1993; Zhou et al. 2002). P450 enzymes bind molecular oxygen and receive electrons from NADPH to introduce an oxygen atom to the substrate. In insects, the diverse functions of P450 enzymes range from the synthesis and degradation of ecdysteroids and juvenile hormones to the metabolism of xenobiotics (Andersen et al. 1994; Feyereisen 1999). P450 enzymes play important roles in adaptation of insects to toxic compounds in their host plants and are involved in metabolism of all commonly used insecticides. Also, they metabolize organophosphorus insecticide compounds to more active toxicants by activation of a P=S bond to a P=O bond (Feyereisen 1999; Sams et al. 2000; Scott and Wen 2001). However, in general, P450 enzymes mediate metabolic detoxification of other insecticides.
Nomenclature of P450 genes has been established to designate all gene members of the P450 super-family with a CYP prefix, followed by a numeral for the family, a letter for the subfamily, and a number for the individual gene (Nelson et al. 1996; Feyereisen 1999). This system defines that members of a family share >40% identity in amino acid sequence, and members of a subfamily share >55% identity (Feyereisen 1999).
The first insect P450 gene (CYP6A1) was isolated from an insecticide-resistant strain of the housefly, Musca domestica (Feyereisen et al. 1989). Subsequently, many P450 genes were cloned. More than 1958 sequences of P450 genes have now been registered in the GenBank database. In China, research on the molecular cloning aspect of P450 genes started recently. In Helicoverpa zea, a xanthotoxin-inducible cytochrome P450 cDNA (CYP6B8) was isolated (Li et al. 2000). Three new full-length cDNAs were cloned from Aedes albopictus (Zhou et al. 2001) and the full length of CYP6BF1 was also obtained from Plutella xylostella through the SMART (Switching Mechanism At 5′ end of the RNA Transcript) technique (Li et al. 2005). Nine CYP4 fragments from Culex pipiens Pallens (Teng et al. 2004), two CYP6 (Ai et al. 2004a) and ten CYP4 (Ai et al. 2004b) fragments from a susceptible strain of Helicoverpa armigera were cloned. In addition, two new P450 cDNA fragments were gained from a deltamethrin resistant strain of Musca domestica using the differential display PCR technique (Ma et al. 2005).
The red flour beetle, Tribolium castaneum (Herbst) (Coleoptera: Tenebrionidae), is a cosmopolitan and destructive pest of raw and processed cereal grains (Sokoloff 1977; Sinha and Watters 1985; Mills and Pedersen 1990). Direct feeding on the host grain enhances mould growth and excretion of hydroxyquinone compounds contaminates and causes damage to the grain. For more than 40 years, this species has shown its ability to develop resistance to insecticides and resistant strains have spread geographically, so that it remains as one of the major pests of stored products (Assié 2007). Occurrence of resistance to insecticide such as phosphine (PH3) in many strains of T. castaneum was reported from many countries in the 1970s (Champ and Dyte 1976). In this study, we undertook efforts to clone the P450 genes because there is evidence that P450 enzymes might be involved in some of the resistance (Miller et al. 1999). Here, we report a novel P450 gene (CYP345D3) from T. castaneum.
Materials and Methods
Insects
The PH3 susceptible strain of T. castaneum was from the Entomology Laboratory of Queensland Department of Primary Industry, Australia. The insects were cultured in the Key Laboratory of Entomology and Pest Management, Southwest University, Chongqing. The beetles were reared at 30 °C in whole wheat flour fortified with 5% (v/v) Brewer's yeast under standard conditions (Beeman and Stuart 1990). The last instar larvae were collected using a sieve to separate the insects from the medium.
Isolation of total RNA and synthesis of first strand cDNA
Total RNA for the amplification of cDNA fragments and the Rapid Amplification of cDNA Ends (RACE) were isolated from the last instar larvae of T. castaneum using TRNzol Reagent (Tiangen, www.tiangen.com). Twenty individuals were homogenized with at least 1 ml TRNzol Reagent in the glass homogenizer. The process of total RNA extraction and purification was carried out following the manufacturer°s instructions including a DNase (Takara, www.takara-bio.co.jp) treatment. Finally the total RNA (A260/A280= 1.8) was dissolved in 40 µl DEPC treated H2O and stored at -80 °C. The first strand cDNA was synthesized using 2 µg of DNase-treated total RNA by RevertAid™ First Strand cDNA Synthesis Kit (Fermentas Life Sciences, www.fermentas.com) with oligo(dT)-adaptors. The total volume of reverse transcriptional system was 25 µl. The reaction condition was performed according to the manufacturer's instructions and the reaction mixture was stored at -20 °C.
Degenerate primers and amplification of cDNA fragment
The degenerate primers used in PCR were designed as described by Kasai et al. (1998) and the nucleotide sequences of synthetic primers were as follows: 5′-CGGARACNHYNMGNAARTAYCC-3′ for the forward primer (DP1) and 5′-CGGGNCCNKCNCCRAANGG-3′ for the reverse primer (DP2). Degenerate PCR was conducted with TGradient PCR Thermal Cycler (Biometra, www.biometra.de) using rTaqTM polymerase (Takara). There was 2 µl cDNA in the total volume of 25 µl. The PCR program included an initial denaturation step of 3 min at 94 °C and then 30 cycles were run as follows: 94 °C for 30 sec, 50 °C for 30 sec and 72 °C for 45 sec with a final extension of 10 min at 72 °C. The PCR products were separated by 1.5 % agarose gel electrophoresis and stained with ethidium bromide (EB). The band of the expected size (243 bp) was excised and the fragment was recovered with the Gel Extraction Mini Kit (Watson Biotechnologies, Inc. Shanghai).
Cloning and sequencing of cDNA fragment
The purified 243 bp fragment was cloned into a pMD-18-T vector (Takara). After transformation into JM109 competent cells (Takara), the DNA inserts of the recombinant clones were amplified by PCR with degenerate primers used above, and sequenced in both directions (Invitrogen Life Technologies, www.invitrogen.com).
Rapid amplification of cDNA ends (3′ RACE and 5′ RACE)
The 3′ RACE was performed using the 3′-Full RACE Core Set Ver. 2.0 (Takara). The 3′ RACE adaptor primer 3AP (anti-sense): 5′-CTGATCTAGAGGTACCGGATCC-3′ and a genespecific primer 3GSP1 (sense): 5′-TTGAACCGGAAGTCAGATGTA-3′ were used for PCR with PrimeSTAR™ HS DNA polymerase (Takara) under the following conditions: an initial denaturation at 98 °C for 5 min, followed by 30 cycles of 98 °C for 10 sec, 55 °C for 20 sec and 72 °C for 1 min and a final extension at 72 °C for 10 min. A nested PCR was conducted under similar conditions using 1 µl of the first PCR product as template, the same 3′RACE adaptor primer and another gene-specific primer, 3GSP2: 5′CCTGAACGGTTTAGTGATGAG-3′. The PCR product was excised and sub-cloned into a pMD-18-T vector. Several recombinant clones were identified by PCR amplification with 3AP and 3GSP2 and then sequenced as described above.
The 5′ RACE was conducted with BD SMART™ cDNA Amplification Kit (Clontech, www.clontech.com). The 5′ region was amplified using a gene-specific antisense primer 5GSP1: 5′-CCTTCCCCAAATGGCAAATACG-3′ and a 5′ RACE adaptor primer, UPM: 5′-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3′. The PCR amplification was performed with Advantage™ 2 PCR Kit (Clontech) under the following conditions: an initial denaturation at 94 °C for 3 min, Mowed by 30 cycles of 94 °C for 30 sec, 65.5 °C for 30 sec and 72 °C for 2 min and a final extension at 72 °C for 10 min. Again, a nested PCR was conducted using 1 µl of the first PCR product as template, the 5′ adaptor primer NUP: 5′-AAGCAGTGGTATCAACGCAGAGT-3′ and the gene-specific primer 5GSP1 used above. The PCR condition was identical with that described above. The PCR product was excised and sub-cloned as described above.
Amplification of full-length cDNA was confirmed with NUP and FGSP1: 5′GAAAGTTTCGAAAATTGTGTG-3′. The cDNA in 5′ RACE was used as template. The PCR amplification was performed with Advantage™ 2 PCR Kit (Clontech) under the following conditions: an initial denaturation at 94 °C for 3 min, followed by 30 cycles of 94 °C for 30 sec, 55 °C for 30 sec and 72 °C for 2 min and a final extension at 72 °C for 10 min. All the gene-specific primers used in 3′ RACE and 5′ RACE was designed utilizing Primer Premier 5.0 (http://www.PremierBiosoft.com).
Sequence analysis
DNA sequence was determined using the ABI-PRISM 3730 sequencer (Invitrogen). Searching of similar sequences was performed using BlastP in the non-redundant protein sequences (nr) database of the NCBI website (http://www.ncbi.nlm.nih.gov). A phylogenetic tree was constructed by MEGA version 3.1 (Kumar et, al. 2004) using the method of Neighbor-Joining.
Results
cDNA and deduced amino acid sequence of the CYP345D3
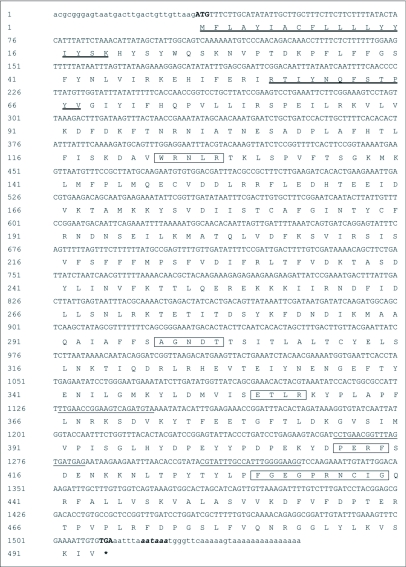
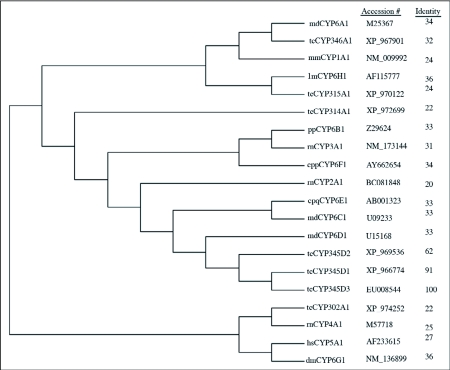
The full-length cDNA sequence with the deduced amino acid sequence below the nucleotide sequence (GenBank accession number, EU008544) is shown in Figure 1. The cDNA sequence was 1554 bp in length with an open reading frame of 1479 bp encoding 493 amino acid residues and had a predicted molecular weight of 57466 Daltons. The putative polydenylation signal AATAAA is shown in bold italic lowercase letters. The deduced protein sequence shares high identity (91% and 62%, respectively) to probable CYP345D1 and CYP345D2 (GenBank accession number, XP_966774 and XP_969536) from T. castaneum. Identity to other members of P450 families is shown in Figure 2. According to the alignment, the heme-binding sequence motif FxxGxxxCxG (residues 430–439) that is universal among P450 enzymes is found in the deduced amino acid sequence. After submission to the P450 nomenclature committee, the sequence was classified into a new family from Tribolium called CYP345, and it was named CYP345D3 using the nomenclature of Nelson et al. (1996) and Nelson (2006).
Figure 1.
Nucleotide and deduced amino acid sequence of CYP345D3. The nucleotides underlined show the positions of gene specific primers used in the experiment. The start codon ATG is indicated with bold and the stop codon TGA is indicated with bold and by an asterisk. Polydenylation signal AATAAA is shown in bold italic lowercase letters. The heme-binding sequence motif FxxGxxxCxG and other sequence motif are indicated by the boxed amino acids. The transmembrane domains are shown in deeply underlined amino acid residues.
Figure 2.
Phylogenetic relationship based on the amino acid sequence comparisons of Cytochrome P450s from various CYP families. GenBank accession numbers are shown followed the P450s' name. Identity is obtained by pairwise alignment of amino acid sequence of CYP345D3 with indicated P450s. Letter designation: cpp, Culex pipiens pollens; md, MUSCCJ domestica; cpq, Culex pipiens quinquefasciatus; lm, Locusta migratoria; tc, T. castaneum; dm; Drosophila melanogaster; pp, Papilio polyxenes; rn, Rattus norvegicus; hs, Homo sapiens; mm, Mus musculus.
Phylogenetic relationship with other P450 families
The deduced amino acid sequence of CYP345D3 contains all important motifs characteristic of the P450 enzymes, particularly the CYP6 family. Using MEGA version 3.1 software (Kumar et, al. 2004), a phylogenetic tree was constructed using the Neighbor-Joining method (Figure 2). The CYP345D3 sequence was found to be more closely related to CYP345D1 and CYP345D2 from T. castaneum and other insect CYP6 members than families of CYP1A1, CYP2A1, CYP3A1, CYP4A1, CYP5A1 from mammals and CYP302A1, CYP314A1, CYP315A1, CYP346A1 from T. castaneum. Not surprisingly, phylogenetic analysis of amino acid sequences from members of various P450 families indicated closer phylogenetic relationship of CYP345D3 with CYP6 members from T. castaneum and other insects than from mammals and a more distant relationship to P450s from other families.
Discussion
Gene cloning by PCR using degenerate primers derived from conserved amino acid sequences from other species has proven to be a powerful method to obtain related DNA sequences from the target species. Although the P450 super-family has a very divergent sequence and the overall homology maybe less than 40% even within the same family, particular in insects (Wang and Hobbs 1995), there are some function-critical sequence motifs preserved during evolution. Typically, the sequence features of P450 genes can be listed as follows. First, the classic heme-binding sequence motif FxxGxxxCxG (residues 430–439) that is conserved among all P450 enzymes (Ranasinghe and Hobbs 1998; He et al. 2002; Nelson 2006), is found in the deduced amino acid sequence, the cysteine of which is present in all P450 sequences. Secondly, the sequence motif (A/G) GxxT (residues 298–302) corresponds to an I-helix which is a conserved alpha helical region in proposed three dimensional models for a member of P450 protein based on the known structure of several bacterial enzymes (Von Wachennfeldt and Johnson 1995). The “T” (Threonine, Thr) is part of the molecular oxygen-binding site (Nelson 2006). Site directed mutagenesis studies have suggested that this highly conserved region around Thr302 could be involved in oxygen binding, while some of the hydrophobic residues preceding the Thr could be involved in substrate binding (Shimizu et al. 1989). Thirdly, the motif WxxxR (residues 123–127) corresponding to a C-helix sequence and conserved in most eukaryotic P450s has been found. Furthermore, the K-helix ExxR sequence motif (residues 355–358), the best conserved feature in P450s, and salt bridge formation in the mature protein has also been reported (Graham-Lorence and Peterson 1996; Nelson 2006). Additionally, the PERF (residues 411–414) sequence motif which is part of the meander before the heme thiolate ligand (Peterson and Graham-Lorence 1995) is present. Analysis with the SOSUI program (http://www.hgsc.bcm.tmc.edu, SearchLauncher, Human Genome Center, Baylor College of Medicine, Houston, TX) for protein secondary structure prediction (Hirokawa et al. 1998) indicates that the CYP345D3 is a membrane protein with two transmembrane domains located near the N-terminus. The primary transmembrane domain contains amino acid residues 1–19 and the secondary one contains residues 56–78. These highly hydrophobic regions serve as the signal peptide for cotranslational insertion of the protein into the target membrane appear in most P450 enzymes (Pernecky and Coon 1996). These results reveal that CYP345D3 contains characteristic functional domains for P450 enzymes.
The deduced amino acid sequence of CYP345D3 shares 91% identity with probable CYP345D1 as computed from the genome of T. castaneum, while the nucleotide sequences share less than 80% identity with each other. However, the CYP345D3 sequence is not present in the Tribolium assembly from the genome project. This suggsts that the Tribolium species are highly polymorphic, and the strain used here is different than that used in the genome project. Analysis of the CYP345D3 sequence, revealed the conserved functional sites. However, the real function of the sequence is still unknown.
Molecular cloning of CYP345D3 is just the first step to study the cytochrome P450 system in the red flour beetle, which has not been very well studied relative to their detoxification of insecticides. In future research, heterogenous expression of CYP345D3 such as in Escherichia coli and Pichia postoris can be used to identify the biochemical characteristics of this enzyme and validate the function of it. In conclusion, the isolation of the CYP345D3 sequence provides some basic knowledge to understand the P450 enzyme system in T. castaneum. However, better understanding of the system needs much further study.
Acknowledgements
We thank David R. Nelson for his assistance with the naming of CYP345D3. This research was funded in part by the National Natural Sciences Foundation (30570231), the Program for New Century Excellent Talents in University (NCET-04-0854), and the Specialized Research Fund for the Doctoral Program of Higher Education (20040625006) of China to Jin-Jun Wang.
References
- Ai Y, Qiu XH, He FQ. Cloning and sequence analysis of cytochrome P450 cDNA fragments CYP4 family from Helicoverpa armigera. Journal of Agricultural University of Hebei. 2004b;27:660–64. [Google Scholar]
- Ai Y, Qiu XH, Zhang LL, Li M, He FQ. Cloning and sequence analysis of cDNA fragments of cytochrome P450 from Helicoverpa armigera. Entomological Knowledge. 2004a;41:3227–231. [Google Scholar]
- Andersen JF, Utermohlen JG, Feyereisen R. Expression of housefly CYP6A1 and NADPH cytochrome P450 reductase in Escherichia coli and reconstitution of an insecticide metabolizing P450 system. Biochemistry. 1994;33:2171–2177. doi: 10.1021/bi00174a025. [DOI] [PubMed] [Google Scholar]
- Assie LK, Francis F, Gengler N, Haubruge E. Response and genetic analysis of malathion-specific resistant Tribolium castaneum (Herbst) in relation to population density. Journal of stored products research. 2007;43:33–44. [Google Scholar]
- Beeman RW, Stuart JJ. A gene for Lindanet Cyclodiene resistance in the red flour beetle Tribolium castaneum (Coleoptera: Tenebrionidae). Journal of Economic Entomology. 1990;83:1745–1751. [Google Scholar]
- Champ BR, Dyte CE. Report on the FAO global survey of pesticide susceptibility of stored grain pests. FAO Plant Protection Production Services No.5; FAO, Rome: 1976. [Google Scholar]
- Feyereisen R, Koener JF, Farnsworth DE, Nebert DW. Isolation and sequence of cDNA encoding a cytochrome P450 from an insecticide-resistant strain of the housefly, Musca domestica. Proceedings of National Academy of Science, USA. 1989;86:1465–1469. doi: 10.1073/pnas.86.5.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyereisen R. Insect P450 enzymes. Annual Review of Entomology. 1999;44:507–533. doi: 10.1146/annurev.ento.44.1.507. [DOI] [PubMed] [Google Scholar]
- Graham-Lorence SE, Peterson JA. Structural alignments of P450s and extrapolations to the unknown. Methods in Enzymology. 1996;272:315–326. doi: 10.1016/s0076-6879(96)72037-2. [DOI] [PubMed] [Google Scholar]
- He H, Chen AC, Davey RB, Ivie GW. Molecular cloning and nucleotide sequence of a new P450 gene, CYP319A1, from the cattle tick, Boophilus microplus. Insect Biochemistry and Molecular Biology. 2002;32:303–309. doi: 10.1016/s0965-1748(01)00091-1. [DOI] [PubMed] [Google Scholar]
- Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinfomatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- Kasai S, Shono T, Yamakawa M. Molecular cloning and nucleotide sequence of a cytochrome P450 cDNA from a pyrethroid-resistant mosquito, Culex quinquefasciatus Say. Insect Molecular Biology. 1998;7:185–190. doi: 10.1046/j.1365-2583.1998.72053.x. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Briefings in Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Li HS, Dai HG, Wei H. Molecular cloning and nucleotide sequence of CYP6BF1 from the diamondback moth, Plutella xylostella. 5pp. Journal of Insect Science. 2005;5:45. doi: 10.1093/jis/5.1.45. available online: http://insectscience.org/5.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XC, Berenbaum MR, Schuler MA. Molecular cloning and expression of CYP6B8: a xanthotoxin-inducible cytochrome P450 cDNA from Helicoverpa zea. Insect Biochemistry Molecular Biology. 2000;30:75–84. doi: 10.1016/s0965-1748(99)00102-2. [DOI] [PubMed] [Google Scholar]
- Ma CX, Li M, Qiu XH, He FQ, Liu HX. Cloning of cytochrome P450 ESTs from a deltamethrin-resistant strain of housefly, Musca domestica, using differential display RT-PCR technique. Acta Parasitology et Medical Entomologica Sinica. 2005;12:4210–215. [Google Scholar]
- Miller RJ, Davey RB, George JE. Characterization of pyrethroid resistance and susceptibility to coumaphos in Mexican Boophilus microplus (Acari: Ixodidae). Journal of Medical Entomology. 1999;36:533–538. doi: 10.1093/jmedent/36.5.533. [DOI] [PubMed] [Google Scholar]
- Mills R, Pedersen J. A Flour Mill Sanitation Manual. Eagan Press; 1990. [Google Scholar]
- Nelson DR, Kamataki T, Waxman DJ, Guengerich FP, Estabrook RW, Feyereisen R, Gonzalez FJ, Coon MJ, Gunsalus IC, Gotoh O, Okuda K, Nebert DW. The P450 superfamily: update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA and Cell Biology. 1993;12:1–51. doi: 10.1089/dna.1993.12.1. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW, Gunsalus IC, Nebert DW. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Phamacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- Nelson DR. Methods in Molecular Biology. Vol. 2. Humana Press; 2006. Cytochrome P450 Nomenclature. pp. 1–10. [DOI] [PubMed] [Google Scholar]
- Pernecky SJ, Coon MJ. N-terminal modifications that alter P450 membrane targeting and function. Methods in Enzymology. 1996;272:25–34. doi: 10.1016/s0076-6879(96)72005-0. [DOI] [PubMed] [Google Scholar]
- Peterson JA, Graham-Lorence SE. Bacterial P450s: Structural similarities and functional differences. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism and Biochemistry. Vol. 2. Plenum Press; 1995. pp. 151–180. [Google Scholar]
- Ranasinghe C, Hobbs AA. Isolation and characterization of two cytochrome P450 cDNA clones for CTP6B6 and CYP6B7 from Helicoverpa amigera (Hubner): Possible involvement of CYP6B7 in pyrethroid resistance. Insect Biochemistry and Molecular Biology. 1998;28:571–580. doi: 10.1016/s0965-1748(98)00045-9. [DOI] [PubMed] [Google Scholar]
- Sams C, Mason HJ, Rawbone R. Evidence for the activation of organophosphate pesticides by cytochromes P450 3A4 and 2D6 in human liver microsomes. Toxicology letters. 2000;116:3217–221. doi: 10.1016/s0378-4274(00)00221-6. [DOI] [PubMed] [Google Scholar]
- Scott JG, Wen Z. Cytochromes P450 of insects: the tip of the iceberg. Pest management science. 2001;57:958–967. doi: 10.1002/ps.354. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Sadeque AJM, Hatano M, Fujii-Kuriyama Y. Binding of axial ligands to cytochrome P-450d mutants: A difference absorption spectral study. Biochimica et Biophysica Acta. 1989;995:116–121. doi: 10.1016/0167-4838(89)90069-1. [DOI] [PubMed] [Google Scholar]
- Sinha RN, Watters FL. Insect pests of flour mills, grain elevators, and feed mills and their control. Agriculture Canada Publication No. 1776. Canadian Government Publishing Centre; Ottawa, Canada: 1985. [Google Scholar]
- Sokoloff A. The Biology of Tribolium. Oxford University Press; 1977. [Google Scholar]
- Teng D, Li XL, Gong MQ, Ma L, Lu XW, Sun Y, Zhu CL. Cloning and sequence analysis of cytochrome family 4 new genes from the deltamethrin-resistant mosquito, Culex pipiens Pallens. Chinese Journal of Parasitic Disease Control. 2004;17:2. [Google Scholar]
- Von Wachenfeldt C, Johnson EF. Structure of eukaryotic cytochrome P450 enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism and Biochemistry. 2nd edition. Plenum Press; 1995. pp. 183–223. [Google Scholar]
- Wang XP, Hobbs AA. Isolation and sequence analysis of a cDNA clone for a pyrethroid resistance in the housefly, Musca domestica. Insect Molecular Biology. 1995;4:135–140. doi: 10.1111/j.1365-2583.1995.tb00018.x. [DOI] [PubMed] [Google Scholar]
- Zhou GL, Huang JL, Wu Y, Wang L, Lao H. Molecular cloning and sequence analysis of three new full-length cDNAs of cytochrome P450 (CYP6N3v1-v3) from Aedes albopictus. Entomologica Sinica. 2001;8:2138–144. [Google Scholar]
- Zhou GL, Huang JL. Diversity and evolution of CYP6 family in insects. Entomological Knowledge. 2002;39:4246–251. [Google Scholar]