Abstract
Hepatocyte nuclear factor-1β(HNF-1β) is a transcription factor required for the expression of several renal cystic genes and whose prenatal deletion leads to polycystic kidney disease (PKD)1. We show here that inactivation of Hnf1b from postnatal day 10 onward does not elicit cystic dilations in tubules after their proliferative morphogenetic elongation is over. Cystogenic resistance is intrinsically linked to the quiescent state of cells. In fact, when Hnf1b deficient quiescent cells are forced to proliferate by an ischemiareperfusion injury, they give rise to cysts, owing to loss of oriented cell division. Remarkably, in quiescent cells, the transcription of crucial cystogenic target genes is maintained even in the absence of HNF-1β. However, their expression is lost as soon as cells proliferate and the chromatin of target genes acquires heterochromatin marks. These results unveil a previously undescribed aspect of gene regulation. It is well established that transcription is shut off during the mitotic condensation of chromatin2,3. We propose that transcription factors such as HNF-1β might be involved in reprogramming gene expression after transcriptional silencing is induced by mitotic chromatin condensation. Notably, HNF-1β remains associated with the mitotically condensed chromosomal barrels. This association suggests that HNF-1β is a bookmarking factor that is necessary for reopening the chromatin of target genes after mitotic silencing.
PKD, one of the most prevalent life-threatening human genetic diseases4, is characterized by the progressive dilation of renal tubules that eventually form cysts, often leading to end-stage renal failure. The homozygous inactivation of most cystic genes leads to a rapid and severe PKD phenotype in mice5,6. Notably, in the last few years several studies have shown that there is a key postnatal developmental switch that defines the onset and the severity of the polycystic phenotype7–11. However, the molecular mechanism at the basis of this differential propensity to develop cysts or not remains poorly defined.
We have previously shown that HNF-1β, a transcription factor expressed in the kidney12, controls the expression of cystic genes, including Pkhd1 (encoding polycystic kidney and hepatic disease-1), Pkd2 (encoding polycystic kidney disease-2) and Umod (encoding uromodulin)1. Hnf1b inactivation in the prenatal mouse kidney results in severe PKD. Mutations in HNF1B are very frequent in individuals suffering from renal cysts or dysplasia13,14, often associated with a particular form of diabetes mellitus, known as maturity onset diabetes of the young type 5 (MODY5).
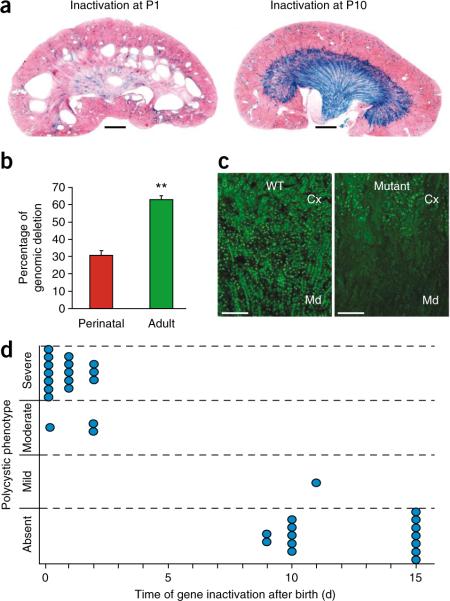
The drastic phenotype elicited by prenatal inactivation of Hnf1b has until now prevented the analysis of the function of this protein in already formed, normally shaped renal tubules. To address this question, we adopted a strategy of a postnatal inducible inactivation. With the use of an interferon (Poly(I)–Poly(C))-inducible Cre system (MxCre; ref. 15) and a floxed allele of Hnf1b16, we analyzed the effect of Hnf1b inactivation at key time points during postnatal renal maturation in mouse pups. To monitor the extent of recombination efficiency, we took advantage of a tester allele of the Cre recombinase activity (ROSA26R; ref. 17). Our results showed that inactivation of Hnf1b in young pups elicited cystic dilations, even with a limited extent of recombination (Fig. 1). In fact, all mice injected with the inducer of the Cre recombinase between 0 d and 2 d after birth developed polycystic disease 30 d later (Fig. 1a,d). These lesions progressed to generate markedly enlarged kidneys 60 d later (Supplementary Fig. 1). In contrast, mice induced on day 10 after birth did not develop any cystic lesions within 1 to 6 months later (Fig. 1a,d and data not shown). This result was not due to a lower extent of gene inactivation, as the degree of recombination in noncystic mice induced in older mice was paradoxically higher than that detected in cystic mice injected at birth (Fig. 1a,b). Moreover, these noncystic mice had lost the expression of HNF-1β protein in the medulla and at the cortico-medullary boundary, as revealed by immunostaining (Fig. 1c). All of these data indicate that HNF-1β function is necessary for maintaining normal tubular morphology only during a narrow time window that corresponds to the first few days after birth. Recent studies have monitored the effects of the inactivation of cystic disease-associated genes (Ift88 (encoding polaris), Kif3a (encoding kinesin-3A) and Pkd1) at various time points in early postnatal life7–11. Similar to what we have observed in this study, mice developed a prompt and dramatic PKD phenotype only when these genes were inactivated shortly after birth, whereas, if the inactivation was carried out later, a much slower onset of the cystic phenotype could be observed (after a minimum of 5–6 months)7–11. Collectively, these data suggest that cyst formation requires more than just the loss of cystic disease–associated gene expression and suggest the existence of unknown cystogenetic triggering events.
Figure 1.
Time-restricted cystogenic competence. (a) Morphology of kidneys from mice where Hnf1b was inactivated at either P1 or P10 and killed 30 d later. X-gal staining reveals the extent of Cre recombination (ROSA26R). Scale bar, 1 mm. (b) Percentage of Hnf1b deletion in mice injected at birth (perinatal) or in adulthood (adult), determined by qRT-PCR on genomic DNA. Data are means ± s.e.m.; n = 7 per group. **P < 0.01. (c) HNF-1β protein immunodetection in kidneys from 30-day old mice where Cre was activated at P10. Cx, cortex; Md, medulla. Scale bar, 100 μm. (d) Diagram showing the time-dependent cystogenic competence of Hnf1b deletion. Each circle represents one mouse.
It has been suggested that cell proliferation participates in cyst formation (reviewed in ref. 18); nevertheless, the dramatic resistance to cyst development from postnatal day 10 (P10) on occurs in a phase when the kidney is still actively growing. To clarify the mechanisms of this restricted cystogenetic competence, we decided to characterize the pattern of renal cell proliferation throughout this transition. Our results demonstrated that cystogenic competence correlates with a particular intense and localized pattern of cell proliferation that characterizes postnatal nephron morphogenesis and elongation (Supplementary Data and Supplementary Fig. 2).
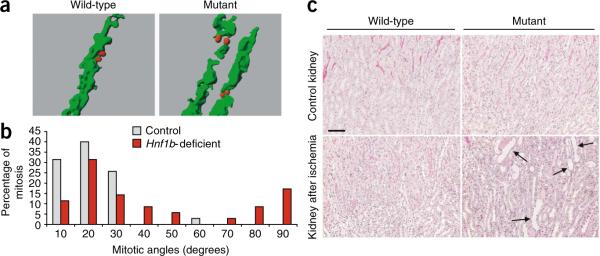
During postnatal kidney maturation, proliferation progressively declines and almost disappears when the organ has reached its final adult size19. Quiescent renal tubular cells in adults can be forced to reenter the cell cycle after ischemia-reperfusion (I/R) injury20. We found that the pattern of cell proliferation after I/R is very similar to that of elongating tubules at P0–P2, with locally intense and cell cycle-synchronized proliferation in the cortico-medullary region (Supplementary Fig. 2). We have previously shown that tubular elongation during early postnatal nephron maturation is linked to oriented cell division21. Notably, when we analyzed the distribution of the mitotic angles of regenerating tubular cells, we observed a strikingly biased orientation comparable to that observed during nephron morphogenesis (Fig. 2a). Ninety-five percent of mitotic angles were smaller than 30°, with an average of 15° (Fig. 2b). These data indicate that the morphogenetic pattern of intense, synchronized and oriented cell proliferation is programmed to be reactivated during renal regeneration.
Figure 2.
Tubular regeneration elicits distorted cell division and dilations in Hnf1b deficiency. (a) Three-dimensional reconstruction of typical examples of longitudinally (left) or perpendicularly (right) oriented mitoses. Tubules are in green; mitoses are in red. (b) Comparison of the distribution of the mitotic angles in regenerating tubules from wild-type (n = 35, from six mice) or Hnf1b-deficient mice (n = 35, from four mice). The Mann-Whitney U test shows a highly statistically significant different distribution of the mitotic angle between mutant and control mice (P < 0.005). (c) Morphology (periodic acid–Schiff staining) of nonischemic control kidneys (top) and kidneys after ischemia (bottom) from wild-type or Hnf1b-inactivated mice, 2 weeks after surgery. Normal-shaped Hnf1b-inactivated tubules underwent dilations (arrows in bottom right image) when forced to reenter the cell cycle. Scale bar, 50 μm.
The correlation between morphogenetic proliferation and cystogenic competence made us wonder whether the locally intense and synchronized proliferation could be the triggering cystogenic event in the context of homozygous deletion of a cystic disease–associated gene. To assess this point, we forced morphologically normal Hnf1b-deficient quiescent tubules (Fig. 2c) to reenter the cell cycle by an I/R-induced injury. Mutant mice, in which the gene was deleted around P60, developed marked dilations and cysts 2 weeks after I/R injury (Fig. 2c). These lesions occurred at the cortico-medullary boundary, the region where the most intense regenerative proliferation takes place in kidneys after ischemia and where Hnf1b is efficiently deleted (Fig. 1a–c). In contrast, we never observed cysts in injured control littermates (Fig. 2c). Hnf1b inactivation in quiescent cells did not elicit a prompt increase in proliferative response or tubular dilation (Supplementary Fig. 3 and data not shown). However, once proliferation was triggered, Hnf1b-deficient kidneys tended to proliferate more than the controls (Supplementary Fig. 3). In addition, we observed that the mitotic orientation of proliferating mutant cells was significantly distorted compared to the controls (Fig. 2a,b). Taken together, our results indicate that the lack of HNF-1β is a permissive (necessary but not sufficient) condition to elicit tubular dilation. In this context, we showed that proliferative bursts, occurring during tubular elongation or regeneration, are the crucial triggering events that can provoke cystic tubular dilations.
We next tried to elucidate the mechanisms underlying the synergistic effect of proliferation on Hnf1b deficiency. HNF-1β controls the expression of several genes, including Pkd2, Pkhd1, Umod and Kif12 (ref. 22). As shown above, Hnf1b gene deletion at P0–P2 resulted in a marked PKD phenotype (Fig. 1). Quantitative RT-PCR (qRT-PCR) showed that this phenotype was accompanied by a dramatic decrease in Umod, Pkhd1, Pkd2 and Kif12 expression (Fig. 3a) similar to that observed with the prenatal inactivation of Hnf1b (ref. 1). The identification of genomic HNF-1 binding sites that are highly conserved throughout evolution allowed us to identify additional potential target genes involved in cell polarity, cystogenesis or both, including Crb3 (encoding crumbs homolog-3), Tcfap2b (transcription factor AP-2β), Tmem27 (encoding transmembrane protein-27) and Bicc1 (encoding bicaudal C homolog 1) (data not shown). All of these genes contained sites that were bound in vivo by HNF-1β (Supplementary Fig. 4). The expression of these genes was markedly downregulated when Hnf1b was inactivated at P0 (Fig. 3a). However, in kidneys where Hnf1b was deleted after P10, we were surprised to see that there was no drop in the expression of these target genes (Fig. 3a). The only exception was Umod, whose expression dropped to a level similar to that observed after perinatal inactivation (Fig. 3a).
Figure 3.
Effect of proliferation on HNF-1β target gene expression and chromatin modifications. (a) Renal expression of Umod, Pkhd1, Pkd2, Kif12, Crb3, Tcfap2b, Tmem27 and Bicc1 mRNA transcripts in wild-type or in mutant adult kidneys 3–4 weeks after the inactivation that was induced in proliferative P0 pups or in quiescent kidneys at P30, respectively. Data are means ± s.e.m.; n = 4–8 for each experimental group. *P < 0.05; **P < 0.01. Ribosomal protein L13 (Rpl13) was used to normalize values. (b) BrdU and Pkd2 (top) or Kif12 (bottom) staining in kidneys of wild-type and mutant mice 2 weeks after ischemia. Colocalization analysis shows that, in wild-type tubules after ischemia, Pkd2 and Kif12 expression (brown cytoplasm) is maintained in cells that have proliferated (blue nuclei). In contrast, in Hnf1b-deficient tubules, Pkd2 and Kif12 expression is selectively turned off in cells that have proliferated. Scale bars, 100 μm. (c) Chromatin modifications in the mIMCD3 cell line expressing a HNF-1β dominant-negative mutant. Chromatin immunoprecipitation of activating (H3K9ac) or inhibiting (H3K9me3 or H3K27me3) histone H3 modifications of the Pkhd1 gene at various locations with respect to the transcriptional start site (indicated on the x axis) in control cells or in cells expressing a dominant-negative mutant of HNF-1β. The results are representative of a set of three independent experiments. Error bars represent s.e.m. *P < 0.05. (d) Chromatin immunoprecipitation of activating modifications (H3K9ac or H3K4me3) of histone H3 on the Pkhd1 gene at various locations with respect to the transcriptional start site (indicated on the x axis) in kidneys from wild-type or Hnf1b-deleted mice inactivated shortly after birth. Immunoprecipitation of total histone H3 is used as a control. Enrichments are calculated with respect to the Rpl13 gene for activating modifications and for the Acta2 gene (encoding smooth muscle α-actin) for repressive modifications. The results are representative of a set of three independent experiments. *P < 0.05.
The paradoxical resilient expression of most target genes could have been explained by a particularly long half-life of messenger RNAs that may have compensated and masked a possible drop in the transcription initiation rate after Hnf1b deletion. To assess this point, we quantified the short-lived heterogeneous-nuclear RNA (hnRNA) that can be detected only in actively transcribed genes, just before splicing has taken place. We found that the transcription initiation rate of these targets was not significantly altered by Hnf1b deletion in quiescent kidneys in vivo (Supplementary Fig. 5). Notably, the decrease in the Pkd2 transcription rate at P0 was smaller than that observed in mature mRNA (Fig. 3a and Supplementary Fig. 5), indicating that the expression of this gene was probably also controlled at the post-transcriptional level. Immunohistochemistry confirmed that in noncystic, Hnf1b deficient tubules (deleted at P10–P15 or in adult mice), Pkd2 and Kif12 expression was not affected (Supplementary Fig. 6 and data not shown). However, when proliferation waves occurred in cells that had lost HNF-1β, either via I/R-induced injury or during tubular elongation, the expression of target genes was markedly reduced (Supplementary Fig. 6). It is noteworthy that Pkd2 or Kif12 expression did not change in the tubules of wild-type mice after ischemia (Supplementary Fig. 6 and data not shown) or in mutant cells that did not proliferate (Supplementary Fig. 7).
To further validate these observations, we subjected Hnf1b-mutant adults to I/R-induced injury followed by a continuous administration of BrdU to label all cells that had undergone proliferation after the inactivation. We inspected the kidneys of mice killed long after the proliferative wave had occurred (14 d) for the presence of BrdU incorporation and Pkd2 or Kif12 expression. A careful examination of the double-stained tissue sections showed that Pkd2 and Kif12 staining tended to be lost in BrdU-positive cells that had been forced to proliferate in the absence of HNF-1β (Fig. 3b). Collectively, these observations indicate that HNF-1β target genes may be classified into two distinct groups. In the first group (class 1 genes), exemplified by Umod, transcription markedly drops after Hnf1b deletion, regardless of the proliferative status of cells. In the second group (class 2 genes, including Pkhd1, Pkd2, Kif12, Crb3, Tcfap2b, Tmem27 and Bicc1), once activated, transcription no longer requires the persistent and redundant action of HNF-1β if cells remain quiescent. However, this resilient transcription does not persist when proliferation occurs.
It is known that when cells enter mitosis, chromatin is condensed and forms the typical heavily packed mitotic chromosomes. Throughout the progression of these events, gene transcription is shut down2,3. At the exit from mitosis, gene expression must be reactivated to restore the typical tissue- and differentiation-specific transcriptional program. Our observations could be ascribed to an essential involvement of HNF-1β in inducing the post-mitotic chromatin reopening and gene transcriptional reactivation of class 2 genes. Notably, Hnf1a, a gene closely related to Hnf1b, also has a key role in the establishment, maintenance or both of an open chromatin structure at the Pah gene locus (encoding phenylalanine hydroxylase) during liver development23. To verify the impact of Hnf1b deletion on chromatin status in proliferative cells, we carried out chromatin immunoprecipitation with antibodies specific for active chromatin modifications including histone H3 Lys4 trimethylation (H3K4me3) and H3 Lys9 acetylation (H3K9ac) and inactive H3 Lys9 (H3K9) or Lys27 (H3K27) trimethylation (H3K9me3 or H3K27me3). To this end, we used a mouse inner medullary collecting duct cell line (mIMCD3) expressing an inducible dominant-negative form of HNF-1β (ref. 24). Class 2 genes underwent silencing and heterochromatinization upon HNF-1β inhibition: the acetylation of H3K9 was significantly reduced, whereas the trimethylation of H3K9 and H3K27 was significantly increased (Fig. 3c). In addition, when we inspected the same sites in vivo, we observed a comparable loss of activating chromatin modifications on target genes in perinatal mutant kidneys (Fig. 3d and Supplementary Fig. 8). In summary, our results suggest that Hnf1b deletion in a proliferative context has a drastic effect on the chromatin status of its target genes.
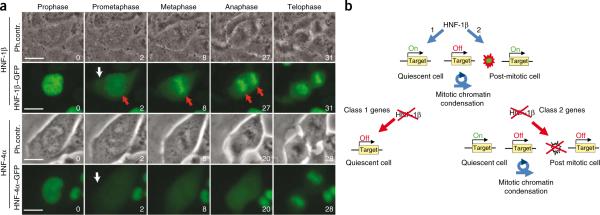
During cell cycle progression in higher eukaryotes, after nuclear envelope breakdown, transcription factors are released from chromatin. This leads to the transcriptional silencing that accompanies mitosis. To further characterize the function of HNF-1β, we monitored its localization during the cell cycle. With the use of a HNF-1β–GFP fusion protein, we discovered that, contrary to the classical behavior of transcription factors (shown here by an HNF4α–GFP fusion protein), HNF-1β is not dissociated from chromatin during mitosis (Fig. 4a). Rather, upon nuclear envelope breakdown, a substantial fraction of HNF-1β remains associated with the mitotic chromatin traveling with the condensed chromosomal barrels throughout the entire mitotic process (Fig. 4a and Supplementary Video 1). Our results support a previously undescribed concept and indicate that transcription factors such as HNF-1β may be involved in gene expression reprogramming after the silencing accompanying mitotic chromatin condensation is relieved. Thus, HNF-1β could act as a bookmarking factor that marks target genes for a rapid transcriptional reactivation after mitosis.
Figure 4.
HNF-1β behaves as a bookmarking factor. (a) Time-lapse microscopy of mIMCD3 cells expressing either HNF1β–GFP or HNF4α–GFP fusion proteins. For each fusion protein, phase contrast (top) and green fluorescence (bottom) are shown at crucial time points. The time (in min) is indicated in the bottom right corner of each frame. The white arrows indicate the sudden appearance of cytoplasmic GFP fusion protein upon nuclear envelope breakdown. The red arrows indicate the persistence of the HNF-1β–GFP fusion protein on mitotically condensed chromosomes. Scale bars, 20 μm. (b) A mitotic transcriptional switch in polycystic kidney disease. HNF-1β activates gene expression via two distinct mechanisms: by recruiting the transcriptional machinery to the promoters of target genes and/or by inducing the reopening of chromatin after a silencing mitotic chromatin condensation. Class 1 genes are silenced whenever HNF-1β is inactivated, independent of the proliferative state of cells. Class 2 gene expression is transiently maintained in quiescence and is lost as soon as cells have progressed through a mitotic chromatin condensation in the cell cycle.
In Hnf1b mutant mice, a severe polycystic kidney phenotype is observed only when this transcription factor is inactivated in highly proliferating tubular cells. These cells are cycling very intensely and probably need to promptly reestablish the expression of cystic disease–associated genes as soon as the cells reenter interphase. These cystic disease–associated genes are probably necessary to maintain the proper alignment of cell division during tubular elongation. The fact that HNF-1β remains associated with mitotic chromatin during mitosis in these proliferating cells might provide a useful and rapid mechanism to reactivate the expression of crucial target genes necessary for preventing tubular dilation. In Hnf1b deficiency, proliferation triggers a positive feedback loop leading to the persistence of a spatially distorted hyperproliferation. It is tempting to speculate that an analogous mechanism might explain the unexpected refractory response to cystic gene inactivation in adult mice7–11,25. The transcription networks downstream of ciliary and cystic genes26,27, similarly to HNF-1β, could maintain the expression of antiproliferative targets in mutant cells in a quiescent metastable state. Notably, it has been recently shown that PKD model mice treated with roscovitine, a cyclin-dependent kinase inhibitor, show long-lasting decreased cystogenesis28, further suggesting that inhibition of proliferative bursts may have a beneficial effect on this pathology.
In conclusion, our study indicates that transcription factors such as HNF-1β may control gene expression via two distinct mechanisms. The first involves the maintenance of active transcription from its target promoters. The second involves a new aspect in the control of gene expression and concerns the reprogramming of transcription by counterbalancing the silencing effect induced by mitotic chromatin condensation (Fig. 4b). The implications of these observations shed light on the crucial connection between genetically determined predispositions and environmentally induced proliferative stress conditions.
ONLINE METHODS
Mice
MxCre mice were as previously described15, as were the ROSA26R strain17 and the Hnf1bflox/flox mice16. We used both Hnf1bflox/+ and Hnnf1b+/+ mice as controls (indicated here as wild type). We conducted procedures in mice in accordance with French government policies (Ministère de l'Agriculture).
Time course of cystogenesis
We injected wild-type and Hnf1bflox/flox (mutant) mice carrying a MxCre transgene intraperitoneally with 20 μg per g body weight of 2 mg ml−1 Poly(I)–Poly(C) (Sigma) on alternate days for a total of three injections. We injected Poly(I)–Poly(C) into either P0–P2 newborns (n = 15 and n = 18 for wild-type and mutants, respectively) or P9–P15 young mice (n = 17 and n = 15 for wild-type and mutants, respectively). We killed the mice 30 d later, except for six mutant and six control mice injected at P0–P2 that we killed 60 d later.
Proliferation pattern
We injected BrdU (100 mg per kg body weight; Sigma) intraperitoneally in either control mice at P0 and P9 or in mice after ischemia, 1 d after surgery. We killed the mice (n = 4–6 per group) 24 h later.
Renal ischemia
We induced renal ischemia in adult male mice as previously described29, with minor modifications. We deliberately chose to carry out the injury in males, as they are more susceptible to I/R than females30. For proliferation studies, six control mice underwent renal ischemia and BrdU injection, and we killed them 48 h after surgery. For Hnf1b inactivation studies, we subjected 8- to 10-week-old Hnf1bflox/+ (wild type; n = 16) and Hnf1bflox/flox (mutant; n = 26) mice to renal ischemia 1 week after Poly(I)–Poly(C) injections. We killed the mice either 2 d (n = 6 for each genotype for mitotic angle measurements) or 14 d (n = 10 and n = 20 for wild-type and mutant mice, respectively, for histological studies) after surgery. In four wild-type and eight mutant mice killed at day 14, we continuously infused BrdU by osmotic minipumps (2002, Alzet; 30 mg per kg body weight per d) over 2 weeks.
Cell culture lines
The mIMCD3 cell line (53A), expressing the inducible HNF1β–ΔC mutant form in fusion with the GFP, as well as the protocol for induction, has been already described24. We generated HNF-4α–GFP by fusing the full-length rat complementary DNA of HNF4α with the EGFP in pEGFP-C1 (Clontech); we made stably transfected clones, resistant to G418, in the mIMCD3 cell line.
Histological analysis and X-gal staining
For histological analysis, we fixed organs in 4% paraformaldehyde and embedded them in paraffin. We stained sections with periodic acid-Schiff. We performed X-gal staining as previously described31.
Immunohistochemistry and immunofluorescence
The antibodies and the procedures used are described in the Supplementary Methods.
Proliferation index
We incubated sections with a rabbit antibody to Ki-67 (Microm) diluted 1 in 50, treated them with a rabbit-specific, peroxidase-linked antibody (Zymed) diluted 1 in 100 and stained them by 3,3'-diaminobenzidine (Dako). We determined the number of Ki-67-positive tubular cells in ten randomly selected fields at the cortico-medullary junction. We calculated the proliferation index as the number of Ki-67-positive nuclei for the total number of tubular nuclei.
Mitotic angle measurement
We removed kidneys 2 d after ischemia. The methodology for mitotic angle measurements has been already described21.
Time-lapse microscopy
For live microscopy, we grew mIMCD3 cells in IBIDI plates (81156) and maintained them at 37 °C and 7% CO2 in a live cell recorder Biostation IM (Nikon). We acquired phase-contrast and fluorescence images every 60 s.
Quantitative reverse transcription PCR
We obtained total RNAs from kidneys by the TRIzol procedure (Invitrogen). We treated the RNAs with DNase (DNase I, RNase-free, Roche) and reverse transcribed them (oligo dT primer) according to the manufacturer's protocols (Superscript II, Invitrogen). We carried out qRT-PCR on a Stratagene system using SYBR Green. We designed primers with PrimerExpress 2.0 software (Applied Biosystems). The primer sequences are detailed in Supplementary Table 1. We measured the levels of Pkd2, Phkd1, Umod, Kif12, Crb3, Tfcap2b, Tmem27 and Bicc1 in control and mutant mice inactivated either perinatally (n = 3–6) or in adulthood (n = 4–6 wild type and 4–9 mutants). We tested for cyclin B1 (Ccnb1) in kidneys after ischemia and contralateral kidneys (n = 3).
Chromatin immunoprecipitation
We prepared chromatin from kidneys as previously described1. We treated mIMCD3 cells (clone 53A) with 10 nM mifepristone (Invitrogen) or vehicle (ethanol) over 5 d and crosslinked them with 1% formaldehyde for 10 min at 24 °C. We prepared chromatin as previously described32. We sonicated the chromatin (Bioruptor) in SBAR buffer (50 mM HEPES, 140 mM NaCl, 1 mM EDTA, 1% Triton, 0.1% sodium deoxycholate and 0.1% SDS with protease inhibitors Complete (Roche) and phenylmethylsulphonyl fluoride 0.5 mM). We then incubated chromatin with antibody to HNF-1β (Santa Cruz, sc22840), antibody to histone H3 (Abcam, ab1791), antibody to histone H3K9me3 (Abcam, Ab8898), antibody to histone H3K27me3 (Abcam, ab6002), antibody to histone H3K9ac (ab4441), antibody to histone H3K4me3 (Abcam, ab46540) and IgG as control. We recovered immunocomplexes with Protein A Dynabeads (Invitrogen). We quantified DNA, purified as previously described32, by qRT-PCR. We calculated enrichments with respect to a site located in the Rpl13 gene (chr8:125.989.647–125.989.703) for activating modifications and with respect to a site located in the Acta2 gene (chr:19:34.319.550–34.319.600) for repressive modifications. Primer sequences are detailed in Supplementary Table 1.
Statistical analyses
Data are means ± s.e.m. We evaluated differences between the experimental groups with the Mann-Whitney U test.
Additional methods
Detailed methodology is described in the Supplementary Methods.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to S. Bettiol, J.B. Weitzman and M. Yaniv for critical advice. We are grateful to R. Sandford (Cambridge Institute of Medical Research) for Pkd2-specific antibodies and to B. Viollet, (Institut Cochin, Paris) for the GFP-HNF-4α fusion construct. MxCre mice were kindly provided by K. Rajewsky (Harvard Medical School, Boston), and the ROSA26R strain was provided by P. Soriano (Fred Hutchinson Cancer Research Center, Seattle).
We thank E. Perret and P. Roux from the “Plate-Forme d'Imagerie Dynamique,” Institut Pasteur, Paris) for assistance and advice. This work was supported by Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, Université Paris Descartes, Polycystic Kidney Disease Foundation, Société de Néphrologie, Agence Nationale Recherche, Fondation de la Recherche Médicale. F.V. was a recipient of a fellowship from Société de Néphrologie and from Fundación La Caixa.
Footnotes
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
Note: Supplementary information is available on the Nature Medicine website.
References
- 1.Gresh L, et al. A transcriptional network in polycystic kidney disease. EMBO J. 2004;23:1657–1668. doi: 10.1038/sj.emboj.7600160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottesfeld JM, Forbes DJ. Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- 3.Komura J, Ikehata H, Ono T. Chromatin fine structure of the c-MYC insulator element/DNase I-hypersensitive site I is not preserved during mitosis. Proc. Natl. Acad. Sci. USA. 2007;104:15741–15746. doi: 10.1073/pnas.0702363104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torres VE, Harris PC, Pirson Y. Autosomal dominant polycystic kidney disease. Lancet. 2007;369:1287–1301. doi: 10.1016/S0140-6736(07)60601-1. [DOI] [PubMed] [Google Scholar]
- 5.Fischer E, Gresh L, Reimann A, Pontoglio M. Cystic kidney diseases: learning from animal models. Nephrol. Dial. Transplant. 2004;19:2700–2702. doi: 10.1093/ndt/gfh533. [DOI] [PubMed] [Google Scholar]
- 6.Guay-Woodford LM. Murine models of polycystic kidney disease: molecular and therapeutic insights. Am. J. Physiol. Renal Physiol. 2003;285:F1034–F1049. doi: 10.1152/ajprenal.00195.2003. [DOI] [PubMed] [Google Scholar]
- 7.Davenport JR, et al. Disruption of intraflagellar transport in adult mice leads to obesity and slow-onset cystic kidney disease. Curr. Biol. 2007;17:1586–1594. doi: 10.1016/j.cub.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lantinga-van Leeuwen IS, et al. Kidney-specific inactivation of the Pkd1 gene induces rapid cyst formation in developing kidneys and a slow onset of disease in adult mice. Hum. Mol. Genet. 2007;16:3188–3196. doi: 10.1093/hmg/ddm299. [DOI] [PubMed] [Google Scholar]
- 9.Piontek K, Menezes LF, Garcia-Gonzalez MA, Huso DL, Germino GG. A critical developmental switch defines the kinetics of kidney cyst formation after loss of Pkd1. Nat. Med. 2007;13:1490–1495. doi: 10.1038/nm1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel V, et al. Acute kidney injury and aberrant planar cell polarity induce cyst formation in mice lacking renal cilia. Hum. Mol. Genet. 2008;17:1578–1590. doi: 10.1093/hmg/ddn045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takakura A, Contrino L, Beck AW, Zhou J. Pkd1 inactivation induced in adulthood produces focal cystic disease. J. Am. Soc. Nephrol. 2008;19:2351–2363. doi: 10.1681/ASN.2007101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coffinier C, Barra J, Babinet C, Yaniv M. Expression of the vHNF-1/HNF-1β homeoprotein gene during mouse organogenesis. Mech. Dev. 1999;89:211–213. doi: 10.1016/s0925-4773(99)00221-x. [DOI] [PubMed] [Google Scholar]
- 13.Lindner TH, et al. A novel syndrome of diabetes mellitus, renal dysfunction and genital malformation associated with a partial deletion of the pseudo-POU domain of hepatocyte nuclear factor-1β. Hum. Mol. Genet. 1999;8:2001–2008. doi: 10.1093/hmg/8.11.2001. [DOI] [PubMed] [Google Scholar]
- 14.Ulinski T, et al. Renal phenotypes related to hepatocyte nuclear factor-1β (TCF2) mutations in a pediatric cohort. J. Am. Soc. Nephrol. 2006;17:497–503. doi: 10.1681/ASN.2005101040. [DOI] [PubMed] [Google Scholar]
- 15.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 16.Coffinier C, et al. Bile system morphogenesis defects and liver dysfunction upon targeted deletion of HNF-1β. Development. 2002;129:1829–1838. doi: 10.1242/dev.129.8.1829. [DOI] [PubMed] [Google Scholar]
- 17.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 18.Igarashi P, Somlo S. Genetics and pathogenesis of polycystic kidney disease. J. Am. Soc. Nephrol. 2002;13:2384–2398. doi: 10.1097/01.asn.0000028643.17901.42. [DOI] [PubMed] [Google Scholar]
- 19.Marquez MG, Cabrera I, Serrano DJ, Sterin-Speziale N. Cell proliferation and morphometric changes in the rat kidney during postnatal development. Anat. Embryol. (Berl.) 2002;205:431–440. doi: 10.1007/s00429-002-0262-9. [DOI] [PubMed] [Google Scholar]
- 20.Nony PA, Schnellmann RG. Mechanisms of renal cell repair and regeneration after acute renal failure. J. Pharmacol. Exp. Ther. 2003;304:905–912. doi: 10.1124/jpet.102.035022. [DOI] [PubMed] [Google Scholar]
- 21.Fischer E, et al. Defective planar cell polarity in polycystic kidney disease. Nat. Genet. 2006;38:21–23. doi: 10.1038/ng1701. [DOI] [PubMed] [Google Scholar]
- 22.Gong Y, et al. HNF-1β regulates transcription of the PKD modifier gene Kif12. J. Am. Soc. Nephrol. 2009;20:41–47. doi: 10.1681/ASN.2008020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pontoglio M, Faust DM, Doyen A, Yaniv M, Weiss MC. Hepatocyte nuclear factor 1alpha gene inactivation impairs chromatin remodeling and demethylation of the phenylalanine hydroxylase gene. Mol. Cell. Biol. 1997;17:4948–4956. doi: 10.1128/mcb.17.9.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Z, et al. Mutations of HNF-1β inhibit epithelial morphogenesis through dysregulation of SOCS-3. Proc. Natl. Acad. Sci. USA. 2007;104:20386–20391. doi: 10.1073/pnas.0705957104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takakura A, et al. Renal injury is a third hit promoting rapid development of adult polycystic kidney disease. Hum. Mol. Genet. 2009;18:2523–2531. doi: 10.1093/hmg/ddp147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhunia AK, et al. PKD1 induces p21(waf1) and regulation of the cell cycle via direct activation of the JAK-STAT signaling pathway in a process requiring PKD2. Cell. 2002;109:157–168. doi: 10.1016/s0092-8674(02)00716-x. [DOI] [PubMed] [Google Scholar]
- 27.Li X, et al. Polycystin-1 and polycystin-2 regulate the cell cycle through the helix-loop-helix inhibitor Id2. Nat. Cell Biol. 2005;7:1202–1212. doi: 10.1038/ncb1326. [DOI] [PubMed] [Google Scholar]
- 28.Bukanov NO, Smith LA, Klinger KW, Ledbetter SR, Ibraghimov-Beskrovnaya O. Long-lasting arrest of murine polycystic kidney disease with CDK inhibitor roscovitine. Nature. 2006;444:949–952. doi: 10.1038/nature05348. [DOI] [PubMed] [Google Scholar]
- 29.Runembert I, et al. Recovery of Na-glucose cotransport activity after renal ischemia is impaired in mice lacking vimentin. Am. J. Physiol. Renal Physiol. 2004;287:F960–F968. doi: 10.1152/ajprenal.00064.2004. [DOI] [PubMed] [Google Scholar]
- 30.Park KM, Kim JI, Ahn Y, Bonventre AJ, Bonventre JV. Testosterone is responsible for enhanced susceptibility of males to ischemic renal injury. J. Biol. Chem. 2004;279:52282–52292. doi: 10.1074/jbc.M407629200. [DOI] [PubMed] [Google Scholar]
- 31.Pontoglio M, et al. Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria and renal Fanconi syndrome. Cell. 1996;84:575–585. doi: 10.1016/s0092-8674(00)81033-8. [DOI] [PubMed] [Google Scholar]
- 32.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.