Abstract
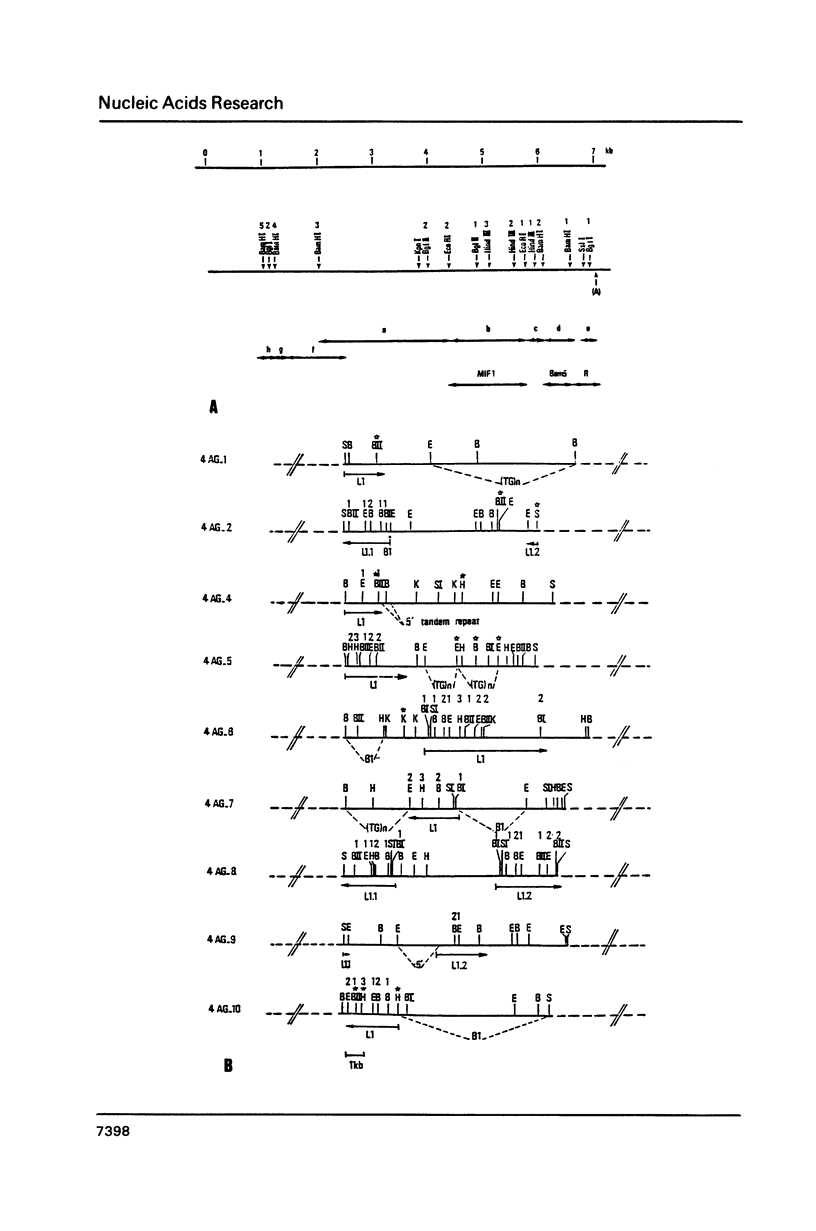
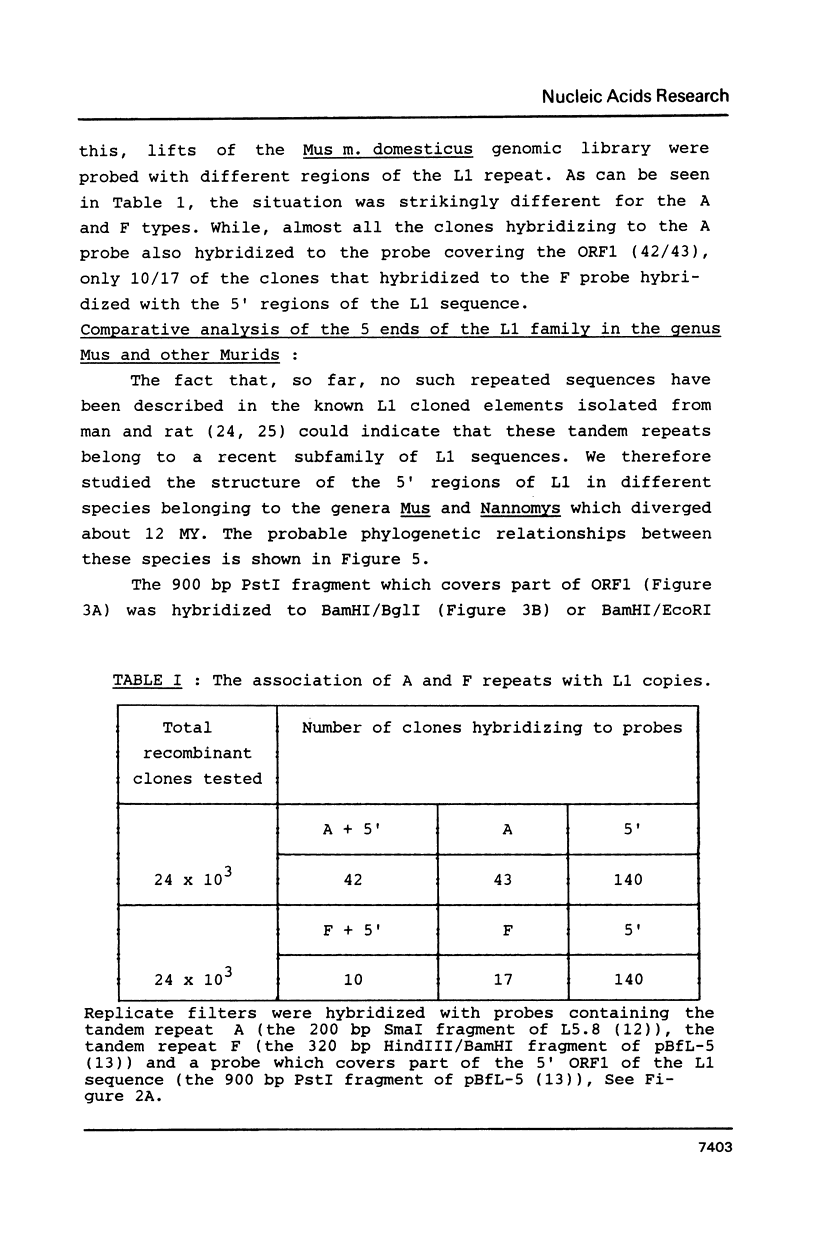
Analysis of a few large L1 elements has revealed two types of tandem repeats at the 5' end: A and F. In this study, the relationships between these repeats and a series of large L1 elements has been analysed. Most of cloned L1 repeats were shown to lack either A or F sequences at their 5' ends. F sequences are found less frequently associated than A sequences to the 5' ends of L1 and an evolutionary comparison shows that the A type was introduced more recently during the evolution of the mouse genome than the F type.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennett K. L., Hastie N. D. Looking for relationships between the most repeated dispersed DNA sequences in the mouse: small R elements are found associated consistently with long MIF repeats. EMBO J. 1984 Feb;3(2):467–472. doi: 10.1002/j.1460-2075.1984.tb01829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Bonhomme F., Catalan J., Britton-Davidian J., Chapman V. M., Moriwaki K., Nevo E., Thaler L. Biochemical diversity and evolution in the genus Mus. Biochem Genet. 1984 Apr;22(3-4):275–303. doi: 10.1007/BF00484229. [DOI] [PubMed] [Google Scholar]
- Brown S. D., Dover G. Organization and evolutionary progress of a dispersed repetitive family of sequences in widely separated rodent genomes. J Mol Biol. 1981 Aug 25;150(4):441–466. doi: 10.1016/0022-2836(81)90374-0. [DOI] [PubMed] [Google Scholar]
- Burton F. H., Loeb D. D., Chao S. F., Hutchison C. A., 3rd, Edgell M. H. Transposition of a long member of the L1 major interspersed DNA family into the mouse beta globin gene locus. Nucleic Acids Res. 1985 Jul 25;13(14):5071–5084. doi: 10.1093/nar/13.14.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton F. H., Loeb D. D., Voliva C. F., Martin S. L., Edgell M. H., Hutchison C. A., 3rd Conservation throughout mammalia and extensive protein-encoding capacity of the highly repeated DNA long interspersed sequence one. J Mol Biol. 1986 Jan 20;187(2):291–304. doi: 10.1016/0022-2836(86)90235-4. [DOI] [PubMed] [Google Scholar]
- D'Ambrosio E., Waitzkin S. D., Witney F. R., Salemme A., Furano A. V. Structure of the highly repeated, long interspersed DNA family (LINE or L1Rn) of the rat. Mol Cell Biol. 1986 Feb;6(2):411–424. doi: 10.1128/mcb.6.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T. G. Size and structure of the highly repetitive BAM HI element in mice. Nucleic Acids Res. 1983 Aug 11;11(15):5073–5091. doi: 10.1093/nar/11.15.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Leavitt J., Kakunaga T. Mutated beta-actin gene: coexpression with an unmutated allele in a chemically transformed human fibroblast cell line. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3634–3638. doi: 10.1073/pnas.78.6.3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Hidaka S., Sakaki Y. Sequence analysis of a KpnI family member near the 3' end of human beta-globin gene. Nucleic Acids Res. 1985 Nov 11;13(21):7813–7827. doi: 10.1093/nar/13.21.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubier-Maurin V., Dod B. J., Bellis M., Piechaczyk M., Roizes G. Comparative study of the L1 family in the genus Mus. Possible role of retroposition and conversion events in its concerted evolution. J Mol Biol. 1985 Aug 20;184(4):547–564. doi: 10.1016/0022-2836(85)90302-x. [DOI] [PubMed] [Google Scholar]
- Krayev A. S., Kramerov D. A., Skryabin K. G., Ryskov A. P., Bayev A. A., Georgiev G. P. The nucleotide sequence of the ubiquitous repetitive DNA sequence B1 complementary to the most abundant class of mouse fold-back RNA. Nucleic Acids Res. 1980 Mar 25;8(6):1201–1215. doi: 10.1093/nar/8.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krayev A. S., Markusheva T. V., Kramerov D. A., Ryskov A. P., Skryabin K. G., Bayev A. A., Georgiev G. P. Ubiquitous transposon-like repeats B1 and B2 of the mouse genome: B2 sequencing. Nucleic Acids Res. 1982 Dec 11;10(23):7461–7475. doi: 10.1093/nar/10.23.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb D. D., Padgett R. W., Hardies S. C., Shehee W. R., Comer M. B., Edgell M. H., Hutchison C. A., 3rd The sequence of a large L1Md element reveals a tandemly repeated 5' end and several features found in retrotransposons. Mol Cell Biol. 1986 Jan;6(1):168–182. doi: 10.1128/mcb.6.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenen W. A., Brammar W. J. A bacteriophage lambda vector for cloning large DNA fragments made with several restriction enzymes. Gene. 1980 Aug;10(3):249–259. doi: 10.1016/0378-1119(80)90054-2. [DOI] [PubMed] [Google Scholar]
- Martin S. L., Voliva C. F., Hardies S. C., Edgell M. H., Hutchison C. A., 3rd Tempo and mode of concerted evolution in the L1 repeat family of mice. Mol Biol Evol. 1985 Mar;2(2):127–140. doi: 10.1093/oxfordjournals.molbev.a040340. [DOI] [PubMed] [Google Scholar]
- Meunier-Rotival M., Soriano P., Cuny G., Strauss F., Bernardi G. Sequence organization and genomic distribution of the major family of interspersed repeats of mouse DNA. Proc Natl Acad Sci U S A. 1982 Jan;79(2):355–359. doi: 10.1073/pnas.79.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottez E., Rogan P. K., Manuelidis L. Conservation in the 5' region of the long interspersed mouse L1 repeat: implications of comparative sequence analysis. Nucleic Acids Res. 1986 Apr 11;14(7):3119–3136. doi: 10.1093/nar/14.7.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J. H. Long interspersed sequences in mammalian DNA. Properties of newly identified specimens. Biochim Biophys Acta. 1985 Feb 20;824(2):113–120. doi: 10.1016/0167-4781(85)90087-9. [DOI] [PubMed] [Google Scholar]
- Rogers J. Molecular biology. CACA sequences - the ends and the means? Nature. 1983 Sep 8;305(5930):101–102. doi: 10.1038/305101a0. [DOI] [PubMed] [Google Scholar]
- Rogers J. The origin of retroposons. 1986 Feb 27-Mar 5Nature. 319(6056):725–725. doi: 10.1038/319725a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voliva C. F., Martin S. L., Hutchison C. A., 3rd, Edgell M. H. Dispersal process associated with the L1 family of interspersed repetitive DNA sequences. J Mol Biol. 1984 Oct 5;178(4):795–813. doi: 10.1016/0022-2836(84)90312-7. [DOI] [PubMed] [Google Scholar]
- Wilson R., Storb U. Association of two different repetitive DNA elements near immunoglobulin light chain genes. Nucleic Acids Res. 1983 Mar 25;11(6):1803–1817. doi: 10.1093/nar/11.6.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo S. L. A sensitive and rapid method for recombinant phage screening. Methods Enzymol. 1979;68:389–395. doi: 10.1016/0076-6879(79)68028-x. [DOI] [PubMed] [Google Scholar]