Abstract
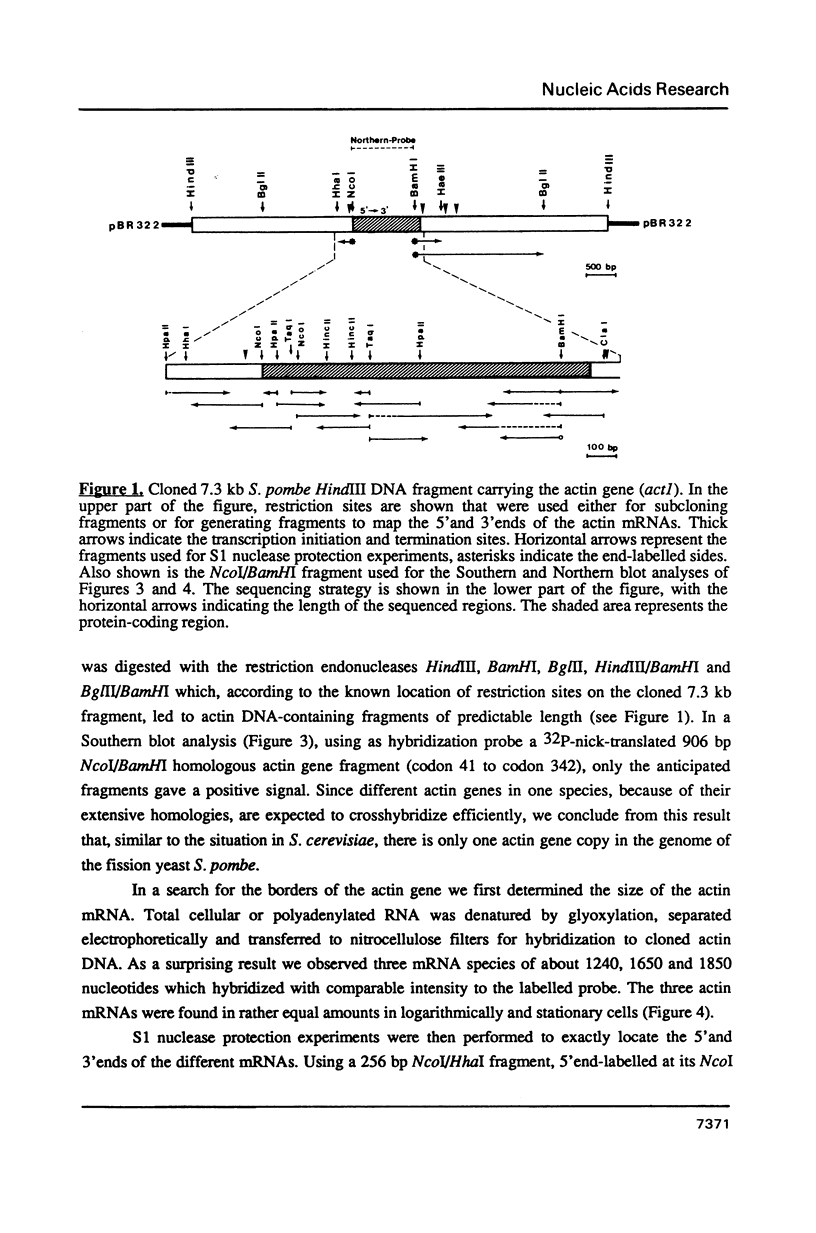
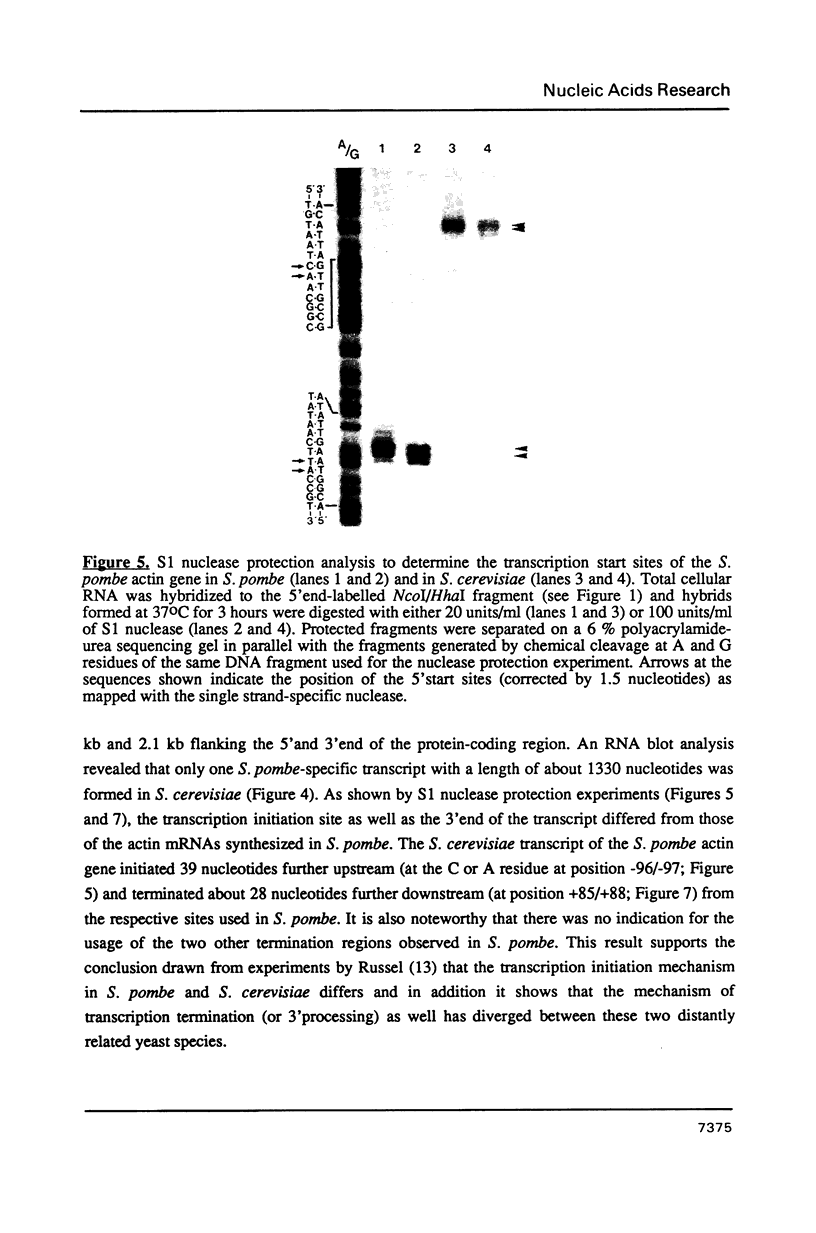
The actin gene of the fission yeast Schizosaccharomyces pombe has been isolated by using as a hybridization probe cloned actin DNA from the budding yeast Saccharomyces cerevisiae. In contrast to most actin genes studied from diverse eukaryotic species, the S. pombe gene is not interrupted by introns. The protein sequence deduced from the nucleotide sequence of the gene shows that the S. pombe actin is more closely related to the mammalian gamma-actin than to the actin of S. cerevisiae. Three transcripts of 1240, 1650 and 1850 nucleotides having the same 5' end but differing in the length of their 3' untranslated region are generated in the fission yeast. Only one messenger RNA of 1330 nucleotides is formed from the S. pombe actin gene in S. cerevisiae. Contrary to the observation made with other S. pombe genes transcribed in the budding yeast, the heterologous actin gene transcript is initiated 39 nucleotides upstream of the initiation start site used in the homologous yeast. The mRNA termination (or 3' processing) mechanism in the two ascomycetes also differs as the 3'end of the S. pombe actin gene transcript in S. cerevisiae does not coincide with either of the three 3'ends mapped in the fission yeast.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aho S., Tate V., Boedtker H. Multiple 3' ends of the chicken pro alpha 2(I) collagen gene. Nucleic Acids Res. 1983 Aug 25;11(16):5443–5450. doi: 10.1093/nar/11.16.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt F. W., Bothwell A. L., Knapp M., Siden E., Mather E., Koshland M., Baltimore D. Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3' ends. Cell. 1980 Jun;20(2):293–301. doi: 10.1016/0092-8674(80)90615-7. [DOI] [PubMed] [Google Scholar]
- Boardman M., Basi G. S., Storti R. V. Multiple polyadenylation sites in a Drosophila tropomyosin gene are used to generate functional mRNAs. Nucleic Acids Res. 1985 Mar 11;13(5):1763–1776. doi: 10.1093/nar/13.5.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallwitz D., Seidel R. Molecular cloning of the actin gene from yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1980 Mar 11;8(5):1043–1059. doi: 10.1093/nar/8.5.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallwitz D., Sures I. Structure of a split yeast gene: complete nucleotide sequence of the actin gene in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 May;77(5):2546–2550. doi: 10.1073/pnas.77.5.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford C. J., Gallwitz D. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983 Jun;33(2):519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- Losson R., Fuchs R. P., Lacroute F. Yeast promoters URA1 and URA3. Examples of positive control. J Mol Biol. 1985 Sep 5;185(1):65–81. doi: 10.1016/0022-2836(85)90183-4. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mertins P., Gallwitz D. Nuclear pre-mRNA splicing in the fission yeast Schizosaccharomyces pombe strictly requires an intron-contained, conserved sequence element. EMBO J. 1987 Jun;6(6):1757–1763. doi: 10.1002/j.1460-2075.1987.tb02428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellen W., Donath C., Moos M., Gallwitz D. The nucleotide sequences of the actin genes from Saccharomyces carlsbergensis and Saccharomyces cerevisiae are identical except for their introns. J Mol Appl Genet. 1981;1(3):239–244. [PubMed] [Google Scholar]
- Nellen W., Gallwitz D. Actin genes and actin messenger RNA in Acanthamoeba castellanii. Nucleotide sequence of the split actin gene I. J Mol Biol. 1982 Jul 25;159(1):1–18. doi: 10.1016/0022-2836(82)90028-6. [DOI] [PubMed] [Google Scholar]
- Ng R., Abelson J. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnes J. R., Robinson R. R., Seidman J. G. Multiple mRNA species with distinct 3' termini are transcribed from the beta 2-microglobulin gene. 1983 Mar 31-Apr 6Nature. 302(5907):449–452. doi: 10.1038/302449a0. [DOI] [PubMed] [Google Scholar]
- Romans P., Firtel R. A. Organization of the actin multigene family of Dictyostelium discoideum and analysis of variability in the protein coding regions. J Mol Biol. 1985 Nov 20;186(2):321–335. doi: 10.1016/0022-2836(85)90108-1. [DOI] [PubMed] [Google Scholar]
- Russell P. R. Evolutionary divergence of the mRNA transcription initiation mechanism in yeast. Nature. 1983 Jan 13;301(5896):167–169. doi: 10.1038/301167a0. [DOI] [PubMed] [Google Scholar]
- Russell P. R. Transcription of the triose-phosphate-isomerase gene of Schizosaccharomyces pombe initiates from a start point different from that in Saccharomyces cerevisiae. Gene. 1985;40(1):125–130. doi: 10.1016/0378-1119(85)90031-9. [DOI] [PubMed] [Google Scholar]
- Russell P., Nurse P. Schizosaccharomyces pombe and Saccharomyces cerevisiae: a look at yeasts divided. Cell. 1986 Jun 20;45(6):781–782. doi: 10.1016/0092-8674(86)90550-7. [DOI] [PubMed] [Google Scholar]
- Scarpulla R. C., Wu R. Nonallelic members of the cytochrome c multigene family of the rat may arise through different messenger RNAs. Cell. 1983 Feb;32(2):473–482. doi: 10.1016/0092-8674(83)90467-1. [DOI] [PubMed] [Google Scholar]
- Scherer S., Davis R. W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setzer D. R., McGrogan M., Schimke R. T. Nucleotide sequence surrounding multiple polyadenylation sites in the mouse dihydrofolate reductase gene. J Biol Chem. 1982 May 10;257(9):5143–5147. [PubMed] [Google Scholar]
- Vandekerckhove J., Lal A. A., Korn E. D. Amino acid sequence of Acanthamoeba actin. J Mol Biol. 1984 Jan 5;172(1):141–147. doi: 10.1016/0022-2836(84)90418-2. [DOI] [PubMed] [Google Scholar]
- Vandekerckhove J., Weber K. At least six different actins are expressed in a higher mammal: an analysis based on the amino acid sequence of the amino-terminal tryptic peptide. J Mol Biol. 1978 Dec 25;126(4):783–802. doi: 10.1016/0022-2836(78)90020-7. [DOI] [PubMed] [Google Scholar]
- Vandekerckhove J., Weber K. The amino acid sequence of Physarum actin. Nature. 1978 Dec 14;276(5689):720–721. doi: 10.1038/276720a0. [DOI] [PubMed] [Google Scholar]
- Vandekerckhove J., Weber K. The complete amino acid sequence of actins from bovine aorta, bovine heart, bovine fast skeletal muscle, and rabbit slow skeletal muscle. A protein-chemical analysis of muscle actin differentiation. Differentiation. 1979;14(3):123–133. doi: 10.1111/j.1432-0436.1979.tb01021.x. [DOI] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]