Summary
RatA (YfjG) is a toxin encoded by the ratA-ratB (yfjG-yfjF) operon on the Escherichia coli genome. Induction of RatA led to the inhibition of protein synthesis, while DNA and RNA synthesis was not affected. The stability of mRNAs was also unchanged as judged by in vivo primer extension experiments and by Northern blotting analysis. The ribosome profile of the cells overexpressing RatA showed that 70S ribosomes as well as polysomes significantly decreased with concomitant increase of 50S and 30S subunits. The addition of purified RatA to a cell-free system inhibited the formation of 70S ribosomes even in the presence of 6 mM Mg2+. RatA was specifically associated with 50S subunits, indicating that it binds to 50S subunits to block its association with 30S subunits leading to the inhibition of formation of 70S ribosomes. However, RatA did not cause dissociation of 70S ribosomes and its anti-association activity was blocked by paromomycin, an inhibitor for IF3, an essential initiation factor, having 21% sequence homology with RatA. Here we demonstrate that RatA is a new E. coli toxin, which effectively blocks the translation initiation step. We propose that this toxin of previously unknown function be renamed as RatA (Ribosome association toxin A).
Introduction
YfjG is a toxin (Brown and Shaw, 2003) encoded by the E. coli genome. The yfjG-yfjF operon maps on 59.32 min of the E. coli chromosome and the yfjG gene has been shown not to be essential as it can be deleted without any detrimental effect on the cell growth (Baba et al., 2006).
To date, Some of toxin-antitoxin (TA) systems have been well characterized, including relBE, (Pedersen et al., 2003; Takagi et al., 2005), chpBIK (Zhang et al., 2005b), mazEF (Kamada et al., 2003; Zhang et al., 2005a; Zhang et al., 2003), yefM-yoeB (Kamada and Hanaoka, 2005; Zhang and Inouye, 2009), dinJ-yafQ (Motiejunaite et al., 2007; Prysak et al., 2009), hipB-hipA (Korch et al., 2003; Schumacher et al., 2009), hicA-hicB (Jorgensen et al., 2009; Makarova et al., 2006), prlF-yhaV (Schmidt et al., 2007), ybaJ-hha (Barrios et al., 2006; Garcia-Contreras et al., 2008), higB-higA (Christensen-Dalsgaard et al., 2010; Hurley and Woychik, 2009), yafN-yafO (Zhang et al., 2009) and mqsR-ygiT (Yamaguchi et al., 2009). Interestingly, RelE by itself has no endoribonuclease activity (Pedersen et al., 2003) and has been demonstrated to interact tightly with several domains of 16S ribosomal RNA, which stimulates the endogenous ribonuclease activity of RelE (Neubauer et al., 2009). On the other hand, MazF is a ribosome-independent endoribonuclease that cleaves mRNAs specifically at ACA sequences, and is thus termed an mRNA interferase (Zhang et al., 2003). ChpBK encoded by the E. coli genome was also found to be another mRNA interferase specifically cleaving mRNAs at UAC sequences (Zhang et al., 2005b). YhaV was also reported to be an mRNA interferase (Schmidt et al., 2007). MqsR was recently found to be a ribosome-independent mRNA interferase cleaving mRNAs at GCU (Yamaguchi et al., 2009). On the other hand, YafQ has been shown to be associated with ribosomes cleaving mRNAs at the lysine (AAA) codon near the initiation codon (Prysak et al., 2009). YoeB is also a ribosome-associating toxin functioning as a ribosome-dependent mRNA interferase (Zhang and Inouye, 2009). It binds to the 50S ribosomal subunit in 70S ribosomes and interacts with the A site which leads to mRNA cleavage. As a result, translation initiation is effectively inhibited (Zhang and Inouye, 2009). YafO is another 50S ribosome-associated mRNA interferase, which inhibits protein synthesis (Zhang et al., 2009). HicA (Jorgensen et al., 2009) has been shown to function as mRNA interferases, HipA is another potent E. coli toxin and the X-ray structure of the complex of HipA and HipB (antitoxin) has been recently determined and suggests that HipA has protein kinase activity (Schumacher et al., 2009).
In the present paper, we demonstrate that YfjG encoded by the gene yfjG belonging to the yfjG-yfjF operon is a new E. coli toxin. We show that YfjG effectively inhibits cell growth upon its induction and binds to the 50S ribosomal subunits to inhibit the 70S ribosome association. This results in blocking of translation initiation. However, it does not cause dissociation of 70S ribosomes. Interestingly, its anti-association activity is blocked by paromomycin, an inhibitor for IF3, an essential initiation factor (Laursen et al., 2005), which is known to have anti-association activity to prevent significant amounts of subunits from associating into 70S ribosomes (Hirokawa et al., 2005). It should be noted that YfjG has 21% sequence homology with IF3, however IF3 associates with 30S ribosome subunits (Allen et al., 2005; Hirokawa et al., 2007) and does not inhibit the 70S ribosome association once the translation initiation step is completed (Laursen et al., 2005). We propose that this toxin, YfjG, of previously unknown function be renamed as RatA (Ribosome association toxin A).
Results
RatA Inhibits Protein Synthesis
To identify the cellular function(s) inhibited by RatA, in vivo protein and DNA synthesis was examined after RatA induction. [35S] methionine incorporation was significantly inhibited at 20 min after RatA induction (Fig. 1B), while the inhibition of 3H-thymidine incorporation was observed at a lower level only at 30 min after RatA induction (Fig. 1C). These results demonstrate that RatA inhibits protein synthesis, but may not directly affect DNA synthesis. Notably, cell growth was completely inhibited 30 min after the addition of arabinose (Fig. 1A). In vivo pulse-labeling experiments with [35S]methionine demonstrated that RatA is a general protein synthesis inhibitor (Fig. 1D). However, RatA shows a much slower inhibitory effect on cell growth in comparison with other E. coli toxins such as MazF or YoeB (Zhang and Inouye, 2009; Zhang et al., 2003). Moreover, protein synthesis was not completely inhibited and a low level of protein synthesis was maintained even after 120 min of RatA induction, suggesting that RatA is not a typical toxin encoded by a canonical TA (toxin-antitoxin) system.
Fig. 1. Effect of RatA on Protein and DNA Synthesis.

(A) Growth Curve of BW25113 cells containing pBAD30-RatA plasmid in the LB medium. Arabinose was added at 150 min as indicated by a solid arrow. (B) Effect of RatA on in vivo 35S-methionine cumulated incorporation in the cells. E. coli BW25113 cells containing pBAD30-RatA were grown at 37°C in glycerol-M9 medium. When the A600 of the culture reached 0.3, arabinose was added to a final concentration of 0.2% and 35S-methionine and 50 μg/ml methionine was added simultaneously. 35S-methionine incorporation into E. coli BW25113 cells containing pBAD30-RatA was measured after RatA induction as indicated in Fig. 2B. (C) Effect of RatA on in vivo 3H-thymidine cumulated incorporation in the cells. E. coli BW25113 cells containing pBAD30-RatA were grown at 37°C in glycerol-M9 medium. When the A600 of the culture reached 0.3, arabinose was added to a final concentration of 0.2% and 3H-thymidine and 10 μg/ml thymidine was added simultaneously. 3H-thymidine incorporation into E. coli BW25113 cells containing pBAD30-RatA was measured after RatA induction as indicated in Fig. 2C. (D) SDS-PAGE analysis of in vivo protein synthesis after the induction of RatA.
RatA Induction does not Cause Cleavage of Cellular mRNAs
Next, we tested if RatA functions as an mRNA interferase to degrade cellular mRNAs. Northern blotting analysis shown in Fig. 2A demonstrates that the induction of RatA does not significantly reduce the stability of cellular mRNAs, since the amounts of the full-length of lpp and ompF mRNAs were almost unchanged even after 120-min induction of RatA. The ompA mRNA is stable up to 30 min after RatA induction and its amount decreases albeit not significantly at 60 min. It should be noted that the 23S and 16S rRNAs were also stable even after 120-min RatA induction (Fig. 2B). These results indicate that RatA does not function as an mRNA interferase like MazF or YoeB.
Fig. 2. Effect of RatA on Cellular mRNAs in vivo.

(A) Total cellular RNAs were extracted from E. coli BW25113 cells containing pBAD30-RatA at various time points as indicated after the addition of arabinose and subjected to Northern blot analysis using radiolabeled lpp, ompA and ompF ORF as probes. (B) The same total cellular RNA in (A) was analyzed by 1% native agarose gel followed by ethidium bromide staining.
Polysome Profiles After RatA Induction
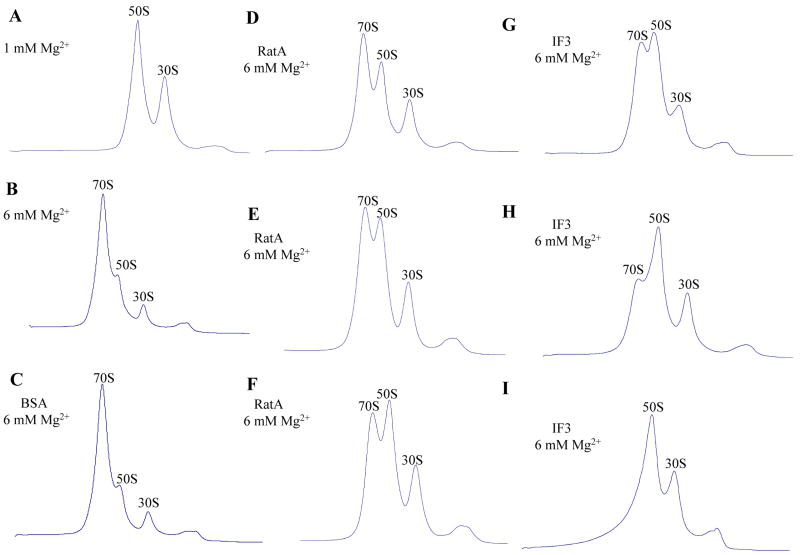
Since RatA inhibits protein synthesis (Fig. 1B and D), we next examined whether RatA affects the polysome profile. As shown in Fig. 3, when the polysome pattern of E. coli BW25113 cells carrying the pBAD-RatA plasmid was analyzed by sucrose density gradient at 30 min and 60 min after induction of RatA. The overexpression of RatA at 30 and 60 min time points resulted in increase of the 50S and 30S peaks, while the 70S peak remained almost constant. Notably however, the polysome peak decreased (compare Fig. 3B and C with Fig. 3A, the ratio of 70S to 50S to 30S ribosome is 2.4:1:1 in Fig. 3A, 2.0:1.8:1 in Fig. 3B and 1.8:2.2:1 in Fig. 3C). It should be noted a similar polysome profile was obtained when IF3 was overproduced (Fig. 3 H-I), which is known to have anti-association activity (Hirokawa et al., 2005), suggesting that RatA may have a similar anti-association activity. However, IF3 expression did not significantly reduce the amount of polysomes. Notably, RatA was detected in both 70S and 50S ribosome fractions (Figs. 3B and C), while IF3 was observed in both 70S and 30S ribosome fractions (Fig. 3H and I), indicating that RatA appears to function differently from IF3.
Fig. 3. Effect of RatA on the polysome profile in vivo.

Ribosome profiles were analyzed by sucrose density gradient centrifugation as described in the Experimental Procedures. (A) Polysome profiles of E. coli BW25113 containing pBAD30-RatA without RatA induction using 10 mM Mg2+ solution. The ratio of 70S to 50S to 30S ribosome is 2.4:1:1. (B) Polysome profiles of E. coli BW25113 containing pBAD30-RatA at 30 min after RatA induction using 10 mM Mg2+ solution. The ratio of 70S to 50S to 30S ribosome is 2.0:1.8:1. (C) Polysome profiles of E. coli BW25113 containing pBAD30-RatA at 60 min after RatA induction using 10 mM Mg2+ solution. The ratio of 70S to 50S to 30S ribosome is 1.6:2.2:1. (D) Polysome profiles of E. coli BW25113 containing pBAD30-RatA without RatA induction using 6 mM Mg2+ solution. (E) Polysome profiles of E. coli BW25113 containing pBAD30-RatA at 30 min after RatA induction using 6 mM Mg2+ solution. (F) Polysome profiles of E. coli BW25113 containing pBAD30-RatA at 60 min after RatA induction using 6 mM Mg2+ solution. (G) Polysome profiles of E. coli BW25113 containing pCN4A-IF3 without IF3 induction using 10 mM Mg2+ solution. (H) Polysome profiles of E. coli BW25113 containing pCN4A-IF3 at 30 min after IF3 induction using 10 mM Mg2+ solution. (I) Polysome profiles of E. coli BW25113 containing pCN4A-IF3 at 60 min after IF3 induction using 10 mM Mg2+ solution. In B, C, H and I, an equal volume of each sucrose density gradient fraction was used to carry out Western blot analysis to detect RatA and IF3 in each gradient fraction using anti-poly-histidine antibodies.
It is known that 70S ribosomes dissociate to 30S and 50S subunits when the concentrations of Mg2+ are in the range of 6 to 10 mM. In order to determine the best Mg2+ concentration to test the anti-association activity of RatA in vitro, we carried out an experiment to determine the in vivo polysome profile in 6 mM Mg2+. As shown in Fig. 3E and F, the 50S and 30S ribosomal peaks increased more significantly compared with Fig. 3B-C, which were carried out in 10 mM Mg2+. These results indicate that the anti-association activity of RatA is more easily observed in 6 mM rather than 10 mM Mg2+.
RatA Blocks 70S Ribosome Association in vitro
In order to test if RatA can block 70S ribosome association, 70S ribosomes were first dissociated into 50S and 30S ribosomal subunits in the buffer containing l mM Mg2+ at 30°C, and then different amounts of purified RatA protein were added. The reaction mixtures were incubated at 30°C for 10 min, and then the Mg2+ concentration was raised to 6 mM. The reaction mixtures were further incubated at 30°C for 10 min. Ribosome profiles were analyzed through a 5%–40% sucrose density gradient ultracentrifugation. As shown in Fig. 4, 70S ribosomes were completely dissociated into 50S and 30S ribosomal subunits in 1 mM Mg2+ (Fig. 4A). When the Mg2+ concentration was increased to 6 mM, almost 80% of 50S and 30S ribosomal subunits associated to form 70S ribosomes (Fig. 4B). However, when RatA were added to the mixture before the Mg2+ concentration was raised to 6 mM, the association of 50S and 30S ribosomal subunits was strongly inhibited (compare Fig. 4B with 4D-F), indicating that RatA blocks 70S ribosome association in a manner similar to that of IF3, which is known to have anti-association activity (Fig. 4G-I) (Hirokawa et al., 2005). However, IF3 seems to exhibit a much more efficient anti-association activity than RatA in vitro. It should be noted that bovine serum albumin (BSA) could not block 70S ribosome association at a concentration similar to that of RatA (Fig. 4C).
Fig. 4. Effect of RatA on the polysome profile in vitro.
70 pmol of 70S ribosomes were incubated in the buffer S at 30°C for 10 min, then different amount of RatA or IF3 was added, the mixture was incubated for another 10 min, then the concentration of Mg2+ was raised to 6 mM and the reaction mixture were further incubated for 10 min before loading to a 5%–40% sucrose density gradient in buffer T following ultracentrifugation and the sedimentation behavior was monitored with the spectrophotometer. (A) Ribosome profile in the presence of 1 mM Mg2+ using a 5%–40% sucrose density gradient in buffer S. (B) Ribosome profile in the presence of 6 mM Mg2+. (C) Ribosome profile in the presence of 6 mM Mg2+ and 10 μg BSA. (D) Ribosome profile in the presence of 6 mM Mg2+ and 0.1 μg RatA. (E) Ribosome profile in the presence of 6 mM Mg2+ and 0.4 μg RatA. (F) Ribosome profile in the presence of 6 mM Mg2+ and 1.25 μg RatA. (G) Ribosome profile in the presence of 6 mM Mg2+ and 0.06 μg IF3. (H) Ribosome profile in the presence of 6 mM Mg2+ and 0.25 μg IF3. (I) Ribosome profile in the presence of 6 mM Mg2+ and 1 μg IF3.
RatA Binds to 50S Ribosomal Subunits and Does not Have Dissociation Activity
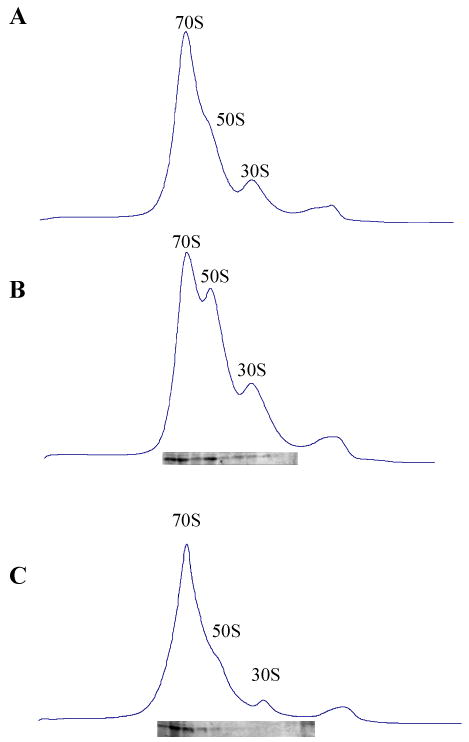
IF3 is known to bind to 30S ribosomal subunits (Allen et al., 2005). In order to examine which ribosomal subuits RatA binds to, we collected each sucrose density gradient fraction in Fig. 3 B and C after RatA induction and carried out Western blot analysis. It was observed that the majority of RatA was associated with both the 70S ribosome and 50S ribosome fractions, but not with the 30S ribosome fraction (Fig. 3 B and C). Next, the cell lysates were prepared in 1 mM Mg2+ to dissociate 70S ribosomes to 50S and 30S ribosomal subunits and analyzed by sucrose density gradient centrifugation. The majority of RatA was detected in the 50S ribosome fraction (Fig. 5A). The identical result was obtained in vitro by Western blot analysis when RatA was incubated with 50S and 30S ribosome in the buffer S at 30°C for 10 min following sucrose density gradient analysis (Fig. 5B), suggesting that RatA is a 50S ribosome associating protein.
Fig. 5. RatA binds to 50S ribosomes and does not have dissociation activity in vitro.

(A) 70S ribosomal fractions of the same lysate used in Fig. 3C were disassociated into 50S and 30S ribosomal subunits using 1 mM Mg2+. Western blot analysis was carried out to detect RatA in each gradient fraction. (B) 70 pmol of 70S ribosomes were incubated in the buffer S at 30°C for 10 min, then 0.55 μg of RatA was added, the mixture was incubated for another 10 min before loading to a 5%–40% sucrose density gradient in buffer S following ultracentrifugation and the sedimentation behavior was monitored using spectrophotometer. Western blot analysis was carried out to detect RatA in each gradient fraction. (C) Ribosome profile in the presence of 6 mM Mg2+. (D) 70 pmol of 70S ribosomes were incubated in the buffer T at 30°C for 10 min, then 0.55 μg of RatA was added, the mixture was incubated for another 10 min before loading to a 5%–40% sucrose density gradient in buffer T following ultracentrifugation and the sedimentation behavior was monitored using spectrophotometer. Western blot analysis was carried out to detect RatA in each gradient fraction using monoclonal anti-polyhistidine antibodies.
It is known that IF3 does not dissociate 70S ribosomes to 30S and 50S ribosomal subunits (Hirokawa et al., 2005). In order to test if RatA has the dissociation activity, the identical amount of RatA used in the antiassociation activity measurement was mixed with 70S ribosomes and the mixture was incubated at 30°C for 10 min. Ribosome profiles were then analyzed through a 5%–40% sucrose density gradient ultracentrifugation. As shown in Fig. 5D, 0.4 μg RatA could not dissociate 70S ribosomes into 50S and 30S subunits (compare Fig. 5C with 5D). This concentration of RatA was able to block 70S ribosomes association (Fig. 4F). However, RatA was still bound to 70S and 50S ribosomes (Fig. 5D).
Paromomycin Interferes with the Inhibitory Function of RatA In Vivo
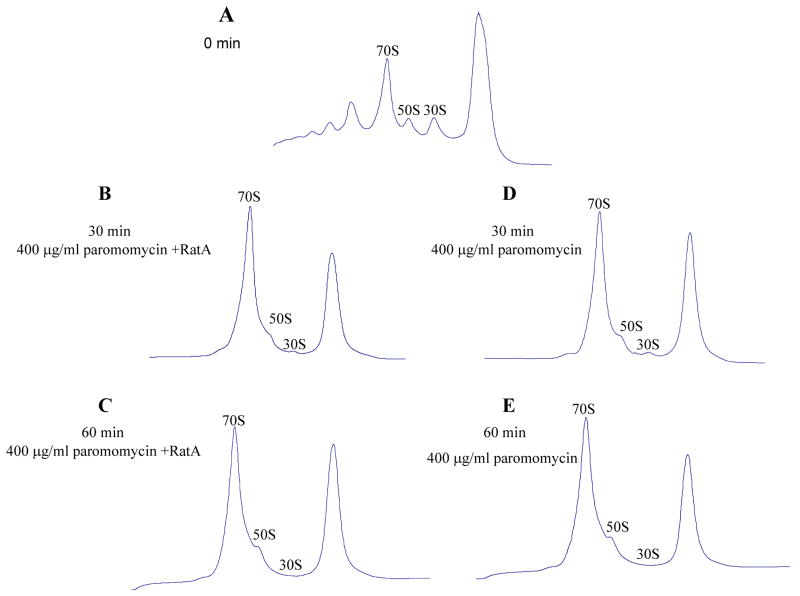
It is known that two paromomycin molecules bind to a ribosome, one to the decoding center on H44 of the small ribosomal subunit to cause miscoding of translation (Carter et al., 2000) and the other to H69 of the large subunit to restrict helical dynamics required for translocation or ribosome recycling (Borovinskaya et al., 2007). Thus, it is speculated that paromomycin may stabilize the subunit interaction by stabilization of the intersubunit B2a bridge formed by these two helices. Moreover, paromomycin inhibits the anti-association activity of IF3 (Hirokawa et al., 2007). Therefore, it is interesting to examine whether the anti-association activity of RatA is inhibited by paromomycin. E. coli BW25113 cells carrying the pBAD-RatA plasmid were treated with 400 μg/ml of paromomycin before RatA induction. The polysomal profiles after RatA induction were similar to those as shown in Fig. 3B-C without paromomycin treatment (data not shown). However, after treatment with paromomycin, the 70S ribosome peak increased, while 50S and 30S ribosomal subunit peaks decreased (compare Fig. 6B-C with 6A). A similar result was observed when the cells were treated with only paromomycin (compare Fig. 6D and E with Fig. 6B and C), suggesting that paromomycin may enhance 70S ribosome association to inhibit RatA anti-association activity in vivo or paromomycin may completely block the inhibitory function of RatA by efficiently inhibiting RatA synthesis.
Fig. 6. Effect of Paromomycin on RatA activity in vivo.
BW25113 cells containing the pBAD30-RatA plasmid were grown at 37°C in 300 ml of LB medium, and at an A600 of 0.6, the cells were divided into two parts, one part was added 400 μg/ml of paromomycin and final concentration of 0.2% arabinose, another part was only added 400 μg/ml of paromomycin. After 30 and 60 min induction, chloramphenicol was added to a final concentration of 200 μg/ml. Extracts were prepared using liquid nitrogen, freezing and thawing four times. About 10 A260 units of the lysate were layered onto a 5-40% sucrose gradient in buffer A (10 mM Tris-Cl pH 7.8, 60 mM NH4Cl, 10 mM MgCl2 and 1 mM DTT), and centrifuged at 35, 000 rpm for 3 h at 4 °C in a Beckman SW41Ti rotor. Gradients were analyzed with continuous monitoring at 254 nm. (A) Polysome profiles of E. coli BW25113 containing pBAD30-RatA without RatA induction. (B) Polysome profiles of E. coli BW25113 containing pBAD30-RatA at 30 min after RatA induction and 400 μg/ml of paromomycin. (C) Polysome profiles of E. coli BW25113 containing pBAD30-RatA at 60 min after RatA induction and 400 μg/ml of paromomycin. (D) Polysome profiles of E. coli BW25113 containing pBAD30-RatA at 30 min after the addition of 400 μg/ml of paromomycin. (E) Polysome profiles of E. coli BW25113 containing pBAD30-RatA at 60 min after the addition of 400 μg/ml of paromomycin.
Paromomycin Inhibits Anti-association Activity of RatA In Vitro
Since paromomycin blocks the inhibitory effect of RatA in vivo, we next tested if paromomycin inhibits the anti-association activity of RatA in vitro. As shown in Fig. 7A, 50S and 30S ribosomal subunits dissociated from 70S ribosomes at 1 mM Mg2+ were able to be re-associated when the concentration of Mg2+ was raised to 6 mM. However, when RatA was added, the reassociation was inhibited (Fig. 7B). On the other hand, when RatA and paromomycin was added together, most of ribosomal subunits were able to be re-associated into 70S ribosomes (compare Fig. 7C with Fig. 7A). These results indicate that paromomycin inhibits the anti-association activity of RatA in vitro. It should be noted the intensity of RatA bands in 70S and 50S ribosome fractions did not change after the addition of paromomycin, indicating that paromomycin did not inhibit RatA binding to 50S ribosomes (compare 7B with 7C). Further study is needed to elucidate the exact mechanism of action of RatA.
Fig. 7. Effect of Paromomycin on RatA activity in vitro.
70 pmol of 70S ribosomes were incubated in the buffer S at 30°C for 10 min, then 0.4 μg of RatA or 400 μg/ml of paromomycin was added, the mixture was incubated for another 10 min, then the concentration of Mg2+ was increased to 6 mM and the mixture were further incubated for 10 min before loading on to a 5%–40% sucrose density gradient in buffer T following ultracentrifugation and the sedimentation behavior was monitored using spectrophotometer. (A) Ribosome profile in the presence of 6 mM Mg2+. (B) Ribosome profile in the presence of 6 mM Mg2+ and 0.4 μg of RatA. (C) Ribosome profile in the presence of 6 mM Mg2+, 0.4 μg of RatA and 400 μg/ml of paromomycin. In B and C, Western blot analysis was carried out to detect RatA in each gradient fraction using anti-RatA antibody.
RatA Mutants Defective in 50S Ribosome Binding
In order to prove that RatA specifically binds to 50S ribosomes to block 70S ribosome association, we created a number of mutants by site-directed mutagenesis such as R78A, R114A, R134A, R150A, Y40F, F75Y, L91A, F96Y, W103L, F105Y, F136Y, F147Y, D93A, K97D, K126D, and K137D, all of which however, retained the toxicity (data not shown). Even with a quadruple mutation with D93A, K97D, K126D and K137D, the RatA toxicity was not eliminated (data not shown). However, when the 10-residue c-myc tag was added to the N-terminal end of RatA (N-myc-RatA), it lost its toxicity (Fig. 8G), was no more able to affect the polysome profile (compare Fig. 8D with E and F) and did not bind to 50S ribosomes (Fig. 8F). It should be noted that in a control experiment with E. coli BL21(DE3), wild-type RatA was still toxic and affected the polysome profile (Fig. 8A-C) and bound to 50S ribosomes (Fig. 8C), indicating that RatA toxicity is not strain specific. Taken together this suggests that, RatA specifically binds to 50S ribosomes to block the 70S ribosome assembly. Interestingly, as shown in Fig. 8I, an identical polysome profile pattern was obtained after RatA induction in pET-21c-RatA E. coli BL21 (DE3) system. Western blotting showed that the level of RatA increased less than 10 times (compare Fig. 8I with H). The ratio of RatA to 50S ribosome is ∼10 (data not shown).
Fig. 8. Addition of N-terminal myc-tag to RatA eliminated its toxicity.
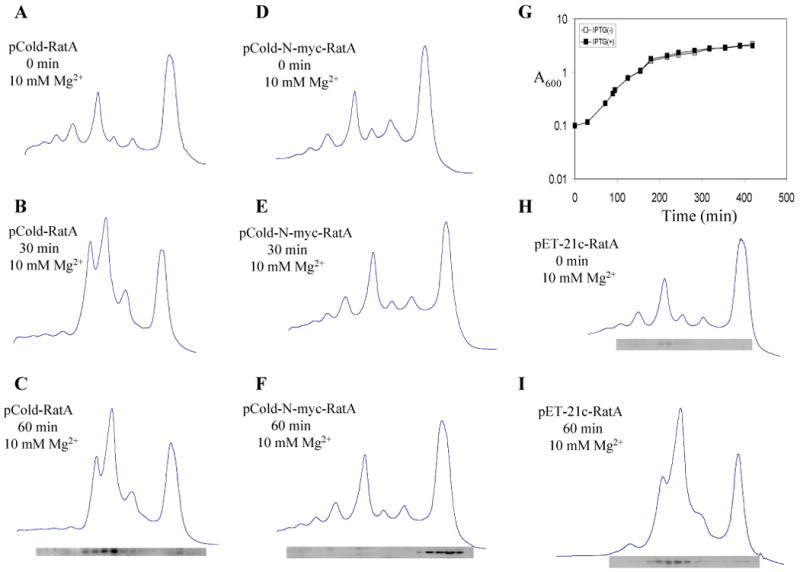
Ribosome profiles were analyzed in 10 mM Mg2+ by sucrose density gradient centrifugation as described in the Experimental Procedures. (A) Polysome profiles of E. coli BL21(DE3) containing pCold-RatA without RatA induction, (B) at 30 min after RatA induction, (C) at 60 min after RatA induction. (D) Polysome profiles of E. coli BL21(DE3) containing pCold–N-myc-RatA without RatA induction, (E) at 30 min after RatA induction, (F) at 60 min after RatA induction. (G) Growth curve of E. coli BL21(DE3) containing pCold–N-myc-RatA. (H) Polysome profiles of E. coli BL21(DE3) containing pET-21c-RatA without RatA induction and (I) at 60 min after RatA induction. In C, F, H and I, Western blot analysis was carried out to detect the RatA protein in each gradient fraction using monoclonal anti-poly-histidine antibodies.
Discussion
In the present paper, we demonstrated that RatA is an inhibitor of 70S ribosome association, which leads to inhibition of protein synthesis. Upon RatA induction, [35S]methionine incorporation into cellular proteins was almost completely inhibited within 60 min (Fig. 1D). Correspondingly, both 50S and 30S ribosomal peaks increase (Fig. 3B-C and Fig. 3E-F). This in vivo anti-association activity of RatA was also confirmed in an in vitro experiment (Fig. 4E-G). It is important to note that RatA does not have the ability to dissociate 70S ribosomes into smaller subunits (Fig. 5D).
It should be also noted that although RatA has a similar activity as that of IF3 to inhibit the association of 30S and 50S ribosomal subunits into 70S ribosomes, they appear to function quite differently. First, they share only 21% sequence homology and secondly RatA associates with 50S ribosomal subunits (Fig 3B-C and Fig. 5A-B), while IF3 interacts with 30S ribosomal subunits (Fig. 3H-I)(Allen et al., 2005; Hirokawa et al., 2007). Furthermore, RatA induction reduces polysomal peaks (compare Fig. 3B and C with Fig. 3A and compare Fig. 3E and F with Fig. 3D), while IF3 induction does not alter the polysomal peaks (compare Fig.3H and I with 3G). Interestingly, site-directed mutagenesis at the eight arginine residues in the IF3C domain has shown that the arginine residues at positions 99, 112, 116, 147, and 168 are important for IF3 binding to the 30S ribosomal subunits (Petrelli et al., 2003). However, the similar site-directed mutagenesis of corresponding arginine residues, R78A, R114A, R134A, R150A and K152A of RatA did not affect the toxicity of RatA (data not shown). Moreover, the site-directed mutagenesis of Y40F, F75Y, L91A, F96Y, W103L, F105Y, F136Y, F147Y, D93A, K97D, K126D, K137D in RatA also did not affect its toxicity (data not shown). Even a quadruple mutation with D93A, K97D, K126D and K137D did not affect the RatA toxicity either. These results indicate that the mechanism of action of RatA may be quite different from that of IF3. It is also interesting to note that the addition of the c-myc sequence at the N-terminal end of RatA (but not at the C-terminal end) inhibited the binding of RatA to 50S ribosomes (Fig. 8F), completely eliminating the RatA toxicity (Fig. 8G). This further supports that RatA specifically binds to 50S ribosomes to inhibit its association with 30S ribosomes and block the protein synthesis.
The RatA gene shares an operon with the downstream yfjF gene overlapping 11 bases in a similar manner as all the other E. coli TA systems on the chromosome, suggesting that YfjF is the antitoxin for RatA and that they form a stable TA complex. We attempted to demonstrate the complex formation between the two proteins using purified RatA and YfiF. However, under the conditions tested we were unable to show the complex formation. Moreover, co-expression of YfjF with RatA did not neutralize the RatA toxicity on plates. It is interesting to note that a yfjG deletion strain has been isolated in the Keio collection (Baba et al., 2006), suggesting that it is not essential. All these results suggest that YfjF may not be the antitoxin for RatA and raise an intriguing question as to how the RatA expression is regulated in the cells. Further studies are needed to elucidate the relationship of RatA with YfjF.
It is quite intriguing that E. coli is equipped with a number of toxins to inhibit protein synthesis by different mechanisms; for example MazF (Zhang et al., 2005a; Zhang et al., 2003), ChpBK (Zhang et al., 2005b), MqsR (Yamaguchi et al., 2009), HicA (Jorgensen et al., 2009) and YgjN (Christensen-Dalsgaard et al., 2010) function as mRNA interferases to cleave mRNAs in a ribosome-independent manner. On the other hand, the other toxins such as RelE (Pedersen et al., 2003), YafO (Zhang et al., 2009), YoeB (Zhang and Inouye, 2009), YafQ (Prysak et al., 2009) and RatA (this study) are ribosome-associating toxins. Among these ribosome-associating toxins, RelE (Pedersen et al., 2003), YafO (Zhang et al., 2009), YoeB (Zhang and Inouye, 2009) and YafQ (Prysak et al., 2009) function as ribosome-dependent mRNA interferases to inhibit translation. However, RatA is unique as it inhibits the association of 30S and 50S ribosomal subunits to form 70S ribosomes and thereby blocking the translation initiation. The diversity of mechanisms targeting translation in E. coli TA systems may be important to form an intricate TA network in the cell for cellular adaptation to ever-changing environmental stresses in nature. It remains to be elucidated how these TA systems play various physiological roles under different stress conditions. On the basis of our finding on the function of YfjG in the present study, we propose to rename the YfjG gene to the ratA gene (ribosome association toxin A) and its gene product to RatA protein.
Experimental procedures
Strains and Plasmids
E. coli BL21(DE3), BW25113 (ΔaraBAD) (Datsenko and Wanner, 2000) and MRE600 (Swaney et al., 1998) were used. The ratA gene was amplified by PCR using the E. coli genomic DNA as template and cloned into the NdeI-BamHI sites of pColdI (Takara). This construction created an in-frame translation fusion with a (His)6 tag at the RatA N-terminal end. The plasmid was designated as pCold-(His)6RatA. The ratA gene was cloned into both pBAD30 and pBAD33 vectors, creating pBAD30-RatA and pBAD33-RatA, respectively, to tightly regulate RatA expression by the addition of arabinose (0.2%). The gene for RatA was cloned into the NdeI-XhoI sites of pET-21c (Novagen), creating pET-21-c-YfjG(His)6. This construction created an in-frame translation fusion with a (His)6 tag at the RatA C-terminal end.
Buffers
Buffer A (10 mM Tris-Cl (pH 7.4), 10 mM MgSO4, 80 mM NH4Cl, 0.2 mM DTT), buffer B (20 mM Tris-HCl (pH 7.5), 10 mM Mg(OAc)2, 500 mM NH4Cl, and 2 mM DTT), buffer C (10 mM Tris-Cl pH 7.8, 60 mM NH4Cl, 10 mM MgCl2 and 1 mM DTT), buffer D (10 mM Tris-HCl pH 7.8, 60 mM NH4Cl, 1 mM MgCl2 and 1 mM DTT), buffer S (10 mM Tris-Cl, pH 7.4, 1 mM MgSO4, 80 mM NH4Cl, and 0.2 mM DTT) and buffer T (10 mM Tris-Cl, pH 7.4, 6 mM MgSO4, 80 mM NH4Cl, and 0.2 mM DTT) (Hirokawa et al., 2005) were used.
Assay of in vivo Protein and DNA Synthesis in the Cells
A 1-ml culture of E. coli BW25113 containing the pBAD30-RatA plasmid was grown at 37°C in M9 medium with 0.5% glycerol (no glucose) and all the amino acids (1 mM each) except for methionine and cysteine. When the A600 of the culture reached 0.3, arabinose was added to a final concentration of 0.2% and 50 μCi of 35S-methionine and 50 μg/ml methionine were added simultaneously. At different time intervals as shown in Fig. 1B, 50-μl samples were removed and the cumulated incorporation of 35S-methionine was measured. For the measurement of in vivo DNA synthesis, arabinose was added to a final concentration of 0.2% and 5 μCi of 3H-thymidine and 10 μg/ml thymidine were added simultaneously (A600, 0.3). At different time intervals as shown in Fig. 1C, 50-μl samples were removed and the cumulated incorporation of 3H-thymidine was measured.
Assay of in vivo Protein Synthesis Rate
E. coli BW25113 cells containing pBAD30-RatA were grown in M9 medium with 0.5% glycerol (no glucose) and all the amino acids (1 mM each) except for methionine and cysteine. When the A600 value of the culture reached 0.4, arabinose was added to a final concentration of 0.2% to induce RatA expression. Cell cultures (0.6 ml) were taken at time intervals as indicated in Fig. 1D and mixed with 30 μCi 35S-methionine. After 1 min incubation at 37°C, the rate of protein synthesis was determined as described previously (Zhang et al., 2003). For SDS-PAGE analysis of the total cellular protein synthesis, samples were removed from the 35S-methionine incorporation reaction mixture (500 μl) at time intervals indicated in Fig. 1D and added into chilled test tubes containing 100 μg/ml each of non-radioactive methionine and cysteine. Cell pellets collected by centrifugation were dissolved into 50 μl loading buffer and subjected to SDS-PAGE followed by autoradiography.
Purification of (His)6 RatA Proteins
(His)6 RatA tagged at the N-terminal end was purified from the strain BL21(DE3) carrying pCold-RatA with use of Ni-NTA resin (Qiagen). The eluted RatA were dissolved in 6 M guanidine-HCl. Denatured (His)6RatA was first trapped on Ni-NTA resin and then refolded by step-by-step dialysis (Zhang and Inouye, 2009).
Preparation of E. coli 70S Ribosomes
Two grams of E. coli MRE 600 cells were suspended in 35 ml of buffer A [10 mM Tris-Cl (pH 7.4), 10 mM MgSO4, 80 mM NH4Cl, 0.2 mM DTT] and lysed by French press. The cell suspension was centrifuged twice at 30,000 × g for 15 min. The supernatant was layered onto four tubes containing 15-ml sucrose solution [20 mM Tris-HCl (pH 7.5), 10 mM Mg(OAc)2, 500 mM NH4Cl, 2 mM DTT, and 1.1 M sucrose] and centrifuged at 45,000 rpm in a 50 Ti rotor for 22 h. The crude ribosome pellets were resuspended in 45 ml of buffer B [20 mM Tris-HCl (pH 7.5), 10 mM Mg(OAc)2, 500 mM NH4Cl, and 2 mM DTT]. After 15-min centrifugation at 30,000 × g, ribosomes were collected by centrifugation at 35,000 rpm in a 50 Ti rotor for 2 h. This step was repeated twice. The washed ribosome pellet was resuspended in storage buffer (buffer A without 50 mM NH4Cl) and stored at −80°C (Hirokawa et al., 2007).
Ribosome Profile Analysis
For polysome profile analysis, cells containing pBAD30-RatA plasmid were grown at 37°C in 150 ml of LB medium, and at an A600 of 0.6, arabinose was added to a final concentration of 0.2%. After 10 min induction, chloramphenicol was added to a final concentration of 200 μg/ml. Cell extracts were prepared using liquid nitrogen, freezing and thawing four times. About 10 A260 units of the lysate were layered onto a 5-40% sucrose gradient in buffer C (10 mM Tris-Cl pH 7.8, 60 mM NH4Cl, 10 mM MgCl2 and 1 mM DTT), and centrifuged at 35, 000 rpm for 3 h at 4°C using Beckman SW41Ti rotor. Gradients were analyzed with continuous monitoring at 254 nm (Zhang and Inouye, 2009). In order to analyze the 50S and 30S ribosome profile, another 10 A260 units of lysates as prepared above were dialyzed overnight against buffer D (10 mM Tris-HCl pH 7.8, 60 mM NH4Cl, 1 mM MgCl2 and 1 mM DTT) (dialysis buffer was changed once). The dialyzed solution was layered onto 5-40% sucrose gradients in buffer D, and then centrifuged at 35,000 rpm for 3 h at 4°C in a Beckman SW41Ti rotor. Gradients were analyzed with continuous monitoring at 254 nm. Western Blot analysis was carried out to detect RatA in each fraction using anti-poly-histidine antibody as shown Fig. 3 using equal volume of each sucrose density gradient fraction.
RNA Isolation and Northern Blot Analysis
E. coli BW25113 containing pBAD30-RatA was grown at 37°C in LB medium. When the A600 value reached 0.6, arabinose was added to a final concentration of 0.2%. The samples were taken at different time intervals as indicated in Fig. 2. Total RNAs were isolated using the hot-phenol method as described previously (Sarmientos and Cashel, 1983). Northern blot analysis was carried out as described previously (Zhang et al., 2003).
Ribosome Dissociation Assay
Ribosomes (0.07 μM) were incubated with RatA or IF3 at 30°C for 15 min in 200 μl of buffer T [10 mM Tris-Cl (pH 7.4), 6 mM MgSO4, 80 mM NH4Cl, and 0.2 mM DTT). Ribosomes were sedimented through a 5%–40% sucrose density gradient in the same buffer used for ultracentrifugation (Beckman SW 41.1 rotor, 35,000 rpm, 3h at 4°C), and the sedimentation profile was monitored using spectrophotometer (Hirokawa et al., 2005).
Anti-association Analysis
Ribosomes (0.07 μM) were incubated at 30°C for 10 min in 200 μl of buffer S [10 mM Tris-Cl (pH 7.4), 1 mM MgSO4, 80 mM NH4Cl, and 0.2 mM DTT). Then, different amounts of RatA or IF3 were added, and the reaction mixture was incubated at 30°C for another 10 min. After incubation, the MgSO4 concentration of the reaction mixture was raised to 6 mM and the final reaction mixture was further incubated at 30°C for 10 min. Ribosomes were sedimented through a 5%–40% sucrose density gradient in buffer T by ultracentrifugation (Beckman SW41.1 rotor, 35,000 rpm, 3 h at 4°C), and the sedimentation profile was monitored with the spectrophotometer (Hirokawa et al., 2005).
Acknowledgments
We thank Dr. Sangita Phadtare for the critical reading of this manuscript. We also thank Guangying Sun for her assistance doing the course of experiments. This work was supported by an NIH grant (1RO1-GM081567) and in part by a grant from Takara Bio Inc.
References
- Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrios AF, Zuo R, Ren D, Wood TK. Hha, YbaJ, and OmpA regulate Escherichia coli K12 biofilm formation and conjugation plasmids abolish motility. Biotechnol Bioeng. 2006;93:188–200. doi: 10.1002/bit.20681. [DOI] [PubMed] [Google Scholar]
- Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JH. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat Struct Mol Biol. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- Brown JM, Shaw KJ. A novel family of Escherichia coli toxin-antitoxin gene pairs. J Bacteriol. 2003;185:6600–6608. doi: 10.1128/JB.185.22.6600-6608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- Christensen-Dalsgaard M, Jorgensen MG, Gerdes K. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol Microbiol. 2010;75:333–348. doi: 10.1111/j.1365-2958.2009.06969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Contreras R, Zhang XS, Kim Y, Wood TK. Protein translation and cell death: the role of rare tRNAs in biofilm formation and in activating dormant phage killer genes. PLoS One. 2008;3:e2394. doi: 10.1371/journal.pone.0002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa G, Kaji H, Kaji A. Inhibition of antiassociation activity of translation initiation factor 3 by paromomycin. Antimicrob Agents Chemother. 2007;51:175–180. doi: 10.1128/AAC.01096-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa G, Nijman RM, Raj VS, Kaji H, Igarashi K, Kaji A. The role of ribosome recycling factor in dissociation of 70S ribosomes into subunits. RNA. 2005;11:1317–1328. doi: 10.1261/rna.2520405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Woychik NA. Bacterial toxin HigB associates with ribosomes and mediates translation-dependent mRNA cleavage at A-rich sites. J Biol Chem. 2009;284:18605–18613. doi: 10.1074/jbc.M109.008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen MG, Pandey DP, Jaskolska M, Gerdes K. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J Bacteriol. 2009;191:1191–1199. doi: 10.1128/JB.01013-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada K, Hanaoka F. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol Cell. 2005;19:497–509. doi: 10.1016/j.molcel.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Kamada K, Hanaoka F, Burley SK. Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol Cell. 2003;11:875–884. doi: 10.1016/s1097-2765(03)00097-2. [DOI] [PubMed] [Google Scholar]
- Korch SB, Henderson TA, Hill TM. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol Microbiol. 2003;50:1199–1213. doi: 10.1046/j.1365-2958.2003.03779.x. [DOI] [PubMed] [Google Scholar]
- Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev. 2005;69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Koonin EV. The HicAB cassette, a putative novel, RNA-targeting toxin-antitoxin system in archaea and bacteria. Bioinformatics. 2006;22:2581–2584. doi: 10.1093/bioinformatics/btl418. [DOI] [PubMed] [Google Scholar]
- Motiejunaite R, Armalyte J, Markuckas A, Suziedeliene E. Escherichia coli dinJ-yafQ genes act as a toxin-antitoxin module. FEMS Microbiol Lett. 2007;268:112–119. doi: 10.1111/j.1574-6968.2006.00563.x. [DOI] [PubMed] [Google Scholar]
- Neubauer C, Gao YG, Andersen KR, Dunham CM, Kelley AC, Hentschel J, Gerdes K, Ramakrishnan V, Brodersen DE. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell. 2009;139:1084–1095. doi: 10.1016/j.cell.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, Ehrenberg M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell. 2003;112:131–140. doi: 10.1016/s0092-8674(02)01248-5. [DOI] [PubMed] [Google Scholar]
- Petrelli D, Garofalo C, Lammi M, Spurio R, Pon CL, Gualerzi CO, La Teana A. Mapping the active sites of bacterial translation initiation factor IF3. J Mol Biol. 2003;331:541–556. doi: 10.1016/s0022-2836(03)00731-9. [DOI] [PubMed] [Google Scholar]
- Prysak MH, Mozdzierz CJ, Cook AM, Zhu L, Zhang Y, Inouye M, Woychik NA. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol Microbiol. 2009;71:1071–1087. doi: 10.1111/j.1365-2958.2008.06572.x. [DOI] [PubMed] [Google Scholar]
- Sarmientos P, Cashel M. Carbon starvation and growth rate-dependent regulation of the Escherichia coli ribosomal RNA promoters: differential control of dual promoters. Proc Natl Acad Sci U S A. 1983;80:7010–7013. doi: 10.1073/pnas.80.22.7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Schuenemann VJ, Hand NJ, Silhavy TJ, Martin J, Lupas AN, Djuranovic S. prlF and yhaV encode a new toxin-antitoxin system in Escherichia coli. J Mol Biol. 2007;372:894–905. doi: 10.1016/j.jmb.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Piro KM, Xu W, Hansen S, Lewis K, Brennan RG. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science. 2009;323:396–401. doi: 10.1126/science.1163806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney SM, Aoki H, Ganoza MC, Shinabarger DL. The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob Agents Chemother. 1998;42:3251–3255. doi: 10.1128/aac.42.12.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi H, Kakuta Y, Okada T, Yao M, Tanaka I, Kimura M. Crystal structure of archaeal toxin-antitoxin RelE-RelB complex with implications for toxin activity and antitoxin effects. Nat Struct Mol Biol. 2005;12:327–331. doi: 10.1038/nsmb911. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Park JH, Inouye M. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J Biol Chem. 2009;284:28746–28753. doi: 10.1074/jbc.M109.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Inouye M. The inhibitory mechanism of protein synthesis by YoeB, an Escherichia coli toxin. J Biol Chem. 2009;284:6627–6638. doi: 10.1074/jbc.M808779200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yamaguchi Y, Inouye M. Characterization of YafO, an Escherichia coli toxin. J Biol Chem. 2009;284:25522–25531. doi: 10.1074/jbc.M109.036624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Hara H, Kato I, Inouye M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J Biol Chem. 2005a;280:3143–3150. doi: 10.1074/jbc.M411811200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell. 2003;12:913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhu L, Zhang J, Inouye M. Characterization of ChpBK, an mRNA interferase from Escherichia coli. J Biol Chem. 2005b;280:26080–26088. doi: 10.1074/jbc.M502050200. [DOI] [PubMed] [Google Scholar]