Abstract
The characterization of factors contributing to the formation and development of surface-associated bacterial communities known as biofilms has become an area of intense interest since biofilms have a major impact on human health, the environment and industry. Various studies have demonstrated that motility, including swimming, swarming and twitching, seems to play an important role in the surface colonization and establishment of structured biofilms. Thereby, the impact of chemotaxis on biofilm formation has been less intensively studied. Pseudomonas aeruginosa has a very complex chemosensory system with two Che systems implicated in flagella-mediated motility. In this study, we demonstrate that the chemotaxis protein CheR1 is a methyltransferase that binds S-adenosylmethionine and transfers a methyl group from this methyl donor to the chemoreceptor PctA, an activity which can be stimulated by the attractant serine but not by glutamine. We furthermore demonstrate that CheR1 does not only play a role in flagella-mediated chemotaxis but that its activity is essential for the formation and maintenance of bacterial biofilm structures. We propose a model in which motility and chemotaxis impact on initial attachment processes, dispersion and reattachment and increase the efficiency and frequency of surface sampling in P. aeruginosa.
Introduction
Biofilms are generally defined as sessile bacterial communities attached to a surface and embedded in a self-produced extracellular matrix. This polymeric matrix acts as a protective shield and - together with cellular appendages - facilitates adherence of the bacteria to each other and/or to surfaces. Biofilm communities represent not only the most prevalent bacterial mode of growth in the environment [1], but they have also become a focus of microbiological research due to their impact on industry and human health [2], [3]. It has been estimated that up to 80% of the bacterial infections in the industrialized countries are biofilm associated infections, refractory to antimicrobial therapy [4]. In order to develop novel strategies for biofilm control, it is critical to understand the adaptive pathways leading to the development and maintenance of bacterial biofilms.
So far, research on global gene or protein biofilm expression patterns did not succeed in the identification of a specific developmental biofilm program and thus putative novel targets for an anti-biofilm strategy. New knowledge about mechanisms involved in biofilm formation have recently been obtained by the use of optical tools to monitor in vitro grown biofilms. From those studies it has become evident that motility can have a profound impact on the colonization of surfaces [5]–[9]. However, the particular aspects of flagella and/or pili biogenesis and function, that are needed for biofilm formation in any species of bacteria, are not clearly defined. Pratt and Kolter have proposed a model for the initiation of Escherichia coli biofilm formation in which chemotaxis is dispensable but motility is required to overcome surface repulsion [9]. In contrast, there is evidence for the impact of chemotaxis on surface interactions and biofilm formation in other bacterial species such as Aeromonas spp. [10], Vibrio cholerae [11], Pseudomonas aeruginosa [12] and Agrobacterium tumefaciens [7].
Genome analysis reveals that a large number of environmental motile bacteria possess several genes involved in chemosensing and chemotatic signal transduction. Motile bacteria sense changes in the concentration of chemicals in their environment and respond in a behavioral manner [13]. The molecular mechanisms underlying bacterial chemotaxis have been studied extensively in the enteric bacteria E. coli and Salmonella enterica serovar Typhimurium [14], [15]. Chemotactic ligands are detected by cell surface chemoreceptors called methyl-accepting chemotaxis proteins (MCPs). Several homologous transmembrane receptors (MCPs) sense extracellular stimuli and produce signals that are transmitted to their cytoplasmic domains. These domains regulate an associated two-component phosphotransfer signal transduction system that controls flagellar rotation. The effect of ligand binding is counterbalanced by reversible MCP methylation providing the ability to detect chemical changes over time. Thereby, the opposing activities of two specific enzymes, CheR, a methyltransferase, and CheB, a methylesterase, control the MCP methylation level. CheR converts specific glutamic acid residues in the MCP cytoplasmic domain to glutamyl methyl esters, using S-adenosylmethionine (SAM) as the methyl donor [16]. CheB hydrolyzes those methylated residues, releasing methanol and restoring glutamic acid [17]. CheR activity is unregulated, whereas CheB is feedback-regulated by MCP output signals [18]. Studies of chemotaxis and MCP methylation in other organisms have revealed both, similarities and differences to the E. coli/S. Typhimurium chemotaxis pathway [19]–[24]. Given the different nature of the chemotactic system in the enteric bacteria and other environmental motile bacteria, it is not surprising that the roles chemotaxis plays in biofilm development are quite distinct.
P. aeruginosa inhabits a wide variety of environmental niches and is capable of locomotion by rotating a single polar flagellum. The bacterium has a very complex chemosensory system with more than 20 chemotaxis (che) genes in five distinct clusters and 26 mcp-like genes [25]. The Che and the Che2 systems, both homologous to the E. coli Che chemotaxis system, have been implicated in flagella-mediated chemotaxis [26]–[29], while genes in Pil-Chp cluster and Wsp cluster are involved in type IV pilus synthesis, twitching motility and biofilm formation, respectively [30]–[33]. Among the 26 MCPs of P. aeruginosa, nine have been identified as MCPs for amino acids, inorganic phosphate, oxygen, ethylene and volatile chlorinated aliphatic hydrocarbons [34], whereas 3 MCPs were demonstrated to be involved in biofilm formation and biosynthesis of type IV pilus [32], [35], [36].
In this study we demonstrate that P. aeruginosa CheR1 is a chemotaxis protein methyltransferase which uses S-adenosylmethionine as a methyl donor to methylate the MCP PctA in response to amino acids. Furthermore, CheR1 activity is shown to be essential for flagella-mediated chemotaxis and involved in bacterial surface sampling which impacts on the biofilm structures.
Results
CheR1 methylates the methyl-accepting chemotaxis protein PctA using S-adenosylmethionine as a methyl donor
The PA01 PA3348 gene product is predicted to be a probable chemotaxis protein methyltransferase (CheR1, www.pseudomonas.com, [37]). This function was proposed based on limited amino acid identity (31%) to the experimentally studied CheR gene product in E. coli and a conserved domain for S-adenosylmethionine (SAM) binding.
In E. coli, CheR methylation activity is strongly enhanced upon binding to a conserved pentapeptide sequence (NWETF) at the C-terminus of highly abundant MCPs [38], [39]. Homologous pentapeptide structures are present in P. aeruginosa only in 2 of the 26 MCPs (CttP and Aer2 [26]). The latter are however organized on the chromosome in the vicinity of the Che2 system, whose predicted gene products (including CheR2) exhibit an even higher overall sequence identity to orthologous E. coli chemotaxis proteins [26]. Thus, we speculated that CheR1 might methylate the MCPs PctA, PctB and PctC despite the lack of the pentapeptide sequence. Those MCPs have been shown to be involved in the detection of amino acids [40], [41].
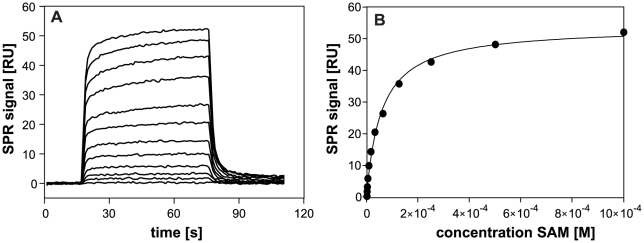
To characterize P. aeruginosa receptor methylation and to gain insight into methylation in MCPs lacking the pentapeptide motif, we performed in vitro methylation assays. We first expressed and purified P. aeruginosa His-tagged CheR1 protein and, since CheR uses S-adenosylmethionine (SAM) as a substrate for receptor methylation, we immobilized purified CheR1-His6 on a sensor chip and analyzed the interaction of CheR1 and SAM by the use of surface plasmon resonance. Figure 1 depicts the specific interaction of the ligand SAM and CheR1 thereby revealing a K D value of 61 µM.
Figure 1. Surface plasmon resonance (SPR) analysis of the interaction of CheR1-His6 and SAM using a Biacore S51.
CheR1 was immobilized on the sensor surface using standard amine coupling chemistry. The CheR1 interaction partner SAM was injected over the sensor surface. (A) SPR signals were detected for association (60 seconds) and dissociation (200 seconds) of SAM to CheR1. The graph shows the kinetic data for 12 SAM concentrations with data for the lowest (500 nM) to the highest (1 mM) SAM concentration displayed from bottom to top. (B) Data from (A) were analyzed with a steady state model. The resulting binding constant (K D) was determined to be 61 µM.
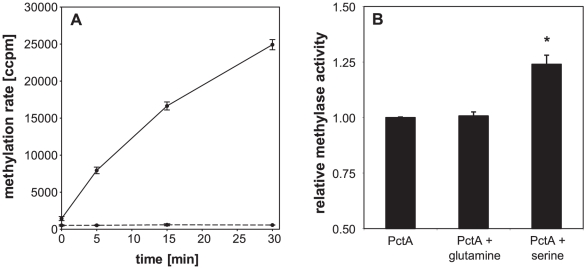
We next performed in vitro methylation assays by the utilization of purified P. aeruginosa His-tagged CheR1 protein, the methyl donor SAM and membranes from an E. coli strain (HCB721) effectively gutted of all the chemotaxis genes [42]. HCB721 cells were transformed with an IPTG inducible plasmid encoding the P. aeruginosa PctA receptor and membranes containing PctA were prepared as described under Materials and Methods. Figure 2A depicts the initial methylation rate of the PctA receptor and demonstrates that PctA was methylated by the methyltransferase CheR1.
Figure 2. Methylation of the methyl-accepting chemotaxis protein PctA by CheR-His6 using [3H-methyl]-SAM as the methyl donor.
(A) Methylation rate of E. coli HCB721 membranes containing no MCP (dashed line) or the P. aeruginosa MCP PctA (continuous line). (B) Methylation reactions carried out for 15 min in the presence of 1 mM serine resulted in an significantly increased methylation rate of the MCP PctA (* p<0.001, paired t-test), whereas the addition of glutamine had no effect. Representative data from one experiment out of at least two are shown. Error bars are standard deviations of three replica.
Previous chemotactic assays of pctA pctB pctC triple mutants supplemented with either one of these MCP genes revealed that PctA, PctB and PctC detected 18 amino acids, 7 amino acids and 2 amino acids, respectively [41]. Among those amino acids, serine was detected by e.g. PctA whereas glutamine was only detected by PctB. We therefore tested the effect of those two amino acids on the methylation rate of PctA. In accordance to the previous chemotactic experiments, the presence of serine but not glutamine increased the methylation rate of PctA by at least 1.2-fold (p<0.001, Figure 2B).
Motility defect of the methyltransferase cheR1 mutant in P. aeruginosa
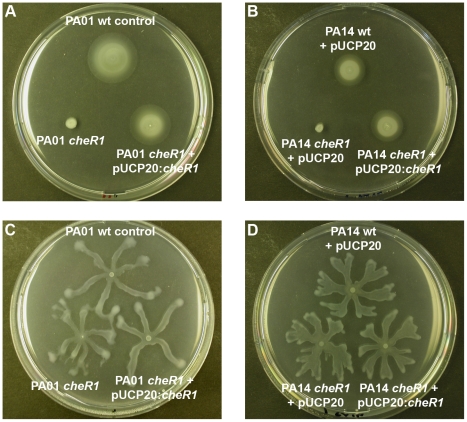
In agreement with results described by Kato et al. [28], our analysis of cheR1 transposon mutants in the PA01 and PA14 strain background (correct insertion of transposon was confirmed by PCR) revealed a severely impaired swimming motility in minimal medium soft agar plates (Figure 3A and B). Both cheR1 transposon mutants formed only small diameter swim rings in contrast to the diffuse large diameter swim rings observed in the wild-type control and the complemented mutant strains. This phenotype defect is characteristic for mutants that cannot respond to a chemical gradient generated upon nutrient consumption and was not due to a defect in the growth rate (data not shown). By contrast, the cheR1 mutant and complemented mutant strain were positive in swarming (Figure 3C and D) and twitching (data not shown) in both, the PA14 and PA01 strain background.
Figure 3. Motility phenotype of the cheR1 mutant and the complemented strain of PA01 (A, C) and PA14 (B, D).
(A, B) Swimming assays were performed on minimal medium soft agar plates supplement with 0.3% agar. The impaired swimming motility of the cheR1 mutant can be complemented by providing the cheR1 gene in trans. (C, D) Swarming assays were performed on minimal medium plates supplemented with 0.5% agar. All strains, the wild-type/wild-type control, the cheR1 mutant and the complemented cheR1 mutant, were capable of swarming.
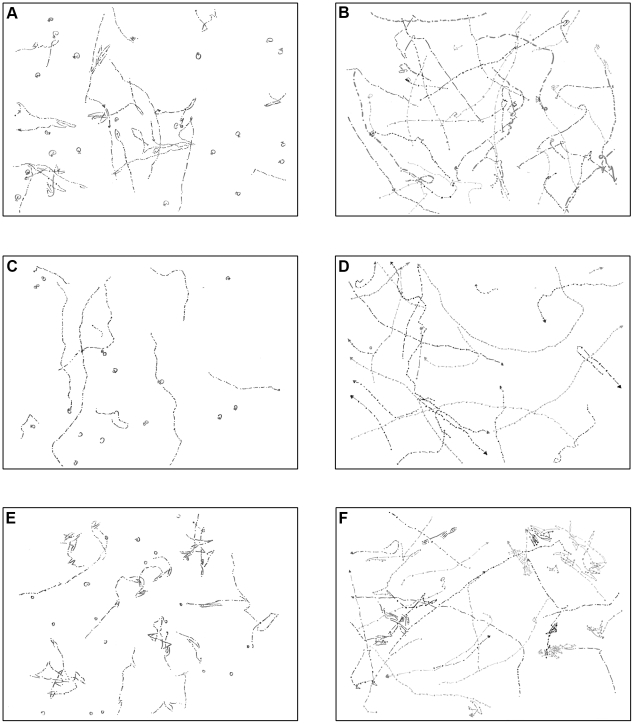
In order to characterize the swimming motility defect in more detail we observed the cells by light microscopy. E. coli bacteria move unidirectional with periodic pauses that involve active tumbling, which reorients the cell prior to continued forward motion. This essential random movement can be biased through taxis mechanisms that modify the frequency of reorientations [14], [15]. Motility by other bacterial species including P. aeruginosa can differ significantly from the E. coli paradigm. As opposed to E. coli which possesses peritrichous flagella, P. aeruginosa possesses a single polar flagellum and swims in a straight, stop, turn mode (described by Pratt et al. [9]), driven by a (i) counter-clockwise rotation of the flagellum, (ii) brief reversal and (iii) reorientation. Both P. aeruginosa strains (PA01 and PA14) were motile and exhibited the run/stop/turn mode of swimming, although obviously the PA14 strain did not exhibit as much stops/reorientations as compared to the PA01 wild-type control under the tested conditions (Figure 4A and B). The cheR1 mutants were also motile, but tented to swim straight and exhibited less frequent stops/reorientations as opposed to the PA01 and PA14 wild-type controls (Figure 4C and D). When the cheR1 mutants were complemented with cheR1 in trans, the effect of CheR became more pronounced. The complemented cheR1 mutants exhibited clearly more reversals as compared with the respective wild-type control and cheR1 mutant in both strain backgrounds (Figure 4E and F).
Figure 4. Motility defect of the cheR1 mutant as observed by light microscopy.
(A, B) The wild-type (A, PA01 wt control and B, PA14 wt) exhibits a straight forward - short backup/reversal - straight forward mode of swimming. (C, D) The cheR1 mutant (C, PA01 and D, PA14) changes its direction less frequently than the wild-type and tends to swim straight. (E, F) The cheR1 mutant complemented with pUCP20:cheR1 in trans (E, PA01 and F, PA14) is oscillating rapidly between forward and backward swimming.
Compromised biofilm formation of the cheR1 mutant
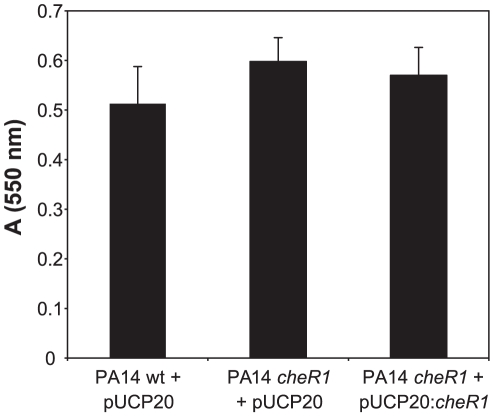
The cheR1 mutant of PA14 was further monitored for attachment and biofilm formation. We used the crystal violet (CV) staining assay for the determination of attached biomass following a 24 h incubation period of static cultures. As depicted in Figure 5, no clear difference in attached biomass could be observed between the PA14 wild-type, the PA14 cheR1 transposon mutant and the complemented strain.
Figure 5. Crystal violet (CV) staining assay for the determination of attached biomass.
PA14 wild-type carrying the vector control (pUCP20), the PA14 cheR1 mutant carrying the vector control (pUCP20) and the complemented PA14 cheR1 mutant (carrying pUCP20:cheR1) were grown in LB for 24 h at 37°C under static conditions before staining with CV. Representative data from one experiment out of five are shown. Error bars are standard deviations of eight replica.
We next examined the formation of biofilms on the bottom of a 96-well plate. Confocal laser scanning microscopy of biofilms stained with the BacLight Live/Dead stain revealed a severe biofilm defect of the PA14 cheR1 mutant after 72 h of growth in LB. A plasmid-borne copy of the deleted wild-type cheR1 gene complemented the mutant phenotype (data not shown). To characterize the differences in biofilm formation in more detail, PA14 wild-type and cheR1 mutant cells were tagged with plasmid-borne GFP and biofilm formation was monitored over time.
After 1 h, only few cells were found at the bottom of a 96-well plate in both, the wild-type and the cheR1 mutant, the majority of which was non-motile. In the following hours, the number of swimming bacteria increased. Thereby, bacteria of the wild-type strain accumulated at the bottom of the well but remained highly motile until the space was completely occupied by bacteria. By contrast, far less bacteria of the cheR1 mutant strain were found in the proximity of the bottom. Surface sampling of motile bacteria proceeded only slowly. The difference in early substratum coverage by the wild-type and the cheR1 mutant is displayed in Figure 6A and Figure S1.
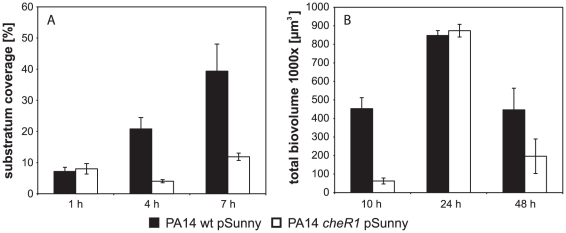
Figure 6. Differences in the development of GFP-expressing PA14 wild-type and cheR1 mutant biofilms.
(A) Quantification of the substratum coverage at the well-bottom of a 96-well plate after 1 h, 4 h and 7 h of growth in LB at 37°C. (B) Quantification of the biovolume after 10 h, 24 h and 48 h of growth in LB at 37°C.
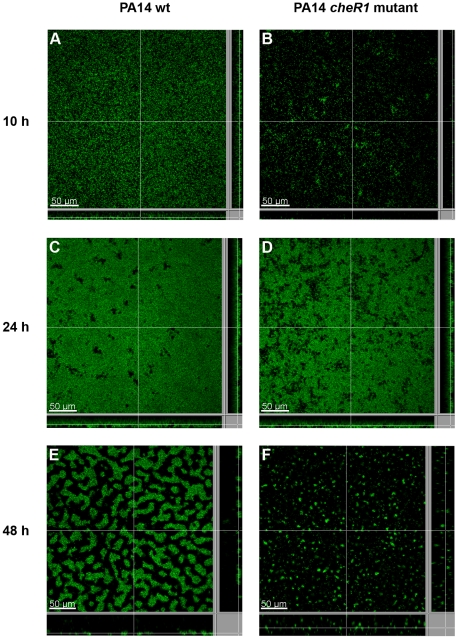
We then monitored the formation of mature biofilms by three dimensional confocal laser scanning microscopy. After 10 h, biofilm formation of the cheR1 mutant was severely delayed as compared with the wild-type (Figure 6B and Figure 7A and B). Nevertheless, after 24 h the structure and biovolume of cheR1 biofilms were found to be very similar to that of wild-type biofilms (Figure 6B and Figure 7C and D). In the following, the wild-type and the cheR1 mutant biofilm underwent structural rearrangement. Whereas the wild-type formed cohesive and densely packed biofilm structures, the mutant biofilm was characterized by a more loosely and in parts disconnected architecture and an overall lower biomass (Figure 6B and Figure 7E andF).
Figure 7. Confocal laser scanning micrographs of P. aeruginosa biofilms grown in LB.
Biofilm development in a well of a 96-well plate at (A, B) 10 h, (C, D) 24 h and (E, F) 48 h is shown for (A, C, E) PA14 wild-type carrying the GFP-expression vector pSunny and (B, D, F) the PA14 cheR1 transposon mutant carrying the GFP-expression vector pSunny. The central image shows a XY section through the biofilm and the flanking pictures show vertical sections. Scale bars, 50 µm.
Discussion
While signaling and chemotaxis have been most extensively explored in E. coli and S. Typhimurium, studies of other organisms revealed much more diversity and complexity in chemotactic signaling than had been previously anticipated. Most motile environmental bacteria have multiple homologues of the E. coli che genes and many more mcp genes than the five found in the enteric bacteria [15]. P. aeruginosa is chemotactic to most of the organic compounds that it can grow on and P. aeruginosa harbors as much as 26 genes that express homology to E. coli mcp genes [34]. P. aeruginosa has furthermore 5 clusters of E. coli-like chemotaxis genes. These include the che clusters cluster I (cheY cheZ cheA cheB motC motD orf1 orf2 cheW), cluster V (cheV cheR) and cluster II (cttP cheY2 cheA2 cheW2 aer2 cheR2 cheD cheB2), furthermore there is cluster III (wspA wspB wspC wspD wspE wspF wspR), as well as cluster IV (pilG pilH pilI pilJ pilK chpA chpB chpC chpD chpE). Whereas cluster I, V and II are evidently involved in flagella-mediated chemotaxis [26]–[29], cluster III is involved in the control of the expression of Pel and Psl exopolysaccharides [32] and cluster IV has been implicated in regulating twitching motility [31], [33].
Given the very different nature of the chemotactic systems present in the enteric bacteria versus environmental bacteria, it is not surprising that the roles the chemotaxis systems play are quite distinct. There are several reports concluding that - depending on the model system - chemotaxis is either required or dispensable for bacterial biofilm formation. P. aeruginosa harbors a complex chemotaxis system that seems to be involved in flagella-driven chemotaxis and in determining the cellular organization within biofilms. This renders P. aeruginosa probably a good model microorganism for investigating the roles of chemotaxis in bacterial adaptation to diverse environmental niches, including the establishment of structured bacterial communities.
In this study we provide evidence that CheR1 is a P. aeruginosa chemotaxis methyltransferase which transfers a methyl group from the methyl donor SAM to a methyl-accepting chemotaxis protein. As determined by surface plasmon resonance studies, CheR1 binds SAM at a K D of approximately 60 µM, which is in the same order of magnitude as has been demonstrated for Salmonella CheR [43]. It has previously been shown that several amino acids are detected by the MCPs PctA, PctB and/or PctC in P. aeruginosa [41]. In accordance with these results, we could demonstrate in this study that in vitro methylation of the MCP PctA by CheR1 could be enhanced by the addition of serine but not of glutamine, the latter of which was shown to be specifically detected by PctB [41]. In E. coli, highly abundant transmembrane receptors harbor a conserved NWETF motif at the extreme C-terminal end of the MCP which recruits CheR to the receptor cluster and is required for efficient methylation and demethylation [38], [39]. Here we demonstrate that the MCP PctA was methylated by CheR1 despite the lack of a conserved C-terminal pentapeptide sequence. Similarly, a pentapeptide-independent methyltransferase has been characterized in the thermophilic bacterium Thermotoga maritima [44]. Co-crystallization of CheR from S. Typhimurium with the conserved NWETF pentapeptide demonstrated that the CheR β-subdomain is the region that interacts with the pentapeptide sequence [45]. β-subdomains can be divided into two groups: those with longer β-loops, found in CheR proteins from organisms containing MCPs with the pentapeptide recognition motif, and those with shorter β-loops, found in organisms that lack MCPs with the pentapeptide recognition motif [44]–[46]. Interestingly, in P. aeruginosa, CheR2 has a long β-loop and CheR1 has a short β-loop [44]. Moreover, only two out of 26 chemoreceptors have the conserved C-terminal pentapeptide motif and both genes encoding those chemoreceptors are located in the close proximity of the che2 system. Overall, this might indicate that CheR2 uses a pentapeptide-dependent methylation mechanism, whereas CheR1 uses a pentapeptide-independent methylation mechanism and that this is one way of preventing crosstalk between different chemotaxis systems in P. aeruginosa.
A recent publication on the influence of motility and chemotaxis in A. tumefaciens revealed an impact of chemotaxis on both, attachment and biofilm formation [7]. In this study, the P. aeruginosa cheR1 mutant did not exhibit a phenotype in the CV assay. Nevertheless, soft-agar plate assays and confocal microscope analysis of static biofilms clearly demonstrate that CheR1 activity is not only essential for flagella-mediated chemotaxis but that it is also involved in the formation of structured biofilms. Interestingly, CheR1 activity seems to be important at two developmental steps within the process of biofilm formation. Firstly, bacterial movement in close proximity to the surface enables surface sampling prior to irreversible attachment which seems to be important in the initial steps of biofilm formation and which seems to be promoted by the chemotaxis system. Secondly, the formation and consolidation of a more structured community seems to involve flagella-mediated chemotaxis. As far as we are aware, the only study examining biofilm formation of a P. aeruginosa che1 chemotaxis mutant (cheY) was performed by Barken et al. [12]. They demonstrated that not type IV pili-driven but flagellum-driven motility is involved in the formation of cap structures in biofilms grown in a flow chamber irrigated with glucose minimal medium. Besides the cheY and fliM mutants, also a P. aeruginosa rhlA mutant deficient in biosurfactant production displayed reduced cap formation [47]. Since swarming motility requires both, the presence of biosurfactants and flagellar activity, Barken et al. [12] suggested that flagellum-driven surface-associated motility (by means of swarming) rather than directed motility (swimming in response to the sensing of certain metabolites) is required for cap formation. In this study, we used different experimental settings (static growth conditions, nutrient-rich medium), nevertheless, we also demonstrated a role of flagellum-mediated motility in the formation of structured biofilms. However, since our cheR1 mutant displayed normal swarming motility on 0.5% agar plates, we rather hypothesize that chemotaxis driven swimming motility is necessary for formation of mature biofilm structures. A role for a substrate gradient that directs the motile bacteria has been suggested before [48].
In conclusion, one reasonable model for the role of motility and chemotaxis in biofilm formation in P. aeruginosa is that the efficiency and frequency of surface sampling may be influenced through chemotactic processes. This is important for initial sampling of bacteria to the surface and as the biofilm matures, chemotactic cues may stimulate dispersion. The released motile bacteria in the planktonic phase have the potential to colonize new sites and promote lateral expansion of the biofilm or to reattach in a coordinated chemotaxis-driven way, thus fine-tuning the architecture of the biofilm structures.
Materials and Methods
Bacterial strains, plasmids and growth conditions
Bacterial strains, plasmids and primers used in this study are listed in Table 1. Of note, transposon mutant P. aeruginosa PA01 ID 8031 from the Washington Genome Center [49] with a transposon insertion in open reading frame (ORF) PA4684 was used as wild-type control (PA01 wt control). ORF PA4684 is most likely coding for a non-functional gene product due to a large gene deletion [50], [51]. P. aeruginosa and E. coli strains were routinely cultured at 37°C in Luria-Bertani (LB) broth unless otherwise indicated. When required for plasmid or transposon selection, 100 µg/ml ampicillin and 50 µg/ml kanamycin were used for E. coli and 400 µg/ml carbenicillin, 15 µg/ml gentamycin, 150 µg/ml streptomycin and 25 µg/ml tetracycline respectively for the selection in P. aeruginosa.
Table 1. Strains, plasmids and primers used in this study.
| Strain, plasmid or primer | Relevant characteristicsa | Source or reference |
| Strains | ||
| E. coli DH5α | F- endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG Φ80dlacZΔM15 Δ (lacZYA-argF)U169, hsdR17(rK − mK +), λ– | [60] |
| E. coli BL21 (DE3) | F- ompT hsdSB(rB −mB −) gal dcm | Stratagene |
| E. coli S17-1 | C600::RP-4 2-(Tc::Mu) (Kn::Tn7) thi pro hsdR hsdM+recA | [61] |
| E. coli HCB721 | Δ (tsr)7021 trg::Tn10 Δ (cheA-cheY)::XhoI(Tn5), Kmr, Tcr | [42] |
| PA01 wt control | PA4684 transposon mutant from the Washington Genome Center PA01 mutant library, ID 8031, Tcr | [49] |
| PA01 cheR1 | cheR1 transposon mutant from the Washington Genome Center PA01 mutant library, ID 47867, Tcr | [49] |
| PA14 wt | Wild-type | [62] |
| PA14 cheR1 | cheR1 transposon mutant from the NR PA14 transposon mutant library, ID 36949, Gmr | [62] |
| Plasmids | ||
| pUCP20 | Shuttle vector, Apr/Cbr | [63] |
| pUCP20:cheR1 | cheR1 (promoter region and gene) cloned into BamHI-XbaI in MCS, Apr/Cbr | This study |
| pET21a(+) | Plasmid for overexpression of proteins with C-terminal His6-tag, Apr | Novagen |
| pET21a(+):cheR1 | cheR1 gene without stop codon cloned into NdeI-HindIII in MCS, Apr | This study |
| pSunny | GFP expressing plasmid, Kmr, Smr | [59] |
| pHSe5:tsr | Plasmid expressing the E. coli MCP Tsr, inducible with IPTG, Apr | [52], [53] |
| pHSe5:pctA | Plasmid expressing the P. aeruginosa MCP PctA, inducible with IPTG, Apr | This study |
| Primers b | ||
| PA3348 fPr1 | 5′-AAAACATATGGTGTCGGCAGCTAATGCG | |
| PA3348 rPr2 | 5′-GATCAAGCTTCTTGGCCCGGTAGAT | |
| PA3348 fPr3 | 5′-GATCGGATCCTTGCATACTTCGTTGTCC | |
| PA3348 rPr3 | 5′-GATCTCTAGACTACTTGGCCCGGTAGATG | |
| pctA fPr2 | 5′-GATCGGATCCATGATCAAAAGTCTGAAGTTCAGC | |
| pctA rPr2 | 5′-GATCAAGCTTTCAGATCTTGAAGCTGTCCAC |
Apr, ampicillin resistant; Cbr, carbenicillin resistant; Gmr, gentamycin resistant; Kmr, kanamycin resistant; Smr, streptomycin resistant; Tcr, tetracycline resistant
Engineered restrictions sites are underlined
PCR amplifications were performed with Pfu polymerase using PA01 genomic DNA as a template. For complementation studies, the cheR1 gene including 210 bp of the upstream region was amplified using the primer pair PA3348 fPr3/rPr3 and cloned into the BamHI-XbaI sites of pUCP20. For expression of CheR-His6, the cheR1 gene was amplified without its stop-codon using primer pair PA3348 fPr1/rPr2 and cloned into the NdeI-HindIII sites of pET21a(+) thereby fusing the gene to a C-terminal His6-tag. Obtained constructs were confirmed by restriction analysis and sequencing before being introduced into P. aeruginosa by electroporation (pUCP20:cheR1) or into E. coli BL21(DE3) by transformation (pET21a(+):cheR1).
The IPTG inducible E. coli Tsr expression plasmid pHSe5 [52], [53], kindly provided by Robert M. Weis (University of Massachusetts, Amherst), was modified for expression of the P. aeruginosa MCP PctA. The tsr sequence was removed by digestion with BamHI and HindIII yielding in a vector backbone (4.8 kb) and a tsr fragment (2.4 kb). The vector backbone was gel-purified and ligated to the PCR amplified and BamHI/HindIII digested pctA gene. The resulting construct, pHSe5:pctA, was transformed into chemically competent E. coli HCB721 cells.
Overexpression and purification of CheR1
E. coli BL21 cells (Stratagene) carrying pET21a(+):cheR1 were grown in LB supplemented with 100 µg/ml ampicillin and expression was induced with 0.1 mM isopropyl 1-thio-β-D-galactopyranoside (IPTG) at an OD600 of 0.5 to 0.7. The culture was shaken for 16 h at 20°C before harvesting the cells by centrifugation. The bacteria were resuspended in lysis buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 10 mM imidazole) containing 1 mM DTT, 1 mg/ml lysozyme, protease inhibitors (Complete mini, EDTA free, Roche) and Benzonase Nuclease (Novagen) and passed through a French pressure cell at 16,000 psi (SLM-Aminco). Unbroken cells were removed by centrifugation at 37,500×g at 4°C for 45 min and the supernatant was incubated with nickel-nitrilotriacetic acid agarose resin (Qiagen) for 1 h at 4°C. The resin was washed with lysis buffer and proteins were eluted with 50 mM NaH2PO4, pH 8.0, 300 mM NaCl and 250 mM imidazole. After SDS-PAGE analysis, fractions containing pure protein were pooled and dialyzed for 16 h at 4°C in 50 mM NaH2PO4, pH 8.0, 300 mM NaCl.
Surface plasmon resonance analysis
Surface plasmon resonance (SPR) interaction analyses were performed in 10 mM Tris, 300 mM NaCl, pH 8.0 (buffer A) at 25°C using a Biacore S51 instrument (GE Healthcare, Biacore). For covalent coupling of CheR-His6, carboxymethylated sensor chip surfaces (Series S CM5) were activated with a 1∶1 ratio of 0.4 M EDC (N-ethyl-N'-(3-dimethylaminopropyl)carbodiimide) and 0.1 M NHS (N-hydroxysuccinimide) for 10 min and purified CheR-His6 (40 µg/ml, 10 mM HEPES, pH 7.0) was injected for 20 min at a flow rate of 5 µl/min. Deactivation of the surface was performed using 1 M ethanolamine-HCl (pH 8.5) for 8 min.
Interaction analyses were performed in buffer A containing 0.005% (vol/vol) surfactant P20 by injection of increasing concentrations (500 nM-1 mM) of S-adenosylmethionine (SAM) at a flow rate of 30 µl/min. Association and dissociation signals were monitored for 60 s and 200 s, respectively. After subtracting the reference spot signal, resulting binding signals were fitted. Data evaluation was performed using the Biacore S51 evaluation software version 1.2.1.
Preparation of membranes containing PctA
E. coli strain HCB721 [42], kindly provided by Howard C. Berg (Harvard University), was used to prepare membranes enriched for the MCP PctA. HCB721 is deficient in all known E. coli MCPs and all cytoplasmic chemotaxis proteins except the CheZ phosphatase and thus ensures that the overexpressed PctA receptor does not undergo posttranslational modification. Cells were grown at 30°C in 500 ml LB supplemented with 100 µg/ml ampicillin. At OD600 0.5–0.7, PctA expression was induced with 1 mM IPTG. After 3 h of induction, cells were harvested by centrifugation and resuspended in buffer containing 100 mM potassium acetate, 50 mM HEPES, pH 7.5, 5 mM magnesium acetate, 0.05% (vol/vol) β-mercaptoethanol, protease inhibitors (Complete mini, EDTA free, Roche) and Benzonase Nuclease (Novagen). Cells were lysed by adding 1 mg/ml lysozyme and by passage through a French Pressure cell at least twice. After removing unbroken cells by centrifugation for 20 min at 7,000×g, the supernatant was loaded on a sucrose step gradient (0.5 M, 1.5 M and 2 M) and centrifuged at 100,000×g for 1 h at 4°C. The second band was removed, diluted 5 times with water containing protease inhibitors (Complete mini, with EDTA, Roche) and centrifuged for 1 h at 100,000×g at 4°C. The pellet was resuspended in a small amount of storage buffer (50 mM NaH2PO4, 1 mM EDTA, 10% (vol/vol) glycerol), analyzed on Coomassie-stained SDS PAGE and the total amount of protein concentration was determined by scanning densitometry against the Low Molecular Weight Calibration Kit (GE Healthcare) as a reference. The membrane samples were adjusted to a final protein concentration of 2 mg protein/ml in storage buffer and stored as single-use aliquots at −70°C.
Receptor methylation assays
In vitro methylation assays were performed as previously described with slight modifications [54], [55]. In brief, the methylation reaction (final volume 100 µl) was carried out at 30°C in 50 mM NaH2PO4, pH 8.0, 300 mM NaCl containing 50 µl receptor-enriched membranes, 0.1 µM CheR1-His6 and with or without 1 mM serine or glutamine. After a pre-incubation step of 5 min, the reaction was started by adding 0.625 µM [3H-methyl]-S-adenosylmethionine (specific activity 80 Ci/mmol, Amersham). At indicated time points, 10 µl aliquots were removed, spotted on filter paper (1 cm2, Rotilabo, Roth) and quenched with 10% (wt/vol) trichloroacetic acid (TCA). The filters were washed twice with 10% (wt/vol) TCA and once with ethanol, for 15 min with gentle shaking. Air-dried filters were transferred into a 24-well sample plate, covered with 1 ml scintillation fluid and incubated over night before radioactivity was quantified using a microplate liquid scintillation counter (1450 MicroBeta TriLux, Wallac).
Motility assays
Swimming and swarming motility assays were performed as previously described [56]. In brief, swimming was evaluated on BM2 glucose plates containing 0.3% agar and swarming on modified BM2 glucose plates containing 0.5% agar supplemented with 0.1% Casamino acids. Plates were inoculated with 1 µl of a pre-culture with OD600>1.0 and incubated over night at 37°C. Twitching assays were performed on LB plates with 1% agar by stab inoculation of single colonies with a toothpick. After 24–48 h incubation at 37°C, the diameter of the twitching zone at the plastic-agar interface was measured.
Motility tracking
Bacterial cells were grown to exponential phase (OD600∼1.0) in LB, diluted 1∶200–1∶1000 in 0.9% NaCl containing 3% Ficoll (Sigma) and transferred into a 96-well plate with a thin glass bottom. Cells were monitored with an inverted microscope (Axiovert 135TV, Zeiss), equipped with a 25×/0.80 oil objective and a CoolSnap HQ2 camera (Visitron Systems) and operated with the MetaMorph software (version 7.5.3, Molecular Devices Corporation). Phase-contrast images visualizing cells near the glass bottom were acquired for 30 s at 5 frames per second, imported as stacks into ImageJ 1.42, where trajectories of swimming bacteria were monitored.
Biofilm and attachment assays
Crystal violet staining of adherent cells was adopted from O'Toole and Kolter [57] with the following modifications. Overnight LB cultures were diluted to an OD600 of 0.02 with fresh medium. The bacterial suspensions were inoculated in PVC 96-well plates (Becton Dickinson Labware) with 100 µl per well (8 replica per strain), sealed with an air-permeable BREATHseal cover foil (Greiner Bio-One) and grown under static conditions at 37°C in an incubator with humid atmosphere. After 24 h of incubation, planktonic cells were removed and wells were washed with water prior to staining with 0.1% (wt/vol) crystal violet for 30 min at room temperature. Wells were carefully washed with water to remove excess staining solution, air-dried and the retained crystal violet was solubilized in 95% ethanol for 30 min at room temperature. For quantification, 125 µl of the resulting solution were transferred into a 96-well polystyrene microtiter plate (Nunc) and absorbance was measured at 550 nm.
Analysis of static biofilms grown at the bottom of 96-well plates was performed as previously described [58]. In brief, overnight LB cultures were adjusted to an OD600 of 0.02 with fresh medium and transferred into a half-area 96-well µClear plate (Greiner Bio-One, 100 µl/well, four replica per strain). The plate was covered with an air-permeable foil and incubated at 37°C in an incubator with humid atmosphere. After 24 h, bacteria were stained with 50 µl diluted staining solution (LIVE/DEAD BacLight Bacterial Viability kit, Molecular Probes/Invitrogen, final concentration of 1.4 µM Syto9 and 8.3 µM propidium iodide) and further incubated for 48 h at 37°C. Microscopic analysis of biofilm formation on the bottom of microtiter plates was performed using an Olympus Fluoview 1000 confocal laser scanning microscope equipped with a 40×/0.90 air objective. Image stacks were acquired in the center of each well with a step size of 2 µm. Images of each experiment were processed and analyzed as previously described [58] and visualized using the IMARIS software package (version 5.7.2, Bitplane).
To monitor biofilm formation in the microtiter plates over time, the constitutively GFP-expressing plasmid pSunny [59] was transferred into P. aeruginosa by conjugation and biofilms were grown, images acquired and data analyzed as described above except that the staining step was omitted. To determine the amount of bacteria attached to the substratum, we acquired single images at the bottom of the wells. We processed the images with a pseudo flat field filter to adjust uneven luminance. The filtered, gray-scale images were thresholded to segment objects' pixel area from background pixels. The percentage of attached bacteria was calculated by determining the ratio of object pixels to the total number of pixels. All processing steps were performed using ImageJ (version 1.43).
Supporting Information
Substratum coverage by GFP-tagged bacteria as monitored by CLSM. The coverage of the well-bottom of a 96-well plate was monitored after (A, B) 1 h, (C, D) 4 h and (E, F) 7 h of growth in LB at 37°C. The cell clusters observed in (A) and (B) are likely to originate from cell clumps of over night grown pre-cultures used for inoculation. (A, C, E) PA14 wild-type and (B, D, F) PA14 cheR1 transposon mutant.
(TIF)
Acknowledgments
We thank Howard C. Berg (Harvard University) for providing the E. coli strain HCB721 and Robert M. Weis (University of Massachusetts, Amherst) for providing the E. coli Tsr expression plasmid pHSe5. We furthermore thank Lena Radamm for excellent technical assistance.
Footnotes
Competing Interests: BZ is an employee of the commercial company Biaffin GmbH & Co KG which contributed to analysis tools. This does not alter the authors′ adherence to all the PLoS ONE policies on sharing data and materials. All other authors declare that no competing interests exist.
Funding: Financial support from the German Research Foundation (DFG) (SFB900) and of the Helmholtz-Gemeinschaft is gratefully acknowledged. Both funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. FWH and BZ are supported by the grant “Affinity Proteome” from EU (222635). The commercial company Biaffin GmbH & Co KG contributed to analysis tools.
References
- 1.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 2.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 3.Costerton JW. Cystic fibrosis pathogenesis and the role of biofilms in persistent infection. Trends Microbiol. 2001;9:50–52. doi: 10.1016/s0966-842x(00)01918-1. [DOI] [PubMed] [Google Scholar]
- 4.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jørgensen A, et al. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol. 2003;48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- 6.Lemon KP, Higgins DE, Kolter R. Flagellar motility is critical for Listeria monocytogenes biofilm formation. J Bacteriol. 2007;189:4418–4424. doi: 10.1128/JB.01967-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merritt PM, Danhorn T, Fuqua C. Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J Bacteriol. 2007;189:8005–8014. doi: 10.1128/JB.00566-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 9.Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 10.Kirov SM, Castrisios M, Shaw JG. Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect Immun. 2004;72:1939–1945. doi: 10.1128/IAI.72.4.1939-1945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moorthy S, Watnick PI. Genetic evidence that the Vibrio cholerae monolayer is a distinct stage in biofilm development. Mol Microbiol. 2004;52:573–587. doi: 10.1111/j.1365-2958.2004.04000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barken KB, Pamp SJ, Yang L, Gjermansen M, Bertrand JJ, et al. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol. 2008;10:2331–2343. doi: 10.1111/j.1462-2920.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- 13.Adler J. Chemotaxis in bacteria. Science. 1966;153:708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- 14.Blair DF. How bacteria sense and swim. Annu Rev Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- 15.Bourret RB, Stock AM. Molecular information processing: lessons from bacterial chemotaxis. J Biol Chem. 2002;277:9625–9628. doi: 10.1074/jbc.R100066200. [DOI] [PubMed] [Google Scholar]
- 16.Springer WR, Koshland DEJ. Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc Natl Acad Sci U S A. 1977;74:533–537. doi: 10.1073/pnas.74.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stock JB, Koshland DEJ. A protein methylesterase involved in bacterial sensing. Proc Natl Acad Sci U S A. 1978;75:3659–3663. doi: 10.1073/pnas.75.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lupas A, Stock J. Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J Biol Chem. 1989;264:17337–17342. [PubMed] [Google Scholar]
- 19.Armitage JP, Schmitt R. Bacterial chemotaxis: Rhodobacter sphaeroides and Sinorhizobium meliloti–variations on a theme?. Microbiology. 1997;143(Pt 12):3671–3682. doi: 10.1099/00221287-143-12-3671. [DOI] [PubMed] [Google Scholar]
- 20.Boin MA, Austin MJ, Häse CC. Chemotaxis in Vibrio cholerae. FEMS Microbiol Lett. 2004;239:1–8. doi: 10.1016/j.femsle.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 21.Kirby JR. Chemotaxis-like regulatory systems: unique roles in diverse bacteria. Annu Rev Microbiol. 2009;63:45–59. doi: 10.1146/annurev.micro.091208.073221. [DOI] [PubMed] [Google Scholar]
- 22.Rao CV, Ordal GW. The molecular basis of excitation and adaptation during chemotactic sensory transduction in bacteria. Contrib Microbiol. 2009;16:33–64. doi: 10.1159/000219372. [DOI] [PubMed] [Google Scholar]
- 23.Szurmant H, Ordal GW. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev. 2004;68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zusman DR, Scott AE, Yang Z, Kirby JR. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat Rev Microbiol. 2007;5:862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
- 25.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 26.Ferrández A, Hawkins AC, Summerfield DT, Harwood CS. Cluster II che genes from Pseudomonas aeruginosa are required for an optimal chemotactic response. J Bacteriol. 2002;184:4374–4383. doi: 10.1128/JB.184.16.4374-4383.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Güvener ZT, Tifrea DF, Harwood CS. Two different Pseudomonas aeruginosa chemosensory signal transduction complexes localize to cell poles and form and remould in stationary phase. Mol Microbiol. 2006;61:106–118. doi: 10.1111/j.1365-2958.2006.05218.x. [DOI] [PubMed] [Google Scholar]
- 28.Kato J, Nakamura T, Kuroda A, Ohtake H. Cloning and characterization of chemotaxis genes in Pseudomonas aeruginosa. Biosci Biotechnol Biochem. 1999;63:155–161. doi: 10.1271/bbb.63.155. [DOI] [PubMed] [Google Scholar]
- 29.Masduki A, Nakamura J, Ohga T, Umezaki R, Kato J, et al. Isolation and characterization of chemotaxis mutants and genes of Pseudomonas aeruginosa. J Bacteriol. 1995;177:948–952. doi: 10.1128/jb.177.4.948-952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D'Argenio DA, Calfee MW, Rainey PB, Pesci EC. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol. 2002;184:6481–6489. doi: 10.1128/JB.184.23.6481-6489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darzins A. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol Microbiol. 1994;11:137–153. doi: 10.1111/j.1365-2958.1994.tb00296.x. [DOI] [PubMed] [Google Scholar]
- 32.Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitchurch CB, Leech AJ, Young MD, Kennedy D, Sargent JL, et al. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol Microbiol. 2004;52:873–893. doi: 10.1111/j.1365-2958.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 34.Kato J, Kim H, Takiguchi N, Kuroda A, Ohtake H. Pseudomonas aeruginosa as a model microorganism for investigation of chemotactic behaviors in ecosystem. J Biosci Bioeng. 2008;106:1–7. doi: 10.1263/jbb.106.1. [DOI] [PubMed] [Google Scholar]
- 35.DeLange PA, Collins TL, Pierce GE, Robinson JB. PilJ localizes to cell poles and is required for type IV pilus extension in Pseudomonas aeruginosa. Curr Microbiol. 2007;55:389–395. doi: 10.1007/s00284-007-9008-5. [DOI] [PubMed] [Google Scholar]
- 36.Morgan R, Kohn S, Hwang S, Hassett DJ, Sauer K. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J Bacteriol. 2006;188:7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winsor GL, Lo R, Sui SJH, Ung KSE, Huang S, et al. Pseudomonas aeruginosa Genome Database and PseudoCAP: facilitating community-based, continually updated, genome annotation. Nucleic Acids Res. 2005;33:D338–43. doi: 10.1093/nar/gki047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okumura H, Nishiyama S, Sasaki A, Homma M, Kawagishi I. Chemotactic adaptation is altered by changes in the carboxy-terminal sequence conserved among the major methyl-accepting chemoreceptors. J Bacteriol. 1998;180:1862–1868. doi: 10.1128/jb.180.7.1862-1868.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu J, Li J, Li G, Long DG, Weis RM. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- 40.Kuroda A, Kumano T, Taguchi K, Nikata T, Kato J, et al. Molecular cloning and characterization of a chemotactic transducer gene in Pseudomonas aeruginosa. J Bacteriol. 1995;177:7019–7025. doi: 10.1128/jb.177.24.7019-7025.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taguchi K, Fukutomi H, Kuroda A, Kato J, Ohtake H. Genetic identification of chemotactic transducers for amino acids in Pseudomonas aeruginosa. Microbiology. 1997;143(Pt 10):3223–3229. doi: 10.1099/00221287-143-10-3223. [DOI] [PubMed] [Google Scholar]
- 42.Wolfe AJ, Conley MP, Berg HC. Acetyladenylate plays a role in controlling the direction of flagellar rotation. Proc Natl Acad Sci U S A. 1988;85:6711–6715. doi: 10.1073/pnas.85.18.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simms SA, Subbaramaiah K. The kinetic mechanism of S-adenosyl-L-methionine: glutamylmethyltransferase from Salmonella typhimurium. J Biol Chem. 1991;266:12741–12746. [PubMed] [Google Scholar]
- 44.Perez E, Stock AM. Characterization of the Thermotoga maritima chemotaxis methylation system that lacks pentapeptide-dependent methyltransferase CheR:MCP tethering. Mol Microbiol. 2007;63:363–378. doi: 10.1111/j.1365-2958.2006.05518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Djordjevic S, Stock AM. Chemotaxis receptor recognition by protein methyltransferase CheR. Nat Struct Biol. 1998;5:446–450. doi: 10.1038/nsb0698-446. [DOI] [PubMed] [Google Scholar]
- 46.Shiomi D, Zhulin IB, Homma M, Kawagishi I. Dual recognition of the bacterial chemoreceptor by chemotaxis-specific domains of the CheR methyltransferase. J Biol Chem. 2002;277:42325–42333. doi: 10.1074/jbc.M202001200. [DOI] [PubMed] [Google Scholar]
- 47.Pamp SJ, Tolker-Nielsen T. Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J Bacteriol. 2007;189:2531–2539. doi: 10.1128/JB.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tolker-Nielsen T, Brinch UC, Ragas PC, Andersen JB, Jacobsen CS, et al. Development and dynamics of Pseudomonas sp. biofilms. J Bacteriol. 2000;182:6482–6489. doi: 10.1128/jb.182.22.6482-6489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dötsch A, Pommerenke C, Bredenbruch F, Geffers R, Häussler S. Evaluation of a microarray-hybridization based method applicable for discovery of single nucleotide polymorphisms (SNPs) in the Pseudomonas aeruginosa genome. BMC Genomics. 2009;10:29. doi: 10.1186/1471-2164-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klockgether J, Munder A, Neugebauer J, Davenport CF, Stanke F, et al. Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J Bacteriol. 2010;192:1113–1121. doi: 10.1128/JB.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rice MS, Dahlquist FW. Sites of deamidation and methylation in Tsr, a bacterial chemotaxis sensory transducer. J Biol Chem. 1991;266:9746–9753. [PubMed] [Google Scholar]
- 53.Lin LN, Li J, Brandts JF, Weis RM. The serine receptor of bacterial chemotaxis exhibits half-site saturation for serine binding. Biochemistry. 1994;33:6564–6570. doi: 10.1021/bi00187a025. [DOI] [PubMed] [Google Scholar]
- 54.Perez E, West AH, Stock AM, Djordjevic S. Discrimination between different methylation states of chemotaxis receptor Tar by receptor methyltransferase CheR. Biochemistry. 2004;43:953–961. doi: 10.1021/bi035455q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simms SA, Stock AM, Stock JB. Purification and characterization of the S-adenosylmethionine:glutamyl methyltransferase that modifies membrane chemoreceptor proteins in bacteria. J Biol Chem. 1987;262:8537–8543. [PubMed] [Google Scholar]
- 56.Yeung ATY, Torfs ECW, Jamshidi F, Bains M, Wiegand I, et al. Swarming of Pseudomonas aeruginosa is controlled by a broad spectrum of transcriptional regulators, including MetR. J Bacteriol. 2009;191:5592–5602. doi: 10.1128/JB.00157-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 58.Müsken M, Di Fiore S, Dötsch A, Fischer R, Häussler S. Genetic determinants of Pseudomonas aeruginosa biofilm establishment. Microbiology. 2010;156:431–441. doi: 10.1099/mic.0.033290-0. [DOI] [PubMed] [Google Scholar]
- 59.Güttler A. Entwicklung eines Rekombinationssystems zum Studium des horizontalen Gentransfer auf unbekannte Bakterien sowie die Konstruktion von Transportvektoren mit gfp als Markergen. PhD Thesis 1998 [Google Scholar]
- 60.Woodcock DM, Crowther PJ, Doherty J, Jefferson S, DeCruz E, et al. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in gram negative bacteria. Biotechnology. 1983;1:784–790. [Google Scholar]
- 62.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.West SE, Schweizer HP, Dall C, Sample AK, Runyen-Janecky LJ. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Substratum coverage by GFP-tagged bacteria as monitored by CLSM. The coverage of the well-bottom of a 96-well plate was monitored after (A, B) 1 h, (C, D) 4 h and (E, F) 7 h of growth in LB at 37°C. The cell clusters observed in (A) and (B) are likely to originate from cell clumps of over night grown pre-cultures used for inoculation. (A, C, E) PA14 wild-type and (B, D, F) PA14 cheR1 transposon mutant.
(TIF)