Abstract
A class of hairpin polyamides linked by 3,4-diaminobutyric acid, resulting in a β-amine residue at the turn unit, showed improved binding affinities relative to their α-amino-γ-turn analogs for particular sequences. We incorporated β-amino-γ-turns in six-ring polyamides and determined whether there are any sequence preferences under the turn unit by quantitative footprinting titrations. Although there was an energetic penalty for G·C and C·G base pairs, we found little preference for T·A over A·T at the β-amino-γ-turn position. Fluorine and hydroxyl substituted α-amino-γ-turns were synthesized for comparison. Their binding affinities and specificities in the context of six-ring polyamides demonstrated overall diminished affinity and no additional specificity at the turn position. We anticipate that this study will be a baseline for further investigation of the turn subunit as a recognition element for the DNA minor groove.
Keywords: Molecular recognition, DNA minor groove, Foldamer
Hairpin pyrrole-imidazole (Py/Im) polyamides are a class of programmable synthetic ligands able to bind a broad repertoire of DNA sequences with affinities and specificities comparable to those of DNA-binding proteins.1,2 They have been shown to localize to the nuclei of living cells3,4 and regulate endogenous gene expression by interfering with transcription factor/DNA interfaces.5–10 Discrimination of the four Watson–Crick base pairs is dependent upon Py/Im ring pairings in the minor groove. Pairing rules have been established whereby N-methyl imidazole/N-methylpyrrole (Im/Py) pairs target G·C, the reverse (Py/Im) target C·G, and Py/Py pairs target A·T and T·A.11–13
The turn unit in the hairpin is a recognition element, favoring T·A/A·T over G·C/C·G.14 There is an energetic penalty for unfavorable steric interaction with the exocyclic amine present at the edge of the G·C base pair (Fig. 1). The question arises whether discrimination between T·A and A·T with the turn unit can be achieved. Previous efforts toward the improvement of hairpin binding affinity have involved modifications of the turn unit. Early studies showed the optimal length of the turn element to be three methylene units, resulting in the use of γ-aminobutyric acid (γ-turn).15 Modification of the α-position of the parent γ-turn with (R)-2,4-diaminobutyric acid (α-amino-γ-turn) results in an approximately 15-fold increase in DNA-binding affinity.16 In contrast, hairpins containing the opposite enantiomer, (S)-α-amino-γ-turn, bind DNA with diminished affinities likely due to a steric clash of the amine with the wall of the minor groove. Polyamides containing α-hydroxy-γ-turns17 and α-diaminobutyric acid18 turns have been reported to impart additional elements of specificity, but with the cost of diminished binding affinities.
Figure 1.
Schematic diagram of six-ring hairpin polyamide (ImImPy-turn-PyPyPy) targeting the DNA sequence 5′-WWGGWW-3′. Dashed lines indicate hydrogen bonds between the polyamide and DNA base pairs.
In a formal sense, a DNA minor groove binding hairpin Py/Im polyamide is an early example of a class of oligomers encoded by the order of monomer units that fold to a desired shape with a specific function, referred to as ‘foldamers.’ 19–21 The ring order of Py/Im polyamides codes in a programmable manner for a specific, contiguous sequence of Watson–Crick base pairs. The turn unit is both a shape element as well as a DNA recognition element, allowing the molecule to fold in a U-conformation. The turn unit deserves more attention and this Letter represents an effort to create a baseline for the field.
Recently, we introduced a new class of hairpin polyamides linked by 3,4-diaminobutyric acid, resulting in a β-amino-γ-turn.22 These molecules showed improved binding affinities relative to their α-amino-γ-turn analogs for A/T-rich sequences. Additionally, polyamides containing the β-amino-γ-turn were found to have improved tolerance for synthetic modification at the amine position presumably due to their more central location on the floor of the minor groove. Due to limitations of quantitative DNase I footprinting titrations23–25 for compounds where Ka ≥ 1010 M–1, relative binding affinities for high affinity molecules are compared by using their thermal stabilization of DNA duplexes. It has been shown previously that increases in melting temperatures (ΔTm) of DNA duplexes bound by hairpin polyamides correlate with DNA-binding affinity.26,27
We report herein a comparison of the sequence specificities of hairpin Py/Im polyamides containing the α-amino-γ-turn and the β-amino-γ-turn. Additionally, we have synthesized both hydroxyl and fluoro-substituted γ-turns and determined their affinities and specificities in polyamides with analogous core ring pairs. By employing six-ring polyamides, which have lower binding affinities compared to eight-ring polyamides,28 we are able to determine reliable equilibrium association constants (Ka) via quantitative DNase I footprinting titrations, and compare them with DNA duplex thermal stabilizations.
Polyamide synthesis
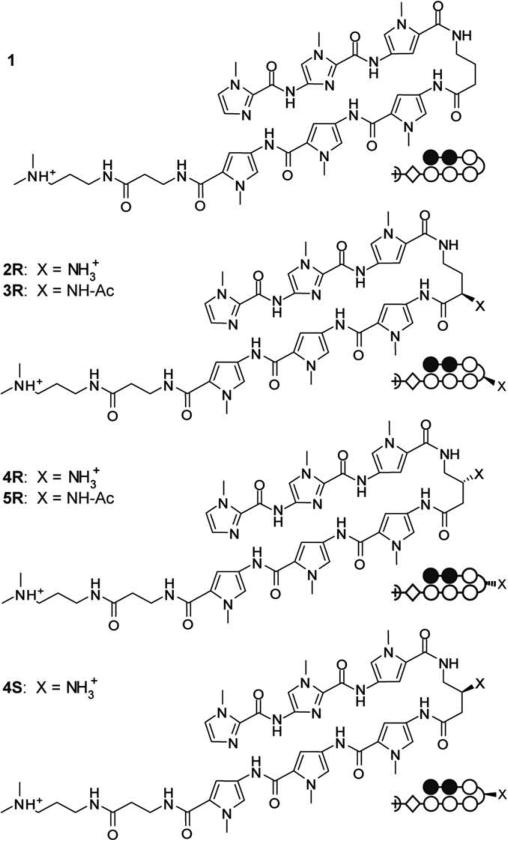
Six-ring hairpin polyamides (ImImPy-turn-PyPyPy) targeting the DNA sequence 5′-WWGGWW-3′ were synthesized by solid phase methods on Pam resin (Fig. 2, Supplementary data 1).22,29 In addition to the parent molecule containing an unsubstituted γ-aminobutyric acid hairpin (1), oligomers containing an amine moiety in the α and β turn positions (2R, 4R, 4S) and two polyamides with acetylated amines (3R, 5R) were also synthesized. The acetylated, or capped, molecules were used to determine tolerance for modifications at the turn. Acetylated turn units have been shown to improve nuclear uptake of polyamides.4
Figure 2.
Chemical and ball-and-stick structures of polyamides containing free and acetylated amines. Ball and stick symbols are defined as follows: pyrrole is denoted by an open circle, imidazole is denoted by a filled circle, and β-alanine is denoted by a diamond shape.
Thermal stabilization of DNA duplexes
Spectroscopic analyses were performed on the 11mer DNA duplex shown in Table 1.22 All hairpins analyzed provided an increase in melting temperature, confirming the formation of DNA/polyamide complexes. The ΔTm values obtained for polyamides containing a free amine were within error of each other. However, acetylated polyamide 3R (α-amino turn) showed a greater decrease in affinity than 5R (β-amino-γ-turn). As had been demonstrated with eight-ring hairpin molecules,22 improvements in binding affinities for β-over α-amino-γ-turn six-ring polyamides are more pronounced with decreasing imidazole content (Supplementary Fig. 1, Table 1).
Table 1.
Melting temperatures of DNA/polyamide complexes
| DNA = 5′-CTATGGTA GAC-3′ | |||
|---|---|---|---|
| Polyamides | Tm (°C) | ΔTm (°C) | |
| – | 45.1 (±0.1) | – | |
| 1 |

|
54.3 (±0.2) | 9.2 |
| 2R |
|
59.8 (±0.3) | 14.7 |
| 4R |
|
60.0 (±0.2) | 14.9 |
| 4S |
|
59.8 (±0.2) | 14.7 |
| 3R |
|
54.9 (±0.1) | 10.1 |
| 5R |
|
57.0 (±0.2) | 12.2 |
All values are derived from at least three melting temperature experiments, with standard deviations indicated in parentheses. ΔTm values are given as Tm (DNA/poiyamide) – Tm (DNA).
DNA binding affinity and sequence selectivity
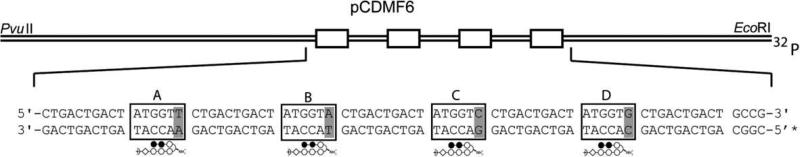
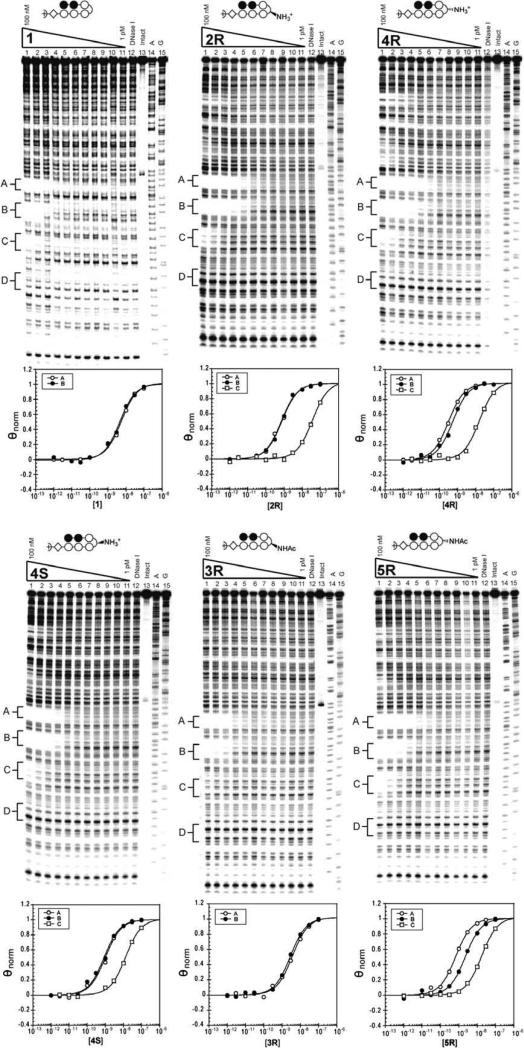
The plasmid pCDMF6 was prepared to characterize polyamides targeting the sequence 5′-WWGGWW-3′ (Fig. 3). The designed insert contains four binding sites, varying the nucleotide base pair present under the turn unit. Quantitative DNase I footprinting titrations were performed with the polyamides in order to measure their binding site affinities and specificities, as previously described.25 As expected, the parent hairpin containing the γ-turn retained the lowest binding affinity, while experiments for 2R, 4R, and 4S (Fig. 4, Table 2) corroborated the similar ΔTm values obtained for duplex stabilization. None of these molecules bound the G·C base pair, and binding to C·G was greatly diminished. Polyamide 4R revealed only a ~2-fold specificity for T·A over A·T (Table 3). Study of an additional polyamide series revealed similar trends (Supplementary Fig. 2, Table 2). Analysis of polyamides 3R and 5R (Fig. 4, Table 2) revealed a greater decrease in binding affinities for the α-amino-γ-turn molecule than the β. Between 2R and 3R there is a five-fold decrease over T·A, and an eight-fold decrease over A·T. For β-amino-γ-turn polyamides 4R and 5R, there are 1.7 and 3.2-fold decreases for T·A and A·T, respectively. 5R shows ~3-fold preference for binding T·A versus A·T (Table 3).
Figure 3.
Illustration of the EcoRI/PvuII restriction fragment derived from plasmid pCDMF6, used to characterize polyamides. The designed polyamide binding sites are indicated by boxes; single base-pair mismatches are indicated by shaded regions.
Figure 4.
Quantitative DNase I footprinting titration experiments for polyamides 1, 2R, and 4R (top, left to right) and 4S, 3R, and 5R (bottom, left to right) on the 5′ end labeled PCR product of plasmid pCDMF6: lanes 1–11, 100 nM, 30 nM, 10 nM, 3 nM, 1 nM, 300 pM, 100 pM, 30 pM, 10 pM, 3 pM, and 1 pM polyamide, respectively; lane 12, DNase I standard; lane 13, intact DNA; lane 14, A reaction; lane 15, G reaction. Respective isotherms are shown below.
Table 2.
Binding affinities (M–1) for polyamides
| 5′-ATGGTT-3′ | 5′-ATGGTA-3′ | 5′-ATGGTC-3′ | 5′-ATGGTG-3′ | |
|---|---|---|---|---|
| 1 | 2.1 (±0.6)× 108 | 2.0 (±0.5) × 108 | <107 | <107 |
| 2R | 1.6 (±0.7) × 109 | 1.7 (±0.9) × 109 | 3.9 (±2.0) × 107 | <107 |
| 4R | 3.3 (±0.1} × 109 | 1.8 (±0.2) × 109 | 5.0 (±1.1) × 107 | <107 |
| 4S | 1.1 (±0.1) × 109 | 1.3 (±0.2) × 109 | 6.0 (± 0,4) × 107 | <107 |
| 3R | 3.2 (±0.4) × 108 | 2.1 {±0.3) × 108 | <107 | <107 |
| 5R | 2.0 (±0.4) × 109 | 5.2 (±1.6) × 108 | 8.9 (±2.8) × 107 | <107 |
Equilibrium association constants are reported as mean values from three DNase I footprinting titration experiments. Standard deviations are shown in parentheses.
Table 3.
Relative binding affinities for polyamides
| R1,R2,R3 | T·A | A·T | C·G | G·C | |
|---|---|---|---|---|---|
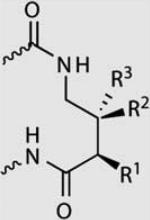
|
H, H, H | 1.0 | 1.0 | <0.01 | <0.01 |
| NH3+, H, H | 1.0 | 0.9 | 0.02 | <0.01 | |
| H, H, NH3+ | 1.0 | 0.5 | 0.02 | <0.01 | |
| H, NH3+, H | 1.0 | 1.2 | 0.05 | <0.01 | |
| NH-Ac, H, H | 1.0 | 0.7 | <0.01 | <0.01 | |
| H, H, NH-Ac | 1.0 | 0.3 | 0.04 | <0.01 |
Relative binding affinities are reported as ratios of binding affinities (Ka) as determined by DNase I footprinting titration experiments for polyamides targeting 5′-WGGWW-3′.
Fluoro and hydroxyl substituted turn units
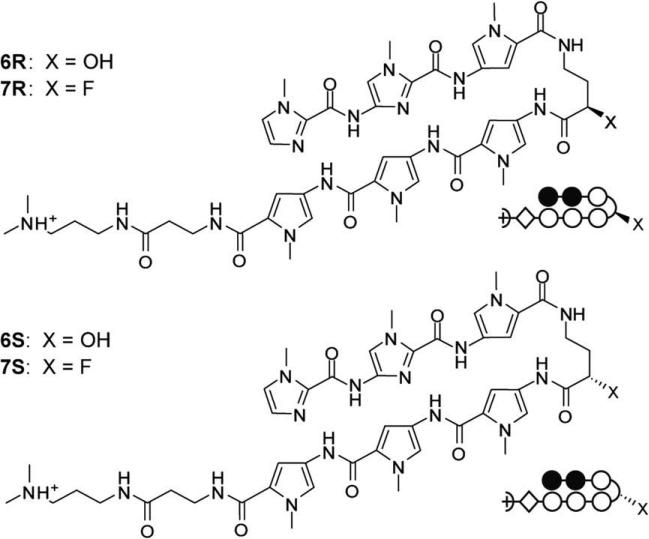
Fluorine and hydroxyl substituted hairpin turns were synthesized (Supplementary Figs. 3 and 4, Supplementary data 130,31) and incorporated in six-ring hairpin polyamides targeting the DNA sequence 5′-WWGGWW-3′ (Fig. 5). Polyamides were synthesized on Pam resin using standard solid phase methods.
Figure 5.
Chemical and ball-and-stick structures of polyamides containing fluoro and hydroxyl substituted hairpin turns. Ball and stick symbols are defined as follows: pyrrole is denoted by an open circle, imidazole is denoted by a filled circle, and β-alanine is denoted by a diamond shape.
Thermal stabilization analysis of the polyamides on an 11mer DNA duplex revealed that fluoro and hydroxyl substituted hairpins resulted in lower stabilizations than the corresponding amine-substituted and acetylated polyamides (Table 4, Supplementary Table 3). DNase I footprinting titrations on the plasmid pCDMF6 (Fig. 3, Supplementary Fig. 5) showed that both enantiomers of the hydroxyl and fluoro-hairpin turns resulted in decreased polyamide binding affinities relative to their amine substituted counterparts (Supplementary Table 4). Additionally, none of these subunits resulted in increased elements of specificity at the turn position of the molecule (Table 5).
Table 4.
Melting temperatures of DNA/ polyamide complexes
| DNA = 5′-CTATGGTA GAC-3′ | |||
|---|---|---|---|
| Polyamide | Tm (°C) | ΔTm (°C) | |
| – | 45.1 (±0.1) | – | |
| 6R |
|
55.1 (±0.2) | 10.0 |
| 7R |

|
53.4 (±0.2) | 8.3 |
| 6S |
|
50.9 (+0.2) | 5.8 |
| 7S |

|
53.0 (±0.2) | 7.9 |
All values are derived from at least three melting temperature experiments, with standard deviations indicated in parentheses. ΔTm values are given as Tm (DNA/polyamide) – Tm (DNA).
Table 5.
Relative binding affinities for polyamides
| R1,R2 | T·A | A·T | C·G | G·C | |
|---|---|---|---|---|---|
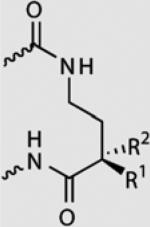
|
OH, H | 1.0 | 1.0 | <0.01 | <0.01 |
| F,H | 1.0 | 0.8 | <0.01 | <0.01 | |
| H, OH | 1.0 | 1.3 | 1.4 | 0.8 | |
| H, F | 1.0 | 0.6 | 0.6 | <0.01 |
Relative binding affinities are reported as ratios of binding affinities (Ka) as determined by DNase I footprinting titration experiments for polyamides targeting 5′-WGGWW-3′ Sites containing G·C at the hairpin position have relative affinities <0.01 (not shown).
By utilizing six-ring hairpin polyamides, we were able to combine DNase I footprinting titration and duplex stabilization analyses in order to fully characterize the binding preferences of various hairpin turn subunits. Although the hairpin turns investigated herein show modest DNA binding specificities, we anticipate that further study will yield moieties enabling discrimination amongst all four Watson–Crick base pairs and add an additional element for DNA recognition.
Supplementary Material
Acknowledgments
We are grateful to the National Institutes of Health for research support. We thank David M. Chenoweth for helpful discussions. Mass spectrometry analyses were performed in the Mass Spec-trometry Facility of the Division of Chemistry and Chemical Engineering at the California Institute of Technology.
Footnotes
Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bmcl.2009.03.072.
References and notes
- 1.Dervan PB. Bioorg. Med. Chem. 2001;9:2215. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 2.Dervan PB, Edelson BS. Curr. Opin. Struct. Biol. 2003;13:284. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 3.Best TP, Edelson BS, Nickols NG, Dervan PB. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12063. doi: 10.1073/pnas.2035074100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edelson BS, Best TP, Olenyuk B, Nickols NG, Doss RM, Foister S, Heckel A, Dervan PB. Nucleic Acids Res. 2004;32:2802. doi: 10.1093/nar/gkh609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olenyuk BZ, Zhang GJ, Klco JM, Nickols NG, Kaelin WG, Dervan PB. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16768. doi: 10.1073/pnas.0407617101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kageyama Y, Sugiyama H, Ayame H, Iwai A, Fujii Y, Huang LE, Kizaka-Kondoh S, Hiraoka M, Kihara K. Acta Oncol. 2006;45:317. doi: 10.1080/02841860500486648. [DOI] [PubMed] [Google Scholar]
- 7.Fukuda N, Ueno T, Tahira Y, Ayame H, Zhang W, Bando T, Sugiyama H, Saito S, Matsumoto K, Mugishima H, Serie K. J. Am. Soc. Nephrol. 2006;17:422. doi: 10.1681/ASN.2005060650. [DOI] [PubMed] [Google Scholar]
- 8.Nickols NG, Dervan PB. Proc. Natl. Acad. Sci. U.S.A. 2007;104:10418. doi: 10.1073/pnas.0704217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nickols NG, Jacobs CS, Farkas ME, Dervan PB. ACS Chem. Biol. 2007;2:561. doi: 10.1021/cb700110z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuda H, Fukuda N, Yao EH, Ueno T, Sugiyama H, Matsumoto K. J. Hypertens. 2008;26:S197. doi: 10.1097/hjh.0b013e3283207fe1. [DOI] [PubMed] [Google Scholar]
- 11.Kielkopf CL, Baird EE, Dervan PD, Rees DC. Nat. Struct. Biol. 1998;5:104. doi: 10.1038/nsb0298-104. [DOI] [PubMed] [Google Scholar]
- 12.White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB. Nature. 1998;391:468. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]
- 13.Pelton JG, Wemmer DE. Proc. Natl. Acad. Sci. U.S.A. 1989;86:5723. doi: 10.1073/pnas.86.15.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu CF, Phillips JW, Trauger JW, Farkas ME, Belitsky JM, Heckel A, Olenyuk BZ, Puckett JW, Wang CCC, Dervan PB. Tetrahedron. 2007;63:6146. doi: 10.1016/j.tet.2007.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mrksich M, Parks ME, Dervan PB. J. Am. Chem. Soc. 1994;116:7983. [Google Scholar]
- 16.Herman DM, Baird EE, Dervan PB. J. Am. Chem. Soc. 1998;120:1382. [Google Scholar]
- 17.Zhang W, Minoshima M, Sugiyama H. J. Am. Chem. Soc. 2006;128:14905. doi: 10.1021/ja064369l. [DOI] [PubMed] [Google Scholar]
- 18.Farkas ME, Tsai SM, Dervan PB. Bioorg. Med. Chem. 2007;15:6927. doi: 10.1016/j.bmc.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gellman SH, Adams BR, Dado GP. J. Am. Chem. Soc. 1990;112:460. [Google Scholar]
- 20.Appella DH, Christianson LA, Klein DA, Powell DR, Huang XL, Barchi JJ, Gellman SH. Nature. 1997;387:381. doi: 10.1038/387381a0. [DOI] [PubMed] [Google Scholar]
- 21.Gellman SH. Accounts Chem. Res. 1998;31:173. [Google Scholar]
- 22.Dose C, Farkas ME, Chenoweth DM, Dervan PB. J. Am. Chem. Soc. 2008;130:6859. doi: 10.1021/ja800888d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brenowitz M, Senear DF, Shea MA, Ackers GK. Methods Enzymol. 1986;130:132. doi: 10.1016/0076-6879(86)30011-9. [DOI] [PubMed] [Google Scholar]
- 24.Senear DF, Dalmaweiszhausz DD, Brenowitz M. Electrophoresis. 1993;14:704. doi: 10.1002/elps.11501401112. [DOI] [PubMed] [Google Scholar]
- 25.Trauger JW, Dervan PB. Methods Enzymol. 2001;340:450. doi: 10.1016/s0076-6879(01)40436-8. [DOI] [PubMed] [Google Scholar]
- 26.Pilch DS, Poklar N, Gelfand CA, Law SM, Breslauer KJ, Baird EE, Dervan PB. Proc. Natl. Acad. Sci. U.S.A. 1996;93:8306. doi: 10.1073/pnas.93.16.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pilch DS, Poklar N, Baird EE, Dervan PB, Breslauer KJ. Biochemistry. 1999;38:2143. doi: 10.1021/bi982628g. [DOI] [PubMed] [Google Scholar]
- 28.Trauger JW, Baird EE, Dervan PB. Nature. 1996;82:559. doi: 10.1038/382559a0. [DOI] [PubMed] [Google Scholar]
- 29.Baird EE, Dervan PB. J. Am. Chem. Soc. 1996;118:6141. [Google Scholar]
- 30.Busby GW. Ph.D. Thesis. Harvard University; 1974. pp. 48–54. [Google Scholar]
- 31.Hoshi H, Aburaki S, Iimura S, Yamasaki T, Naito T, Kawaguchi H. J. Antibiot. 1990;43:858. doi: 10.7164/antibiotics.43.858. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.