Structured Abstract
Objective
To analyze DNA from women with premature ovarian failure (POF) for genome-wide copy number variations (CNVs), focusing on novel autosomal microdeletions.
Design
Case-control genetic association study.
Setting
Department of Obstetrics and Gynecology, Baylor College of Medicine, Houston, TX, USA.
Patients
Of 89 POF patients, eight experienced primary amenorrhea while 81 exhibited secondary amenorrhea prior to age 40.
Interventions
Genomic DNA from peripheral blood samples was analyzed for CNVs using high-resolution single nucleotide polymorphism (SNP) arrays.
Main outcome measures
Identification of novel CNVs in 89 POF cases, using the Database of Genomic Variants as a control population.
Results
A total of 198 autosomal CNVs were detected by SNP arrays, ranging in size from 0.1 Mb to 3.4 Mb. These CNVs included seventeen novel microduplications and seven novel microdeletions larger than 0.1 Mb, six of which contained coding regions: 8q24.13, 10p15-p14, 10q23.31, 10q26.3, 15q25.2, and 18q21.32. Most of the novel CNVs were derived from autosomes rather than the X chromosome.
Conclusions
The present pilot study revealed novel microdeletions/microduplications in POF. Two novel microdeletions caused haploinsufficiency for SYCE1 and CPEB1, genes known to cause ovarian failure in knockout mouse models. Chromosomal microarrays may be a useful adjunct to conventional karyotyping when evaluating genomic imbalances in women with POF.
Keywords: premature ovarian failure, POF, copy number, CNV, microdeletion and microduplication, SNP microarray, POI, array CGH, chromosomal microarray
INTRODUCTION
Menopause, or the permanent cessation of menses, naturally occurs in most American women between 46 and 54 years of age (1). Approximately 1–5% of women in the world experience cessation of their menstrual cycle prior to age 40 (2), a condition known as premature ovarian failure (POF) or premature ovarian insufficiency (POI). POF encompasses a broad spectrum of individuals, ranging from women with primary amenorrhea to women who have reproduced but experience loss of menstrual cycles prior to age 40, coupled with menopausal serum FSH levels (3–4). Unlike menopause, however, this condition does not always result in permanent loss of ovarian function, as nearly 5–10% of women are able to conceive after the onset of POF (5).
Most POF cases are classified as idiopathic, and these cases are further divided into sporadic and familial forms. Genetics is known to contribute significantly to idiopathic POF cases (6), and approximately 10–15% of diagnosed women have an affected first degree relative (5). Initial genetic interest has focused on the X chromosome due to Turner syndrome, as well as published associations of POF cases with X chromosome deletions and translocations (7–10). Genes specifically expressed from the X chromosome that have been implicated in POF include FMR1 (premutation; Fragile X Syndrome), BMP15, and POF1B (11–14).
In addition to X chromosome abnormalities, POF has been associated with autosomal gene aberrations. FSHR mutations are associated with POF in Finnish women, although such mutations are rare outside of Finland (15). Other autosomal genes that have been more widely associated with POF include GDF9, FIGLA, FOXO1a, FOXO3a, and NOBOX (13, 16–18).
Current genetic guidelines in the evaluation of POF cases recommend karyotyping and FMR1 premutation carrier screening. Conventional karyotyping has been a mainstay to determine genomic imbalances in POF individuals; however, it cannot detect genomic imbalances smaller than 3–5 Mb in size (microdeletions and microduplications, also known as copy number variations or CNVs) due to banding resolution. Recent studies utilizing oligonucleotide and single nucleotide polymorphism (SNP) microarray technologies have shown that genomic imbalances involving as few as 1,000 base pairs can be detected (19). It is possible that some of the idiopathic POF cases are due to CNVs. Therefore, we utilized a SNP based array to determine if women with POF exhibit novel CNVs.
MATERIALS AND METHODS
Participant population, blood collection and genomic DNA isolation
The study was composed of 89 women with POF: eight women who exhibited primary amenorrhea and 81 who exhibited secondary amenorrhea for more than six months prior to the age of 40, with FSH serum levels greater than 40 IU/L. Women with a history of pelvic surgery, cancer, radiation exposure, smoking, and genetic syndromes were excluded from the study. Peripheral blood samples were collected from all women, and genomic DNA (gDNA) was extracted from 5 mL of whole blood (Qiagen Puregene protocol, Valencia, CA, USA), as previously described (20). One of the eight women with primary amenorrhea failed genotyping quality control and was excluded from analysis. Therefore, a total of 88 women were available for analysis. This study was approved by the Institutional Review Board at Baylor College of Medicine.
Single nucleotide polymorphism arrays and data analysis
Illumina’s (San Diego, CA, USA) HumanCNV370-Duo DNA Analysis BeadChip with more than 318,000 tag SNP markers and 52,000 markers targeting additional CNV regions (5.5 kb median marker spacing) was used to examine CNVs among gDNA for 38 individuals on the BeadArray 500GX. Illumina’s Human660W-Quad v1 DNA Analysis BeadChip with 550,000 tag SNPs and an additional 100,000 markers targeting common CNV regions (2.5 kb median marker spacing) was used for an additional 51 individuals on the iScan System.
The array data was analyzed using Illumina’s GenomeStudio Genotyping Module software. A call rate of >99% was accepted as the validity cutoff for each sample on both chips, which indicates >99% of the probes successfully identified a genotype. Using the GenomeStudio software, the log of the signal intensity (log R ratio) highlighted CNVs as an increase or decrease from the baseline intensity which was standardized to zero. The B-allele frequency, standardized at 0.5, was coupled with the log R ratio values to examine areas of DNA loss/gain. The concurrent use of these two tools enabled CNVs to be differentiated from regions of copy-neutral loss of heterozygosity.
All CNVs were screened against polymorphisms noted in the Database of Genomic Variants (DGV; http://projects.tcag.ca/variation); those already listed with a high population frequency in the DGV were regarded as benign polymorphisms and subsequently excluded from further analyses.
Quantitative real-time PCR
Quantitative real-time PCR (qPCR) was utilized to corroborate SNP array findings (20). Emphasis was placed on unique microdeletions that were not present in the DGV database. Where possible, a relevant protein-coding gene within each deleted region was chosen as a target, and oligonucleotides were designed for qPCR analysis (Table 1). Primers were custom-designed through Primer3Plus (http://frodo.wi.mit.edu/primer3).
Table 1.
Oligonucleotides designed for quantitative real-time PCR corroboration of novel autosomal microdeletions with coding regions
| Microdeletion locus | Patient | Targeted gene | Forward primer (5′ -> 3′) | Reverse primer (5′ -> 3′) |
|---|---|---|---|---|
| 8q24.13 | POF-65 | FER1L6 | CGA ACC ACA GTG CAG AAG AA | AGG GCT ACG TCA TTA ATG CT |
| 10p15-p14 | POF-105 | AKR1C1 | TGT GGA TGG TGA CAC AGA GG | TCT TTC CTT TCT GGC CAA TG |
| 10q23.31a | POF-58 | STAMBPL1 | TAA GCC CAG AAG AGC GAG TC | CGC CAT CCT CTC CAT CTC TA |
| 10q26.3 | POF-108 | SYCE1 | CCA GTG ACA TGG TGG AGT TG | GGT GTC TTC ATT GCC ACT CA |
| 15q25.2 | POF-87 | CPEB1 | CCT GGG TAT TAG CCG ACA GT | AAT CCC GGC ATA CAC CAC T |
| 18q21.32 | POF-119 | PMAIP1 | CAC CGT GTG TAG TTG GCA TC | CCT TCT TCC CAG GCA TCT |
Unable to corroborate via quantitative real-time PCR
Quantitative real-time PCR was conducted using 100 ng of gDNA, 0.4 μM forward and reverse primers, 1X iQTM SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) and nuclease free water, with a total volume of 20 μL per well. Five replicates were run for each POF individual for the targeted gene within each respective deleted region. A pooled human gDNA sample from five women was used as a control (Promega, Cat # G1521). Threshold cycle (Ct) values were calculated and averaged for each set of five replicates; these values were then used to calculate the standard error of the mean. The 2−Δ Δ Ct method of quantification was used to analyze fold changes in copy number between control (2n) and deleted regions (1n or absent).
RESULTS
We analyzed the genomic DNA of 89 women with POF utilizing high-resolution SNP arrays to determine the prevalence of novel CNVs (microdeletions/microduplications) which would not be detected by conventional karyotype methods. Eighty-eight samples were successfully arrayed. Microarray data revealed a total of 50 microduplications and 148 microdeletions on autosomal chromosomes, ranging in size from 0.1 Mb to 3.4 Mb. These CNVs occurred in 72 out of 88 genotyped POF individuals, and they spanned every autosome. In order to distinguish novel CNVs from seemingly benign, non-pathogenic imbalances, we compared our data to the Database of Genomic Variants. The DGV is a compilation of common CNVs detected in more than 10,000 healthy individuals, and it has been used as a control population in several studies (21–23). We found 33/50 microduplications and 141/148 microdeletions in the control population of the DGV. Therefore, 24 autosomal CNVs in a total of nineteen women were considered novel. There were seven novel autosomal microdeletions (Table 2), one in each of seven different women, and seventeen novel autosomal microduplications (Table 3), one in each of eleven women and two in each of three women.
Table 2.
Novel hemizygous microdeletions in cohort of women with POF
| Microdeletion locus (hg18) | Patient | Length (Mb) | Age at POF onset (years) | Genes within deleted region |
|---|---|---|---|---|
| chr 8: 124,964,990-125,101,913 | POF-65 | 0.14 | 36 | FER1L6 |
| chr 10: 4,867,522-5,196,096 | POF-105 | 0.33 | 35 | AKR1E2, AKR1C1, AKR1C2, AKR1C3, AKR1CL1 |
| chr 10: 90,517,549-90,648,588 | POF-58 | 0.13 | 36 a | LIPN, LIPM, ANKRD22, STAMBPL1 |
| chr 10: 135,092,227-135,256,027b | POF-108 | 0.16 | 21 | CYP2E1, SYCE1 |
| chr 14: 40,060,168-40,305,161 | POF-66 | 0.24 | 31 | None |
| chr 15: 80,972,574-82,640,853 | POF-87 | 1.67 | PA | CPEB1, AP3B2, FSD2, HOMER2, WHAMM, FAM103A1, C15orf40, BTBD1, TM6SF1, HDGFRP3, BNC1, SH3GL3, ADAMTSL3 |
| chr 18: 55,460,039-55,783,281 | POF-119 | 0.32 | PA | CCBE1, PMAIP1 |
| chr X: 48,104,013-52,083,957 | POF-37 | 3.98 | 34 | SSX4..........BMP15......... AK09637 c |
Age at onset of POF unknown; age at time of POF diagnosis indicated
Pearson’s chi square test was used to show statistical significance:
3/2,802 control individuals showed a variation of this deletion in Database of Genomic Variants at the indicated locus (43-44); P < 0.0001
The deletion encompasses a total of 65 genes; the first and last genes in the deleted interval are listed, as well as the BMP15 gene, known to play critical roles in folliculogenesis
PA = primary amenorrhea (absence of menarche)
Table 3.
Novel microduplications in cohort of women with POF
| Microduplication locus (hg18) | Patient | Length (Mb) | Genes within duplicated region |
|---|---|---|---|
| chr 1: 119,652,659-119,966,935 | POF-106 | 0.31 | HAO2, HSD3B2, HSD3B1 |
| chr 1: 119,719,096-119,953,605 | POF-21 | 0.23 | HAO2, HSD3B2, HSD3B1 |
| chr 1: 237,422,778-237,602,136 | POF-56 | 0.18 | None |
| chr 2: 51,536,418-51,865,782 | POF-87 | 0.33 | None |
| chr 3: 7,979,828-8,127,315 | POF-87 | 0.15 | None |
| chr 4: 4,705,258-4,939,891 | POF-124 | 0.23 | MSX1 |
| chr 4: 6,003,730-6,247,313 | POF-124 | 0.24 | CRMP1, C4orf50, JAKMIP1 |
| chr 6: 57,377,166-57,503,129 | POF-34 | 0.13 | PRIM2 |
| chr 7: 70,257,034-70,592,167 | POF-37 | 0.34 | WBSCR17, CALN1 |
| chr 7: 75,984,206-79,394,983 | POF-83 | 3.41 | PMS2L11, CCDC146, FL2, PION, PTPN12, RSBN1L, TMEM60, PHTF2, MAGI2, RPL13AP17 |
| chr 7: 97,252,046-97,393,215 | POF-10 | 0.14 | ASNS |
| chr 8: 509,919-835,807 | POF-16 | 0.33 | ERICH1 |
| chr 8: 2,442,385-3,126,569 | POF-45 | 0.68 | CSMD1 |
| chr 8: 47,626,500-48,043,110 | POF-13 | 0.42 | BEYLA |
| chr 9: 71,619,570-72,483,319 | POF-119 | 0.86 | C9orf135, MAMDC2, SMC5, KLF9, TRPM3 |
| chr 11: 48,556,067-51,447,829 | POF-10 | 2.89 | FOLH1, OR4C13, OR4C12, OR4A5, OR4C46 |
| chr 16: 14,983,979-16,367,263 | POF-11 | 1.38 | PCXDC1, NTAN1, RRN3, MPV17L, C16orf45, NDE1, MIR484, MYH11, C16orf63, ABCC1, ABCC6, NOMO3 |
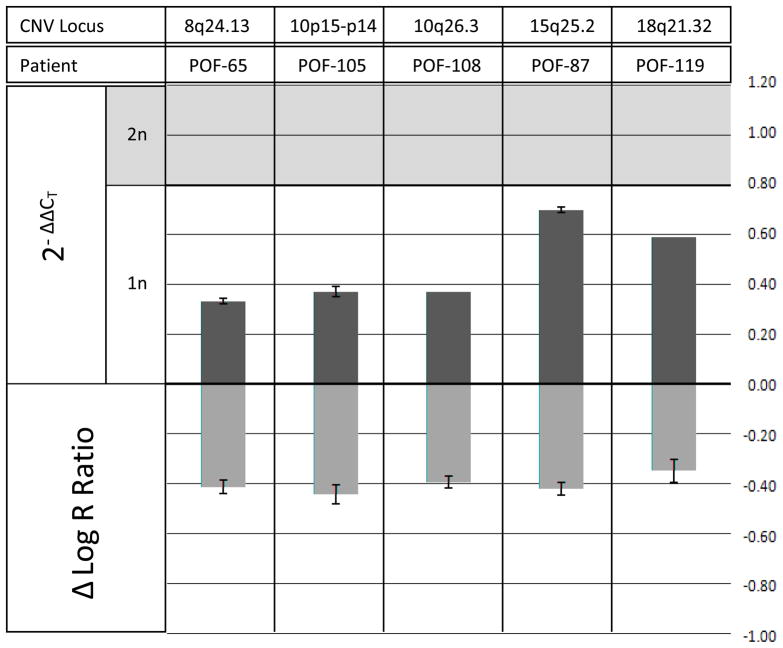
We specifically focused on microdeletions, as genomic losses are more likely to result in a clinical phenotype than genomic gains (24). Seven women with POF exhibited novel hemizygous microdeletions greater than 0.1 Mb in size, across the autosomes. Six of the novel autosomal microdeletions contained coding regions, and five of the deletions were subsequently verified by qPCR analysis: 8q24.13, 10p15-p14, 10q26.3, 15q25.2, and 18q21.32 (Table 2). Control amplifications for regions of no change in copy number did not show detectable Ct shifts. A 2-Δ ΔCt (fold change) quantification analysis was also performed (Figure 1), with the standard error of the mean Ct difference for each group of replicates given as the error bars. The fold change was plotted against the mean intensity shift (log R ratio), as measured on the array. All plotted microdeletions showed considerable fold reductions, as indicated by the top bars that lie below the shaded region (2n = diploid). SYCE1 and CPEB1 were genes included in the verified microdeletions. These genes are known to cause ovarian failure in knockout mouse models (25–26).
Figure 1. Data from SNP arrays and qPCR plotted as Δ log R ratio and 2−Δ Δ Ct, respectively, for five autosomal microdeletions in women with POF.
Regions of interest were initially determined based on log R ratio shifts, where reported copy number reductions in gDNA from peripheral blood samples are indicated above by a negative Δ log R ratio. 2−Δ Δ Ct values (fold changes) were then calculated from qPCR output for the target regions. Fold change values lying within the shaded region are indicative of a normal diploid copy number of 2, whereas those below the shaded region are indicative of a copy number reduction of 1, a fold change of approximately 0.5 (1/2). Hemizygous deletions were corroborated on 8q24.13 in POF-65, 10p15-p14 in POF-105, 10q26.3 in POF-108, 15q25.2 in POF-87 and 18q21.32 in POF-119.
Interestingly, microdeletions were enriched in women with primary amenorrhea; two of the seven genotyped women with primary amenorrhea (29%) had novel microdeletions as compared to 6/81 women with secondary amenorrhea (7%). Although our study focused on autosomes, we detected one novel interstitial microdeletion on the X chromosome in a woman with secondary amenorrhea, which mapped to Xp11.23-p11.22 where the BMP15 gene is located (Table 2).
DISCUSSION
Animal models indicate that the majority of candidate genes for ovarian failure are located on autosomes (13, 16–18). Although initial genetic screening studies for POF focused on the X chromosome, recent genome-wide studies in humans have found associations with autosomal chromosomes, as well (21, 27–29). These findings offer further motivation to continue looking more closely across the entire genome for genomic imbalances that could contribute to the phenotypic spectrum of POF.
Previous studies have highlighted the possible role of CNVs in the pathogenesis of several common and rare diseases (30–32). Haploinsufficiency encountered in microdeletions is a well-known cause of human genetic disorders; these microdeletions can occur sporadically, or they can be inherited. Sporadic cases likely arise from non-allelic homologous recombination or unequal crossing-over between large, highly identical segmental duplications. It has also become clear that the severity of clinical presentations varies widely, with identical microdeletions causing no apparent symptoms in carrier parents, yet severe clinical presentation in the offspring (33). The causes of such variable expression are unclear but likely include predisposing polymorphisms in the affected individuals. The objective of our study was to utilize a genome-wide approach to detect microdeletions and microduplications in a cohort of 89 women diagnosed with POF. We hypothesized that novel CNVs associate with POF.
We identified seven novel autosomal microdeletions among 88 successfully arrayed POF women. Quantitative real-time PCR was used to corroborate five of the six novel autosomal microdeletions that contained known coding regions. In the case of POF-66, the deletion (0.24 Mb) was in a genomic region devoid of known genes, miRNAs or other non-coding RNAs. Although it is easy to dismiss genomic regions that do not contain known transcriptional units, recent data shows that transcriptionally inactive regions can account for an abnormal phenotype when disrupted (34).
Among the genes found within the novel autosomal microdeletions, two have been strongly linked to abnormal reproductive phenotypes in knockout mouse models – SYCE1 (10q26.3) and CPEB1 (15q25.2). SYCE1 has been shown to disrupt the repair of DNA double-strand breaks during meiosis, which leads to apoptosis of germ cells and subsequent infertility of both male and female knockout mice (25). Based on this null phenotype in female mice, SYCE1 haploinsufficiency in humans may have a similar effect, leading to loss of oocytes and POF (35). The woman who presented with a hemizygous deletion of SYCE1 had menarche at 14 and experienced cessation of menstrual cycles at age 21. Another gene, CPEB1, is preferentially expressed in the oocyte and encodes a protein that regulates the translation of mRNAs for proteins involved in synaptenomal complex formation during oocyte maturation (35). Adult female mice deficient in CPEB1 lack follicles due to loss of oocytes, while mid-gestation female embryos show oocytes arrested at the pachytene stage (36). CPEB1 has also been found to regulate mitotic cell progression in the S and M phases, which are especially important during embryonic cell divisions (26). CPEB1 haploinsufficiency in humans may accelerate germ cell loss during the reproductive lifespan, as is the case in POF. This hypothesis could support the fact that the woman who presented with a hemizygous deletion of CPEB1 exhibited ovarian failure at an earlier age; she never experienced menarche (primary amenorrhea).
Other microdeletions identified in our screen harbor genes that are not known to play a role in reproductive biology. POF-119 exhibited a deletion that resulted in haploinsufficiency of CCBE1 and PMAIP1. We used microarray data generated from wild-type newborn mouse ovaries (37), as well as NCBI EST databases (Unigene and Gene Expression Omnibus), to determine whether CCBE1 and PMAIP1 are expressed in mammalian ovaries. Based on these data sets, both CCBE1 and PMAIP1 are expressed in mouse ovaries. Similarly, CTNNA3 (POF-85), ANKRD22 and STAMBPL1 (POF-58) transcripts are present in the murine ovarian transcriptome. It is possible that such genes may play a yet-to-be-defined role in ovarian development.
We also identified seventeen autosomal microduplications which have not been reported in the DGV. The role of duplications in human disorders is less compelling, since corresponding animal models are lacking or difficult to interpret. Two duplications on chromosome one were nearly identical in two distinct POF individuals (POF-21 and POF-106), and they included genes for hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 (HSD3B2) and hydroxyacid oxidase 2 (HAO2). The role of HA02 in ovarian biology is unknown, despite its expression in the mouse ovary. Conversely, HSD3B2 is preferentially expressed in the adrenal glands and ovaries of mice, according to the Unigene and BioGPS databases (38–39). HSD3B2 is essential for steroid hormone production in humans, and deficiencies cause a severe form of congenital adrenal hyperplasia (40). Furthermore, additional studies report increased expression of HSD3B2 in theca cells of women with polycystic ovary syndrome, a frequent cause of infertility in women (41). It is possible that duplication of HSD3B2 may lead to androgen and estrogen imbalances within the ovary with subsequent disruption of folliculogenesis, but this hypothesis remains to be tested.
We discovered seven novel autosomal microdeletions and seventeen novel autosomal microduplications; in addition, we identified one novel microdeletion located on the X chromosome. Our results add to the growing body of recent literature supporting our hypothesis that CNVs associate with POF. A recent study on French women used a lower resolution BAC array (0.7 Mb mean spatial resolution) to identify CNVs that statistically associate with POF (21). As with our study, there was an excess of CNVs on the autosomes as compared to the X chromosome. Eight CNVs showed statistical association with POF in their study, (seven autosomal and one X chromosomal), but no novel CNVs were presented. A second study focused only on the X chromosome (22). A BAC tiling array for X/Y chromosomes was utilized, and the authors discovered new, discrete X chromosome intervals associated with POF. It is important to note that there may be additional CNVs that went undetected by our arrays and those from previous studies, as even the highest resolution arrays may only tag approximately 50% of CNVs in the genome (42). In addition to technological limitations, our study was limited by the lack of family members and the small sample size. However, due to the high variability in the clinical phenotype of carriers and affected family members with microdeletions (33), the presence of such microdeletions in a fertile parent would not rule out the pathogenic role of such genomic imbalances in ovarian function in the offspring. Future studies with larger populations will be necessary to determine whether recurring genomic imbalances are present in women with POF and to discern the utility of molecular karyotyping methods, such as array CGH, in replacing conventional karyotyping. Moreover, animal models that mimic novel human deletions and/or duplications may be useful in further delineating the importance of such regions in mammalian reproductive biology.
Acknowledgments
We acknowledge Lisa Marsh, RN, for patient recruitment and data collection.
Financial Support: This study was supported by the NIH grant R21 HD058125.
Footnotes
Where the work was done:
Magee-Womens Research Institute, University of Pittsburgh, Pittsburgh, PA, USA, and Baylor College of Medicine, Houston, TX, USA
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.van Noord PA, Dubas JS, Dorland M, Boersma H, te Velde E. Age at natural menopause in a population-based screening cohort: the role of menarche, fecundity, and lifestyle factors. Fertil Steril. 1997;68:95–102. doi: 10.1016/s0015-0282(97)81482-3. [DOI] [PubMed] [Google Scholar]
- 2.Coulam CB, Adamson SC, Annegers JF. Incidence of premature ovarian failure. Obstet Gynecol. 1986;67:604–6. [PubMed] [Google Scholar]
- 3.Kalantaridou SN, Nelson LM. Premature ovarian failure is not premature menopause. Ann N Y Acad Sci. 2000;900:393–402. doi: 10.1111/j.1749-6632.2000.tb06251.x. [DOI] [PubMed] [Google Scholar]
- 4.Rebar RW, Connolly HV. Clinical features of young women with hypergonadotropic amenorrhea. Fertil Steril. 1990;53:804–10. [PubMed] [Google Scholar]
- 5.van Kasteren YM, Schoemaker J. Premature ovarian failure: a systematic review on therapeutic interventions to restore ovarian function and achieve pregnancy. Hum Reprod Update. 1999;5:483–92. doi: 10.1093/humupd/5.5.483. [DOI] [PubMed] [Google Scholar]
- 6.Coulam CB, Stringfellow S, Hoefnagel D. Evidence for a genetic factor in the etiology of premature ovarian failure. Fertil Steril. 1983;40:693–5. doi: 10.1016/s0015-0282(16)47433-9. [DOI] [PubMed] [Google Scholar]
- 7.Bione S, Rizzolio F, Sala C, Ricotti R, Goegan M, Manzini MC, et al. Mutation analysis of two candidate genes for premature ovarian failure, DACH2 and POF1B. Hum Reprod. 2004;19:2759–66. doi: 10.1093/humrep/deh502. [DOI] [PubMed] [Google Scholar]
- 8.Rizzolio F, Bione S, Sala C, Goegan M, Gentile M, Gregato G, et al. Chromosomal rearrangements in Xq and premature ovarian failure: mapping of 25 new cases and review of the literature. Hum Reprod. 2006;21:1477–83. doi: 10.1093/humrep/dei495. [DOI] [PubMed] [Google Scholar]
- 9.Rossetti F, Rizzolio F, Pramparo T, Sala C, Bione S, Bernardi F, et al. A susceptibility gene for premature ovarian failure (POF) maps to proximal Xq28. Eur J Hum Genet. 2004;12:829–34. doi: 10.1038/sj.ejhg.5201186. [DOI] [PubMed] [Google Scholar]
- 10.Toniolo D, Rizzolio F. X chromosome and ovarian failure. Semin Reprod Med. 2007;25:264–71. doi: 10.1055/s-2007-980220. [DOI] [PubMed] [Google Scholar]
- 11.Di Pasquale E, Rossetti R, Marozzi A, Bodega B, Borgato S, Cavallo L, et al. Identification of new variants of human BMP15 gene in a large cohort of women with premature ovarian failure. J Clin Endocrinol Metab. 2006;91:1976–9. doi: 10.1210/jc.2005-2650. [DOI] [PubMed] [Google Scholar]
- 12.Ennis S, Ward D, Murray A. Nonlinear association between CGG repeat number and age of menopause in FMR1 premutation carriers. Eur J Hum Genet. 2006;14:253–5. doi: 10.1038/sj.ejhg.5201510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laissue P, Christin-Maitre S, Touraine P, Kuttenn F, Ritvos O, Aittomaki K, et al. Mutations and sequence variants in GDF9 and BMP15 in patients with premature ovarian failure. Eur J Endocrinol. 2006;154:739–44. doi: 10.1530/eje.1.02135. [DOI] [PubMed] [Google Scholar]
- 14.Riva P, Magnani I, Fuhrmann Conti AM, Gelli D, Sala C, Toniolo D, et al. FISH characterization of the Xq21 breakpoint in a translocation carrier with premature ovarian failure. Clin Genet. 1996;50:267–9. doi: 10.1111/j.1399-0004.1996.tb02642.x. [DOI] [PubMed] [Google Scholar]
- 15.Doherty E, Pakarinen P, Tiitinen A, Kiilavuori A, Huhtaniemi I, Forrest S, et al. A Novel mutation in the FSH receptor inhibiting signal transduction and causing primary ovarian failure. J Clin Endocrinol Metab. 2002;87:1151–5. doi: 10.1210/jcem.87.3.8319. [DOI] [PubMed] [Google Scholar]
- 16.Qin Y, Choi Y, Zhao H, Simpson JL, Chen ZJ, Rajkovic A. NOBOX homeobox mutation causes premature ovarian failure. Am J Hum Genet. 2007;81:576–81. doi: 10.1086/519496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watkins WJ, Umbers AJ, Woad KJ, Harris SE, Winship IM, Gersak K, et al. Mutational screening of FOXO3A and FOXO1A in women with premature ovarian failure. Fertil Steril. 2006;86:1518–21. doi: 10.1016/j.fertnstert.2006.03.054. [DOI] [PubMed] [Google Scholar]
- 18.Zhao H, Chen ZJ, Qin Y, Shi Y, Wang S, Choi Y, et al. Transcription factor FIGLA is mutated in patients with premature ovarian failure. Am J Hum Genet. 2008;82:1342–8. doi: 10.1016/j.ajhg.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter NP. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat Genet. 2007;39:S16–21. doi: 10.1038/ng2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowden W, Skorupski J, Kovanci E, Rajkovic A. Detection of novel copy number variants in uterine leiomyomas using high-resolution SNP arrays. Mol Hum Reprod. 2009;15:563–8. doi: 10.1093/molehr/gap050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aboura A, Dupas C, Tachdjian G, Portnoi MF, Bourcigaux N, Dewailly D, et al. Array comparative genomic hybridization profiling analysis reveals deoxyribonucleic acid copy number variations associated with premature ovarian failure. J Clin Endocrinol Metab. 2009;94:4540–6. doi: 10.1210/jc.2009-0186. [DOI] [PubMed] [Google Scholar]
- 22.Quilter CR, Karcanias AC, Bagga MR, Duncan S, Murray A, Conway GS, et al. Analysis of X chromosome genomic DNA sequence copy number variation associated with premature ovarian failure (POF) Hum Reprod. 2010;25:2139–50. doi: 10.1093/humrep/deq158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pangas SA, Choi Y, Ballow DJ, Zhao Y, Westphal H, Matzuk MM, et al. Oogenesis requires germ cell-specific transcriptional regulators Sohlh1 and Lhx8. Proc Natl Acad Sci U S A. 2006;103:8090–5. doi: 10.1073/pnas.0601083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee C, Iafrate AJ, Brothman AR. Copy number variations and clinical cytogenetic diagnosis of constitutional disorders. Nat Genet. 2007;39:S48–54. doi: 10.1038/ng2092. [DOI] [PubMed] [Google Scholar]
- 25.Bolcun-Filas E, Hall E, Speed R, Taggart M, Grey C, de Massy B, et al. Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet. 2009;5:e1000393. doi: 10.1371/journal.pgen.1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novoa I, Gallego J, Ferreira PG, Mendez R. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nat Cell Biol. 2010;12:447–56. doi: 10.1038/ncb2046. [DOI] [PubMed] [Google Scholar]
- 27.Kang H, Lee SK, Kim MH, Song J, Bae SJ, Kim NK, et al. Parathyroid hormone-responsive B1 gene is associated with premature ovarian failure. Hum Reprod. 2008;23:1457–65. doi: 10.1093/humrep/den086. [DOI] [PubMed] [Google Scholar]
- 28.Oldenburg RA, van Dooren MF, de Graaf B, Simons E, Govaerts L, Swagemakers S, et al. A genome-wide linkage scan in a Dutch family identifies a premature ovarian failure susceptibility locus. Hum Reprod. 2008;23:2835–41. doi: 10.1093/humrep/den278. [DOI] [PubMed] [Google Scholar]
- 29.Skillern A, Rajkovic A. Recent developments in identifying genetic determinants of premature ovarian failure. Sex Dev. 2008;2:228–43. doi: 10.1159/000152039. [DOI] [PubMed] [Google Scholar]
- 30.Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, Smith J, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–5. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 31.Gijsbers AC, D'Haene B, Hilhorst-Hofstee Y, Mannens M, Albrecht B, Seidel J, et al. Identification of copy number variants associated with BPES-like phenotypes. Hum Genet. 2008;124:489–98. doi: 10.1007/s00439-008-0574-9. [DOI] [PubMed] [Google Scholar]
- 32.Ye L, Santarpia L, Cote GJ, El-Naggar AK, Gagel RF. High resolution array-comparative genomic hybridization profiling reveals deoxyribonucleic acid copy number alterations associated with medullary thyroid carcinoma. J Clin Endocrinol Metab. 2008;93:4367–72. doi: 10.1210/jc.2008-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antonacci F, Kidd JM, Marques-Bonet T, Teague B, Ventura M, Girirajan S, et al. A large and complex structural polymorphism at 16p12.1 underlies microdeletion disease risk. Nat Genet. 2010;42:745–50. doi: 10.1038/ng.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, et al. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–12. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng P, Griswold MD, Hassold TJ, Hunt PA, Small CL, Ye P. Predicting meiotic pathways in human fetal oogenesis. Biol Reprod. 2010;82:543–51. doi: 10.1095/biolreprod.109.079590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tay J, Richter JD. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev Cell. 2001;1:201–13. doi: 10.1016/s1534-5807(01)00025-9. [DOI] [PubMed] [Google Scholar]
- 37.Choi Y, Qin Y, Berger MF, Ballow DJ, Bulyk ML, Rajkovic A. Microarray analyses of newborn mouse ovaries lacking Nobox. Biol Reprod. 2007;77:312–9. doi: 10.1095/biolreprod.107.060459. [DOI] [PubMed] [Google Scholar]
- 38.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krone N, Arlt W. Genetics of congenital adrenal hyperplasia. Best Pract Res Clin Endocrinol Metab. 2009;23:181–92. doi: 10.1016/j.beem.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelson VL, Qin KN, Rosenfield RL, Wood JR, Penning TM, Legro RS, et al. The biochemical basis for increased testosterone production in theca cells propagated from patients with polycystic ovary syndrome. J Clin Endocrinol Metab. 2001;86:5925–33. doi: 10.1210/jcem.86.12.8088. [DOI] [PubMed] [Google Scholar]
- 42.Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat Genet. 2008;40:1199–203. doi: 10.1038/ng.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pinto D, Marshall C, Feuk L, Scherer SW. Copy-number variation in control population cohorts. Hum Mol Genet. 2007;16(2):R168–73. doi: 10.1093/hmg/ddm241. [DOI] [PubMed] [Google Scholar]
- 44.Shaikh TH, Gai X, Perin JC, Glessner JT, Xie H, Murphy K, et al. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res. 2009;19:1682–90. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]