Abstract
Two pathways for DNA recombination, AddAB (RecBCD-like) and RecRO, were identified in Helicobacter pylori, a pathogenic bacterium that colonizes human stomachs resulting in a series of gastric diseases. In this study, we examined the physiological roles of H. pylori RecRO pathway in DNA recombinational repair. We characterized H. pylori single mutants in recR and in recO, genes in the putative gap repair recombination pathway, and an addA recO double mutant that is thus deficient in both pathways that initiate DNA recombinational repair. The recR or recO single mutants showed the same level of sensitivity to mitomycin C as the parent strain, suggesting that the RecRO pathway is not responsible for the repair of DNA double strand breaks. However, H. pylori recR and recO mutants are highly sensitive to oxidative stress and separately to acid stress, two major stress conditions that H. pylori encounters in its physiological niche. The complementation of the recR mutant restored the sensitivity to oxidative and acid stress to the wild type level. By measuring DNA transformation frequencies, the recR and recO single mutants were shown to have no effect on inter-genomic recombination, whereas the addA recO double mutant had a greatly (~12-fold) reduced transformation frequency. On the other hand, the RecRO pathway was shown to play a significant role in intra-genomic recombination with direct repeat sequences. Whereas the recA strain had a deletion frequency 35-fold lower than that of background level, inactivation of recR resulted in a 4-fold decrease in deletion frequency. In a mouse infection model, the three mutant strains displayed a greatly reduced ability to colonize the host stomachs. The geometric means of colonization number for the wild type, recR, recO, and addA recO strains were 6 × 105, 1.6 × 104, 1.4 × 104 and 4 × 103 CFU/g stomach, respectively. H. pylori RecRO-mediated DNA recombinational repair (intra-genomic recombination) is thus involved in repairing DNA damage induced by oxidative and acid stresses and plays an important role in bacterial survival and persistent colonization in the host.
Keywords: Helicobacter pylori, Gastric pathogen, Oxidative stress, DNA repair, RecRO pathway, Intra-genomic recombination
1. Introduction
DNA recombinational repair is crucial for maintaining genomic integrity by repairing DNA double strand (ds) breaks and stalled replication forks [1]. DNA recombinational repair requires a large number of proteins that act at various stages of the process [2]. The central step in DNA recombination is the DNA strand exchange (synapsis) mediated by the nucleoprotein filament formed by RecA on single stranded (ss) DNA [3]. In the pre-synapsis stage, complex systems (pathways) are required for DNA damage recognition, processing of DNA substrate and recruiting of RecA. In the well-studied bacterial model system Escherichia coli, RecFOR and RecBCD pathways are required to initiate homologous DNA recombination [3, 4]. Whereas the RecBCD pathway is responsible for dsDNA break repair, the RecFOR pathway is mainly used for ssDNA gap repair. The RecFOR pathway can also repair dsDNA breaks when the RecBCD pathway is inactivated. The RecF, RecO and RecR proteins act together to promote loading of RecA onto ssDNA [5].
Helicobacter pylori colonizes the stomachs of about half the human population and is implicated in the etiology of gastric ulcers and cancer [6]. In its physiological environment, H. pylori frequently suffers both oxidative and acid stress, leading to DNA damage [7–10]. Therefore, DNA recombinational repair plays an important role in H. pylori’s survival and persistence in the host [11, 12]. It is also well known that H. pylori exhibits a high frequency of DNA recombination [13] which plays a critical role in generating genetic diversity by allowing genomic DNA rearrangements and integration of exogenous DNA into the genome through transformation [14].
Although a limited number of genes in the H. pylori genome were predicted to be involved in DNA recombinational repair [15–17], recent studies revealed the existence of both pathways for DNA recombinational repair in H. pylori. An AddAB class of helicase-nuclease enzymes, related to the E. coli RecBCD enzyme, was shown to be functional in H. pylori [18, 19], and the RecB-like helicase (now termed AddA) was also characterized in other studies [20, 21]. Although RecF historically served as a reference for RecFOR pathway, it is absent from genomes of many bacteria including H. pylori [4]. By bioinformatics analysis, Marsin et al [20] identified a novel RecO orthologue, suggesting the presence of the RecRO pathway in H. pylori. Recent studies in E. coli indicated that RecOR in the absence of RecF can perform recombination by loading RecA[22, 23]. Whereas the RecO protein can displace ssDNA-binding protein (SSB) and bind to ssDNA, RecR is the key component for loading RecA onto ssDNA [22, 24].
In characterizing DNA recombination mutants of H. pylori, there were discrepancies in measuring DNA recombination rate from different studies using different DNA substrates [18, 20, 21, 25, 26]. While the RecRO is supposed to have a major role in intragenomic recombination at repeat sequences [27], the effect of H. pylori RecR and RecO on the intragenomic recombination reported by Marsin et al [20] was marginal (less than 2-fold difference compared to the wild type). Thus, the roles of RecRO pathway in H. pylori DNA repair and recombination remain to be clarified, and the relative contributions of the two pathways (RecRO and AddAB) in DNA recombinational repair in H. pylori are unknown. More importantly, the in vivo role of H. pylori RecRO as a DNA recombinational repair pathway remain to be determined. In this study we further examined the roles of RecR and RecO in H. pylori DNA repair and recombination using different types of DNA substrates. Furthermore, we examined the contribution of the RecRO pathway to bacterial survival in vivo by using a mouse infection model.
2. Materials and Methods
2.1. H. pylori strains and culture conditions
H. pylori strain 26695, 43504 or X47 was cultured on Brucella agar (Difco) plates supplemented with 10% defibrinated sheep blood or 4% fetal bovine serum (called BA plates). Cultures of H. pylori were grown microaerobically at 37°C in an incubator under continuously controlled levels of oxygen (5% partial pressure O2, 5% CO2, and the balance was N2). For assessing the susceptibility to mitomycin C, H. pylori strains were grown on BA plates containing different concentration of mitomycin C under microaerobic conditions, and the growth was recorded after 2 days.
2.2. Construction of H. pylori isogenic mutants and complementation
To construct the recR mutant, a DNA fragment containing the recR gene was amplified by polymerase chain reaction (PCR) from genomic DNA using the primer pair listed in Table 1, and the PCR product was cloned into pGEM-T vector. Subsequently, a part of the target gene sequence in the recombinant plasmid was replaced by insertion of a chloramphenicol acetyl transferase (cat) cassette. The disrupted gene was then introduced into H. pylori wild type strain by natural transformation via allelic exchange and chloramphenicol (50 μg/ml) resistant colonies were isolated. The recO mutant was constructed by an overlapping PCR method in which the major portion of the gene was replaced by a kanamycin resistance cassette (aphA). The primers used for this procedure are listed in Table 1. The appropriate PCR product was used to transform the H. pylori wild type strain by selecting on kanamycin-containing (40 μg/ml) BA plates that were incubated at low O2 (1% partial pressure) condition. Each resistance cassette (cat or aphA) contains its own promoter but lacks a transcription terminator and in all cases was inserted in the same direction of transcription as that of the native gene. After transformation, four to eight clones were selected and evaluated by PCR to confirm replacement of the wild type allele with the null allele, and a single clone was selected for use in further experiments. To construct the recR recO and addA recO double mutants, the PCR fragment of recO::aphA was used to transform recR::cat or addA::cat mutant, respectively.
Table 1.
Primers used in this study.
| Name | Sequence (5′ – 3′) | Description |
|---|---|---|
| PrecR-F | AACGCTTCATTAAGCGCTTT | To construct recR::cat mutant |
| PrecR-R | ACAAAGGCACTTCATGCAC | |
| recR-1 | GTTGAATCTCCTGGCACGAGTTTGATATTG | For recR complementation |
| recR-2 | ACGGCCACTATCGCATGGTTATAAGCATGC | |
| rdxA-L | CACAACCAAGTAATCGCATC | |
| rdxA-1 | CTCGTGCCAGGAGATTCAACCACAGCATGC | |
| rdxA-2 | AACCATGCGATAGTGGCCGTGTGGTAACAA | |
| rdxA-R | AGCGCCATTCTTGCAAGATG | |
| PrecO-F | CATGCGGAAATGGTAATGGC | To construct recO::aphA mutant |
| PrecO-R | GCTTGTTCTGTGTTTTCGCC | |
| PrecO-11 | CGAATCACCTAGTATCCGCTATGGCGTTTGCCATAG | |
| PrecO-22 | GATTCAGGAACTGATCGCATTAGAGCGTTCTGTCGC | |
| KanF-11 | CGGATACTAGGTGATTCGAGACGATAAATGCGTCGG | Forward primer for aphA cassette |
| KanR-22 | GCGATCAGTTCCTGAATCGACATCTAAATCTAGGTAC | Reverse primer for aphA cassette |
| D350P1 | TTATCGAGCTGTATGCGGAG | To construct IDS350 |
| D350P2 | TCCATACGATACCGGGTACCTGCCATCTTTCACAAAGATG | |
| D350P3 | CTCCGCATACAGCTCGATAA | |
| D350P4 | GGTACCCGGTATCGTATGGA | |
| Hp405-F | CTCCCTTACTCAACCCTAAC | To construct hp405::IDS100 and hp405::IDS350 |
| Hp405-R | CGTGAAGTGCAAGCTCACTC | |
| Hp405-11 | CGAATCACCTAGTATCCGCCATATCTTTCAAATACGGC | |
| Hp405-22 | GATTCAGGAACTGATCGCATACGCTAATCGCACACGGC | |
| RecR-F | CCTCATCTATAATGACCACA | To construct recR::IDS100 and recR::IDS350 |
| RecR-R | GAGATGAATTGAGTCGTCAG | |
| RecR-11 | CGAATCACCTAGTATCCGACCCAAATGATACGCCA | |
| RecR-22 | GATTCAGGAACTGATCGCGACTCAGTTTCGCTCTCA | |
| RecA-F | TTGCGCTCACATCATAAGTG | To construct recA::IDS100 and recA::IDS350 |
| RecA-R | CTTGATTGATCGCTTCAAGC | |
| RecA-11 | CGAATCACCTAGTATCCGGAGCCTGTAGAAATAGAGTC | |
| RecA-22 | GATTCAGGAACTGATCGCCTCTAATGAAGAGATCATGC |
The complemented recR+ strain was constructed by inserting a wild-type copy of the recR gene in the rdxA locus of the recR::cat chromosome. PCR products corresponding to the 3′ end of the rdxA gene (266 bp), 810 bp of the full-length recR gene (including 186 bp of upstream sequence containing its promoter), and the 5′ end of the rdxA gene (256 bp) were amplified in three separate PCRs and then stitched together in subsequent PCR using the primers listed in Table 1. The final PCR product was used to disrupt the rdxA locus of the recR::cat strain by selecting for metronidazole (16 μg/ml) resistant colonies (96 bp at the center of the rdxA gene was deleted and replaced by an intact recR gene).
2.3. Oxygen sensitivity (air survival) assay
H. pylori strains were grown on BA plates to late log-phase, and the cells were suspended in PBS at a concentration of ~108 cells/ml. The cell suspensions were incubated at 37°C under normal atmospheric conditions (21% O2, without alteration of CO2 partial pressure) with moderate shaking. Samples were then removed at various time points (2, 4, 6, 8, and 10 hours), serially diluted, and spread onto BA plates. Colony counts are recorded after 4 days of incubation in a microaerobic atmosphere (5% partial pressure O2) at 37°C.
2.4. Assessment for sensitivity to low pH condition
H. pylori strains were grown on BA plates to late log-phase, and the cells were suspended at a concentration of ~108 cells/ml in the buffer (20 mM Tris-HCl, 150 mM NaCl, pH 7.0), or, for acid stress condition, in the same solution pre-adjusted to pH 3.0 by adding appropriate amount of HCl. The cell suspensions were incubated under a microaerobic condition (4% O2) at 37°C for 1 hour. The samples were serially diluted and plated for CFU counts (after 4 days incubation under microaerobic growth condition, see text above). The percentage of cell survival in pH 3.0 relative to that in pH 7.0 was calculated.
2.5. DNA transformation assay to assess inter-genomic recombination frequency
The donor DNA used in this study included: (i) a 330 bp PCR fragment of H. pylori rpoB gene fragment containing a site-specific mutation (at the center of the fragment) conferring rifampicin resistance, (ii) a linear DNA fragment containing a kanamycin resistance cassette (1.4 kb) flanked by H. pylori acnB gene sequences (about 550 bp on each side of the Kan cassette), and (iii) pHP1, a H. pylori-E. coli shuttle plasmid carrying a Kan cassette [28].
H. pylori strains were grown on BA plates to late log-phase, and the cells were suspended in PBS at a concentration of ~ 108/ml (recipient cells for transformation). A 30 μl cell suspension sample was mixed with 100 ng of donor DNA and spotted onto a BA plate. After incubation for 18 hours under microaerobic condition at 37°C, the transformation mixture was harvested and suspended in 1 ml PBS. 100 μl portions of the suspension (or appropriate dilution as needed) were plated onto either BA plates or BA plates containing selective antibiotic (20 μg/ml rifampicin or 40 μg/ml kanamycin, depending on the donor DNA used). The plates were incubated for 4 days under the microaerobic condition at 37°C, and the numbers of colonies were counted. The transformation frequency was determined by the number of resistant colonies divided by the total number of CFU. In a normalized DNA transformation assay, the frequency of transformation is expressed as the number of transformants per 108 recipient cells. As negative controls, H. pylori strains with no DNA added were tested under this assay condition; no antibiotic-resistant colonies were observed.
2.6. Construction of the intra-genomic recombination substrates (deletion cassettes)
A plasmid (pUre100) was kindly provided by Dr. Martin Blaser. pUre100 contains a deletion cassette (IDS100) and was described and used for determination of the intra-genomic recombination frequency (deletion frequency) resulting from recombination on direct repeat sequences [26, 29, 30]. The deletion cassette IDS100 contains a chloramphenicol resistance gene (CAT) with flanking identical repeat segments (100 bp long) of the kanamycin resistance gene (aphA). By using a multiplex overlapping PCR method (primers listed in Table 1) and IDS100 as the starting template, we constructed IDS350 in which the length of the identical repeat segments is 350 bp. The complete nucleotide sequence of IDS350 was confirmed by DNA sequencing. Subsequently, the recR::IDS and recA::IDS were constructed by overlapping PCR method (primers listed in Table 1) in which the major portion of the gene was replaced by the deletion cassette (IDS100 or IDS350). The appropriate PCR product was used to transform H. pylori wild type strain by selecting Cm-resistant colonies. As controls, the deletion cassettes were inserted into hp405 locus which has no known effect on DNA recombination or any other characteristics.
2.7. Determination of the intra-genomic recombination (deletion) frequency
Strains to be tested were grown on BA plates containing chloramphenicol (50 μg/ml) under normal microaerobic condition (4% O2) to late log phase. The cells were suspended in PBS at a concentration of 108 – 109 cells/ml and serial dilutions were made. Appropriate dilutions were plated onto BA plates with or without kanamycin (40 μg/ml), and the plates were incubated for 4 to 5 days followed by counting the number of colonies. The frequencies of intra-genomic recombination (deletion of cat cassette and recovery of intact aphA cassette) were calculated as the number of KanR cells divided by the number of total cells. The average frequencies and their standard deviations were calculated from at least five independent experiments.
2.8. Mouse colonization
Mouse colonization assays were performed essentially as described previously [11, 21, 31]. Briefly, the wild type X47 or isogenic rec mutant cells were harvested after 48 h of growth on BA plates (37°C, 4% oxygen) and suspended in PBS to an OD600 of 1.7. Headspace in the tube was sparged with argon gas to minimize oxygen exposure, and the tube was tightly sealed. The bacterial suspensions were administered to C57BL/6J mice (3 × 108 H. pylori cells/mouse) twice, with each of the oral deliveries made 2 days apart. Three weeks after the first inoculation, the mice were sacrificed and the stomachs were removed, weighed, and homogenized in argon-sparged PBS [32] to avoid O2 exposure. Stomach homogenate dilutions (dilutions conducted in tubes in argon-sparged buffer) were plated on BA plates supplemented with bacitracin (100 μg/ml), vancomycin (10 μg/ml) and amphotericin B (10 μg/ml), and the plates were rapidly transported into an incubator containing sustained 4% partial pressure O2. After incubation for 5 to 7 days the fresh H. pylori colonies were enumerated and the data expressed as CFU per gram of stomach.
3. Results and Discussion
3.1 Sensitivity of H. pylori recR and recO mutants to DNA damaging agents
Previously, we characterized H. pylori recN and addA mutants [11, 21]. To focus on the physiological roles of the RecRO pathway herein, we constructed recR and recO mutants. The mutants were constructed in strain X47, a mouse-colonizing strain, to also determine the effects on mouse colonization ability. In addition, we constructed an addA recO double mutant that is deficient in both pathways of DNA recombinational repair. All three mutant strains, X47 recR::cat, X47 recO::aphA, and X47 addA::cat recO::aphA, grew much slower than the wild type X47 under the normal microaerobic conditions (the growth rate is about half that of the WT). In the report of Marsin et al [20], the recR and recO single mutants (in the strain 26695 background) did not show obvious growth impairment by measuring the colony size after 4 days growth on plates. To determine whether these are strain-dependent difference, we constructed recR mutants in both background strains 26695 and X47, and measured the growth curves (data not shown). We observed that both recR mutants grew at a slower rate than the wild type in exponential growth phase, but can eventually reach a similar yield to the wild type at stationary phase (after 3–4 days).
In the study of Marsin et al [20], the recR and recO single mutants showed marked sensitivity to DNA damaging agents metronidazole and UV light, indicating roles of RecR and RecO in DNA repair. On the other hand, the recR and recO single mutants did not show significant sensitivity to ionizing radiation (IR), suggesting that all the recombinational repair of IR-induced damage is mediated by the AddA (RecB)-dependent pathway. We examined UV sensitivity of our strains; similar results were obtained, i.e. the recR and recO single mutants are more sensitive to UV light than the wild type (data not shown), confirming the role of H. pylori RecRO pathway in repairing UV-induced DNA damage.
Mitomycin C (MMC) causes predominantly DNA intra-strand cross-links, leading ultimately to DNA double strand breaks. In this study, we determined the sensitivity of the mutant strains to MMC by growing H. pylori strains on BA plates containing different concentrations of MMC. The wild type H. pylori strain can grow on the plates containing 5 ng/ml MMC, but not on 10 ng/ml MMC. The recN mutant strain is more sensitive than the wild type, as it tolerated 2.5 ng/ml MMC but could not grow on the plates containing 5 ng/ml MMC [11]. The addA mutant strain showed even higher sensitivity with no growth on plates containing MMC at 2.5 ng/ml or higher concentrations [21]. In contrast, the MMC sensitivity of the recR or recO single mutant was the same as the wild type, and the addA recO double mutant showed the same level of sensitivity to MMC as the addA single mutant (data not shown). These results indicate that the RecRO pathway is not responsible for the repair of MMC-induced DNA damage.
3.2. H. pylori recR and recO mutants are sensitive to oxidative stress and acid stress
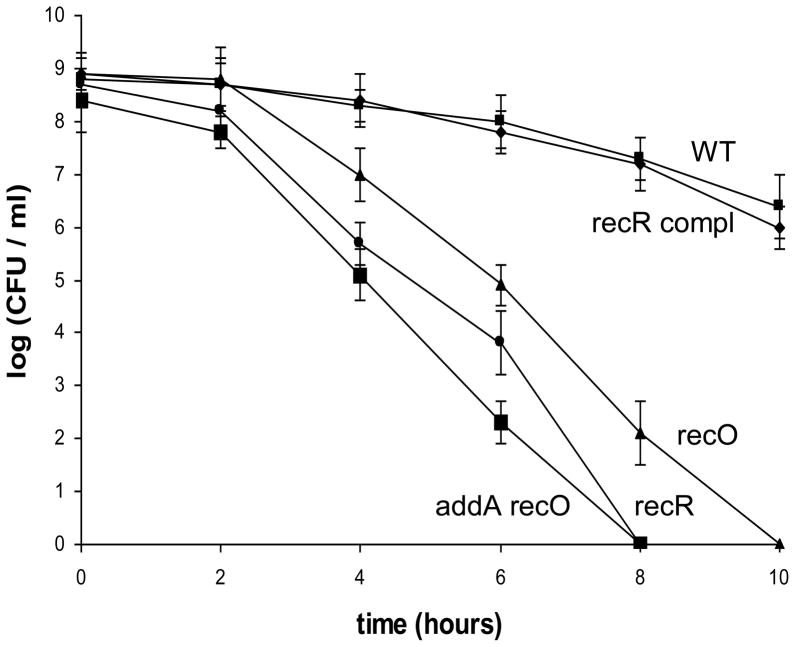
Oxidative stress is a major stress condition that H. pylori encounters in its physiological niche. H. pylori induces strong host inflammatory responses that involve recruitment of neutrophils, lymphocytes and macrophages; these release reactive oxygen species that damage DNA. H. pylori DNA was shown to be a target for host-generated oxidative stress based on studies of H. pylori nth strains that are unable to repair oxidized pyrimidines [7]. Further studies showed that mutant cells of ruvC [12], recN [11] or addA (recB) [21] were more sensitive to oxidative stress, indicating an important role of DNA recombinational repair in H. pylori for the bacterial survival of oxidative damage. To examine the role of the RecRO pathway in repairing DNA damage derived from oxidative stress, we examined the sensitivity of the H. pylori strains to oxidative stress by an air survival assay. The cell suspensions (~ 5×108 cells/ml) were exposed to air, and the numbers of surviving cells were determined at various time points (Fig. 1). The number of wild type cells decreased slowly; and at the 10 h time point, about 5×106 cells (~1% of that at the time zero) survived. The recR complemented strain behaved like the wild type. In contrast, the recO, recR, and addA recO mutants showed a greater sensitivity to air exposure. Two hours after exposing cells to air, the number of surviving mutant cells started to decrease at a rate much faster than that of the wild type cells. At the 10 h time point, the recO mutant cells were completely killed (i.e. no viable cells recovered), and the recR mutant cells were completely killed at the 8 h time point. The addA recO double mutant is statistically significantly more air-sensitive than the recO single mutant and is slightly more sensitive than the recR single mutant. The results indicated that both RecRO and AddAB pathways are important for survival of oxidative damage. Similar roles of the RecBCD and the RecFOR pathways for survival of oxidative (H2O2) damage were also observed in E. coli [3, 33] and in Neisseria gonorrhoeae [34]. In those bacteria, however, the RecBCD appeared to be the predominant (over the RecFOR) repair pathway for oxidative damage. Our results suggest that the two pathways in H. pylori play similarly important roles in repairing oxidative stress-derived DNA damage.
Fig. 1. Survival of H. pylori cells upon exposure to air.
H. pylori cell suspensions in PBS were incubated at 37°C under normal atmospheric conditions (21% partial pressure O2, and no alteration of CO2 partial pressure). Samples were removed at the times indicated in the x axis and were used for plate count determinations in a 5% oxygen environment. The data are the means of three experiments with standard deviation as indicated. Symbols: square, wild type; diamond, recR complementation strain; triangle, recO::aphA; circle, recR::cat; large square, addA::cat recO::aphA. Based on statistical analysis (Student t-test), the cell survival differences between the WT and the mutant strains are significant (P<0.01) for all the data points except for the 2 h time point.
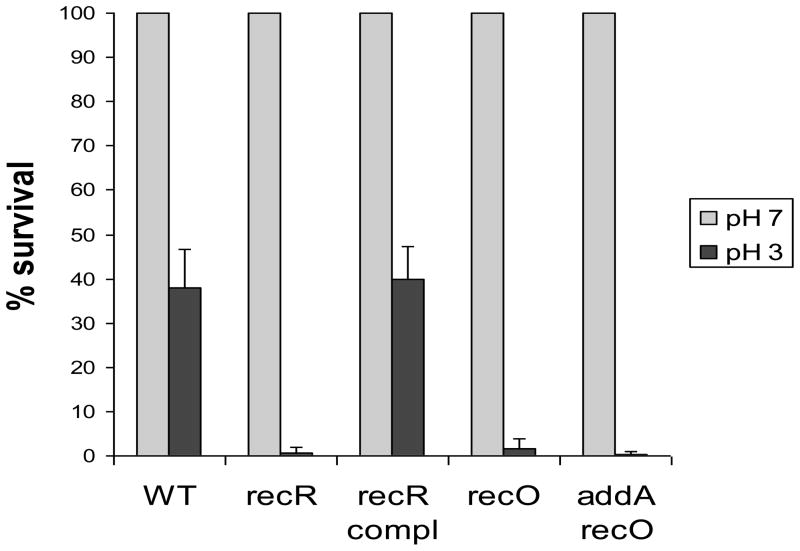
H. pylori appears to colonize an acidic niche on the gastric surface [10]. Therefore, low pH is another stress condition that H. pylori encounters in its physiological niche. Despite the existence of sophisticated pH homeostasis systems and acid tolerance mechanisms, bacteria may still suffer DNA damage from acid stress. Indeed, E. coli O157:H7 chromosomal DNA was shown to be significantly damaged by acid stress, and the Dps protein plays an important role in protecting acid-induced damage [35]. Notably, the acid tolerance of the recA mutant was significantly lower than that of the dps mutant and parent strain, suggesting that DNA recombinational repair might play an even more important role in acid tolerance than Dps [35]. Previously, we showed that H. pylori RecN is involved in repair of acid-induced DNA damage [11]. In this study, we characterized the recR, recO, and addA recO mutants for their sensitivity to low pH conditions. The cells of wild type H. pylori or the rec mutants were treated for 1 hour at different pH (pH 7.0 or pH 3.0) conditions, and the survival was determined. As shown in Fig. 2, about 40% of the wild type H. pylori cells survived the pH 3.0 condition for 1 hour. In contrast, only 1–2% of the recR cells or the recO cells survived the same stress condition. The complementation of the recR mutant restored the acid sensitivity to the wild type level. The addA recO double mutant cells were almost completely killed (>99.9% lethality) by the same treatment. The results clearly indicated that the RecRO pathway plays an important role in repairing DNA damage derived from acid stress.
Fig. 2. Acid sensitivity.
H. pylori cell suspensions were treated in solutions at different pH levels (pH 7.0 or pH 3.0) under a microaerobic condition (5% O2) at 37°C for 1 hour, and the numbers of surviving cells were determined. Numbers shown are the percentages of cell survival after treatment in pH 3.0 relative to that in pH 7.0. Data are means ± standard errors from three independent determinations.
3.3. Disruption of H. pylori recR and recO has no effect on inter-genomic recombination
H. pylori is naturally competent for DNA transformation, and has a highly efficient system for recombination of short-fragment involving multiple recombination events within a single locus [25, 36]. Natural transformation involves uptake of DNA into the cell followed by recombination into the genome at the site of homologous sequence (inter-genomic recombination). DNA uptake in H. pylori is mediated by a composite system involving proteins at the comB locus and ComEC [37, 38]. Assuming the wild type H. pylori and its isogenic rec strains are equally competent for DNA uptake, the frequency of natural transformation is an indicator of DNA recombination frequency. Previously, different results regarding the effect of rec genes on transformation frequency were reported by different research groups using different H. pylori strains and different DNA substrates. For example, Marsin et al [20] concluded that DNA integration into the chromosome following transformation is AddA (RecB)-independent. In contrast, studies from our group [21] and Kulick et al [25] indicated that inactivation of addA (recB) significantly reduced transformation frequency. In this study, we examined the effect of recR and recO on DNA transformation. As described previously [11, 21], we used two different types of DNA for examining DNA transformation of H. pylori. A specific A-to-G mutation in the H. pylori rpoB gene (rpoB3 allele) confers rifampicin resistance [39]. A 330-bp PCR fragment containing this specific mutation at the center of the fragment was used to transform H. pylori strains by using rifampicin resistance as a selective marker. Another type of DNA used for transformation was the sequence of H. pylori acnB gene (a housekeeping gene, 1.1 kb) in which a kanamycin resistance cassette (Kan, 1.4 kb) was inserted at the center (acnB:Kan).
The results for transformation are shown in Table 2. The recR and recO single mutants had a transformation frequency at a level similar to the wild type strain. According to Student t-test, the differences between the wild type and the mutants were not statistically significant. In contrast, the addA recO double mutant had a greatly reduced transformation frequency (2.13 × 10−5 transformants/recipient cell), which was ~12-fold lower than that of the wild type strain (2.56 × 10−4 transformants/recipient cell). According to Student t-test, this difference was highly significant (P<0.001). The effect of the addA recO double mutation was similar (no significant difference) to that observed for the addA single mutation (3.09 × 10−5 transformants/recipient cell). These results indicated that recombination of exogenous DNA into H. pylori genome in the process of transformation is dependent on AddA, but not on RecRO-pathway.
Table 2.
Transformation frequency with different types of donor DNA
| H. pylori strains | Donor DNA |
|
|---|---|---|
| rpoB3 (330 bp) | acnB:Kan (2.5 kb) | |
| X47 WT | 2.56 ± 0.38 × 10−4 | 7.20 ± 0.84 × 10−6 |
| X47 recR::cat | 3.25 ± 0.76 × 10−4 | 5.60 ± 0.32 × 10−6 |
| X47 recO::aphA | 3.04 ± 0.82 × 10−4 | ND |
| X47 addA::cat recO::aphA | 2.13 ± 0.68 × 10−5 | ND |
| X47 addA::cat | 3.09 ± 0.62 × 10−5 | 2.90 ± 0.30 × 10−7 |
The transformation frequencies are presented as the number of transformants (resistant colonies) per recipient cell. Data are means ± standard errors from three independent determinations. ND: When a strain contains a KanR marker (aphA gene), the transformation frequency with acnB:Kan cannot be determined.
3.4. Roles of RecR and RecA on intra-genomic recombination at repetitive sequence
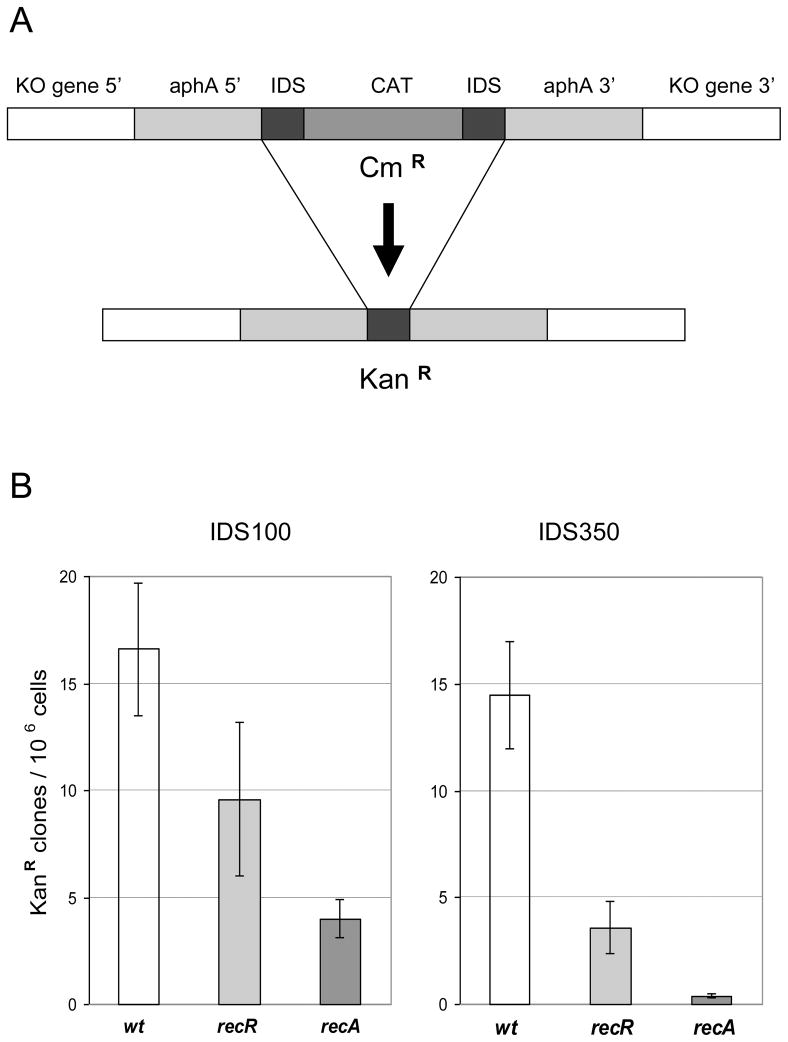
The E. coli paradigm of recombinational repair is that the RecBCD pathway acts on double-strand breaks and the RecFOR pathway acts on single-strand DNA (ssDNA) gaps. The transformation assays described above mainly measure the recombination activity involving DNA double strand breaks for which the RecBCD pathway is responsible in the organisms that have been well studied. For testing the role of the RecFOR pathway, an assay is required to measure intra-genomic recombination with repeat sequences [27]. We adopted an assay developed by Aras et al [26] and Kang et al [29, 30] to assess the deletion frequency resulting from recombination on direct repeat sequences, as described in Materials and Methods and in Fig. 3A. We made two DNA constructs (deletion cassettes) that contain identical repeat sequences of different length (IDS100 and IDS350).
Fig. 3. Intra-genomic recombination frequency in H. pylori recR and recA mutant strains.
(A) Description of the experimental system. A target knockout (KO) gene (recR, recA or hp405 representing WT) on H. pylori genome was disrupted by insertion of a deletion cassette (gray bars). The deletion cassette contains the aphA gene interrupted by cat gene with two flanking identical repeat sequences (IDS, black bars, 100 bp or 350 bp long). Recombination between the two IDS sequences gives rise to a functional aphA gene conferring kanamycin resistance. (B) Recombination frequencies determined for different KO genes with IDS100 or IDS350. Recombination frequencies were determined as described in Materials and Methods. Shown data are average frequency and standard deviation of at least 5 independent determinations.
The deletion cassettes were inserted into the target rec gene locus in the H. pylori genome. As a control, the deletion cassettes were inserted into hp405 locus which has no known effect on DNA recombination. Using this assay, we measured the deletion frequency of H. pylori recR and recA mutants (Fig. 3B). For IDS100, the deletion frequency in the wild type background was approximately 1.7×10−5 KanR clones/cell. Inactivation of recR or recA resulted in a 2- or 4-fold decrease in deletion frequency, respectively. According to Student’s t-test, the decreases are significant (P<0.01). These results indicated that the intra-genomic recombination of 100 bp-long direct repeat sequences in H. pylori is partially dependent on RecR and RecA. However, a big portion of the recombination event is RecR- and RecA-independent. This is basically in agreement (with small variance) with the results of Aras et al [26] who reported that the repeat sequences of 100 bp or shorter recombined through a RecA-independent pathway. Currently we are trying to identify such a RecA-independent recombination pathway in H. pylori.
For IDS350, the deletion frequency in the wild type background was approximately 1.4×10−5 KanR clones/cell. In contrast to IDS100, recombination on IDS350 was almost completely dependent on RecA, as the deletion frequency in recA strain was only 4×10−7 KanR clones/cell, 35-fold lower than that of background level. Inactivation of recR resulted in a significant (P<0.01) 4-fold decrease in deletion frequency, indicating that the intra-genomic recombination of IDS350 is RecR-dependent. The earlier report from Marsin et al [20] also observed that the H. pylori recO and recR mutants had a reduced recombination frequency compared to the wild type; however the difference was much smaller (less than 2-fold) than what we observed here. The discrepancy from the two studies could be due to the difference of the H. pylori strains and the assay systems used. Thus, our results here clearly indicate that RecR plays a significant role in recombination of IDS350. It is interesting to note that the deletion frequencies in the recR strain for both IDS100 and IDS350 were higher than those observed for the recA strain. Thus, the RecA-dependent IDS recombination (for both IDS100 and IDS350) is only partially dependent on RecR. Most likely, the AddAB pathway is also involved in intra-genomic recombination.
3.5. H. pylori recR and recO mutants have attenuated ability to colonize mouse stomachs
Repair of damaged DNA is known to be important for H. pylori survival and pathogenesis [7, 31]. Defects in DNA recombinational repair due to loss of RuvC, RecN, or AddAB function in H. pylori resulted in reduced ability to colonize the host stomach [11, 12, 18, 21]. As H. pylori recR and recO mutants showed sensitivity to both oxidative stress and acid stress, the condition that H. pylori encounters in its physiological niche, we sought to determine the effect of RecRO pathway on H. pylori colonization in the host. We performed an assay using a mouse infection model as described previously [11, 21, 31].
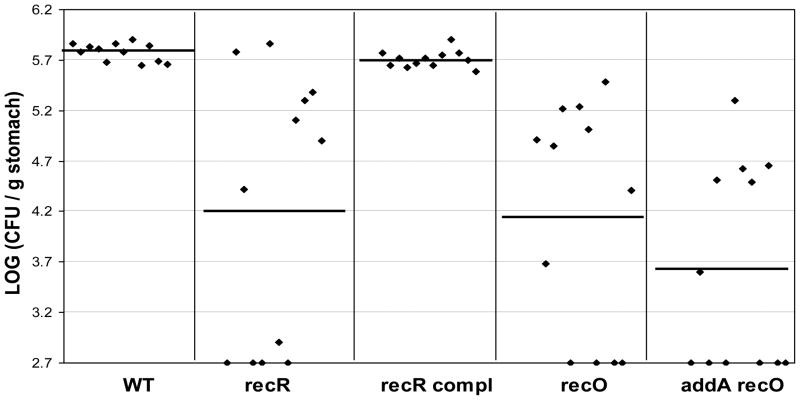
The wild type X47 or the isogenic mutant strains of recR, recO, and addA recO, as well as the recR complementation strain, were inoculated into 12 C57BL/6J mice, and the colonization of H. pylori cells in the mouse stomachs was examined 3 weeks after inoculation (Fig. 4). H. pylori was recovered from all 12 mice that had been inoculated with the wild type strain, with a geometric mean number of 6.0 ×105 CFU/g stomach. Similar results were obtained for the recR complementation strain (mean of 5.1 ×105 CFU/g). In contrast, 8 of 12 mice that were inoculated with the recR or recO strain were found to harbor H. pylori. The geometric mean of the colonization number for the recR or recO strain in the 12 mice was 1.6 ×104 or 1.4 ×104 CFU/g stomach, respectively. According to Wilcoxin rank test analysis, the ranges of colonization values of the recR or recO strain are significantly lower than that of the wild type at the 99% confidence level (P<0.01). Therefore, the RecRO pathway plays a significant role in H. pylori survival/colonization in the mouse stomachs. Of the 12 mice that were inoculated with the addA recO double mutant strain, only 6 mice had detectable H. pylori in their stomachs. The geometric mean of the colonization number for the addA recO strain in the 12 mice was 4 ×103 CFU/g stomach. Based on the previous data [21], the geometric mean of the colonization number for the addA single mutant (14 mice tested) was 1 ×104 CFU/g stomach. According to Wilcoxin rank test analysis, the ranges of colonization values of the addA recO double mutant is significantly lower than that of the addA or recO single mutant at the 95% confidence level (P<0.05). Thus, the effect of the double mutation on attenuation of mouse colonization is more severe than those observed for addA or recO single mutation.
Fig. 4. Mouse colonization results for H. pylori rec mutants.
The mice were inoculated with H. pylori two times (two days apart) with a dose of 1.5 × 108 viable cells administered per animal each time. Colonization of H. pylori in mouse stomachs was examined 3 weeks after the first inoculation. Conditions were used during stomach homogenization and homogenate dilution to minimize oxygen exposure (see text). Data are presented as a scatter plot (log scale) of colony forming units per gram of stomach as determined by plate counts. Each point represents the CFU count from one mouse stomach, and the solid horizontal lines represent the geometric means of the colonization numbers for each group. The base line [log10 (CFU/g) =2.7] is the detection limit of the assay, which represents a count below 500 CFU/g stomach.
3.6. Conclusions
H. pylori RecRO pathway is not responsible for repairing DNA double strand breaks. In accord with this, it is not involved in the integration of exogenous DNA fragments into the genome during the transformation process. Similarly to the RecFOR pathway in E. coli and other bacteria, the RecRO pathway in H. pylori plays a significant role in intra-genomic recombination involving direct repeat sequences as substrate. H. pylori recR and recO mutants were shown to be much more sensitive to oxidative stress and acid stress than the wild type strain, indicating that H. pylori RecRO pathway is involved in repairing DNA damage induced by these stress conditions. Furthermore, we demonstrated that RecRO-mediated DNA recombinational repair in H. pylori plays an important role in bacterial survival and persistent colonization in the host.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R21AI076569 to GW, R01AI077569 to RJM, and by the University of Georgia Foundations. We thank Sue Maier for her expertise and assistance on mouse colonization assays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404(6773):37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 2.Cromie GA, Connelly JC, Leach DR. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol Cell. 2001;8(6):1163–74. doi: 10.1016/s1097-2765(01)00419-1. [DOI] [PubMed] [Google Scholar]
- 3.Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev. 1999;63(4):751–813. doi: 10.1128/mmbr.63.4.751-813.1999. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rocha EP, Cornet E, Michel B. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 2005;1(2):e15. doi: 10.1371/journal.pgen.0010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morimatsu K, Kowalczykowski SC. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol Cell. 2003;11(5):1337–47. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 6.Dunn BE, Cohen H, Blaser MJ. Helicobacter pylori. Clin Microbiol Rev. 1997;10(4):720–41. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Rourke EJ, Chevalier C, Pinto AV, Thiberge JM, Ielpi L, Labigne A, Radicella JP. Pathogen DNA as target for host-generated oxidative stress: role for repair of bacterial DNA damage in Helicobacter pylori colonization. Proc Natl Acad Sci U S A. 2003;100(5):2789–94. doi: 10.1073/pnas.0337641100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang G, Alamuri P, Maier RJ. The diverse antioxidant systems of Helicobacter pylori. Mol Microbiol. 2006;61(4):847–60. doi: 10.1111/j.1365-2958.2006.05302.x. [DOI] [PubMed] [Google Scholar]
- 9.Ding SZ, Minohara Y, Fan XJ, et al. Helicobacter pylori infection induces oxidative stress and programmed cell death in human gastric epithelial cells. Infect Immun. 2007;75(8):4030–9. doi: 10.1128/IAI.00172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott DR, Marcus EA, Wen Y, Oh J, Sachs G. Gene expression in vivo shows that Helicobacter pylori colonizes an acidic niche on the gastric surface. Proc Natl Acad Sci U S A. 2007;104(17):7235–40. doi: 10.1073/pnas.0702300104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G, Maier RJ. Critical role of RecN in recombinational DNA repair and survival of Helicobacter pylori. Infect Immun. 2008;76(1):153–60. doi: 10.1128/IAI.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loughlin MF, Barnard FM, Jenkins D, Sharples GJ, Jenks PJ. Helicobacter pylori mutants defective in RuvC Holliday junction resolvase display reduced macrophage survival and spontaneous clearance from the murine gastric mucosa. Infect Immun. 2003;71(4):2022–31. doi: 10.1128/IAI.71.4.2022-2031.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suerbaum S, Smith JM, Bapumia K, Morelli G, Smith NH, Kunstmann E, Dyrek I, Achtman M. Free recombination within Helicobacter pylori. Proc Natl Acad Sci U S A. 1998;95(21):12619–24. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kraft C, Suerbaum S. Mutation and recombination in Helicobacter pylori: mechanisms and role in generating strain diversity. Int J Med Microbiol. 2005;295(5):299–305. doi: 10.1016/j.ijmm.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Tomb JF, White O, Kerlavage AR, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388(6642):539–47. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 16.Alm RA, Ling LS, Moir DT, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397(6715):176–80. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 17.Kang J, Blaser MJ. Bacterial populations as perfect gases: genomic integrity and diversification tensions in Helicobacter pylori. Nat Rev Microbiol. 2006;4(11):826–36. doi: 10.1038/nrmicro1528. [DOI] [PubMed] [Google Scholar]
- 18.Amundsen SK, Fero J, Hansen LM, Cromie GA, Solnick JV, Smith GR, Salama NR. Helicobacter pylori AddAB helicase-nuclease and RecA promote recombination-related DNA repair and survival during stomach colonization. Mol Microbiol. 2008;69(4):994–1007. doi: 10.1111/j.1365-2958.2008.06336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amundsen SK, Fero J, Salama NR, Smith GR. Dual nuclease and helicase activities of Helicobacter pylori AddAB are required for DNA repair, recombination, and mouse infectivity. J Biol Chem. 2009;284(25):16759–66. doi: 10.1074/jbc.M109.005587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marsin S, Mathieu A, Kortulewski T, Guerois R, Radicella JP. Unveiling novel RecO distant orthologues involved in homologous recombination. PLoS Genet. 2008;4(8):e1000146. doi: 10.1371/journal.pgen.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G, Maier RJ. A RecB-like helicase in Helicobacter pylori is important for DNA repair and host colonization. Infect Immun. 2009;77(1):286–91. doi: 10.1128/IAI.00970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakai A, Cox MM. RecFOR and RecOR as distinct RecA loading pathways. J Biol Chem. 2009;284(5):3264–72. doi: 10.1074/jbc.M807220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Handa N, Morimatsu K, Lovett ST, Kowalczykowski SC. Reconstitution of initial steps of dsDNA break repair by the RecF pathway of E. coli. Genes Dev. 2009;23(10):1234–45. doi: 10.1101/gad.1780709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inoue J, Honda M, Ikawa S, Shibata T, Mikawa T. The process of displacing the single-stranded DNA-binding protein from single-stranded DNA by RecO and RecR proteins. Nucleic Acids Res. 2008;36(1):94–109. doi: 10.1093/nar/gkm1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kulick S, Moccia C, Didelot X, Falush D, Kraft C, Suerbaum S. Mosaic DNA imports with interspersions of recipient sequence after natural transformation of Helicobacter pylori. PLoS One. 2008;3(11):e3797. doi: 10.1371/journal.pone.0003797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aras RA, Kang J, Tschumi AI, Harasaki Y, Blaser MJ. Extensive repetitive DNA facilitates prokaryotic genome plasticity. Proc Natl Acad Sci U S A. 2003;100(23):13579–84. doi: 10.1073/pnas.1735481100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galitski T, Roth JR. Pathways for homologous recombination between chromosomal direct repeats in Salmonella typhimurium. Genetics. 1997;146(3):751–67. doi: 10.1093/genetics/146.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleanthous H, Clayton CL, Tabaqchali S. Characterization of a plasmid from Helicobacter pylori encoding a replication protein common to plasmids in gram-positive bacteria. Mol Microbiol. 1991;5(10):2377–89. doi: 10.1111/j.1365-2958.1991.tb02084.x. [DOI] [PubMed] [Google Scholar]
- 29.Kang J, Blaser MJ. UvrD helicase suppresses recombination and DNA damage-induced deletions. J Bacteriol. 2006;188(15):5450–9. doi: 10.1128/JB.00275-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang J, Tavakoli D, Tschumi A, Aras RA, Blaser MJ. Effect of host species on recG phenotypes in Helicobacter pylori and Escherichia coli. J Bacteriol. 2004;186(22):7704–13. doi: 10.1128/JB.186.22.7704-7713.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang G, Alamuri P, Humayun MZ, Taylor DE, Maier RJ. The Helicobacter pylori MutS protein confers protection from oxidative DNA damage. Mol Microbiol. 2005;58(1):166–76. doi: 10.1111/j.1365-2958.2005.04833.x. [DOI] [PubMed] [Google Scholar]
- 32.Seyler RW, Jr, Olson JW, Maier RJ. Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect Immun. 2001;69(6):4034–40. doi: 10.1128/IAI.69.6.4034-4040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imlay JA, Linn S. Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J Bacteriol. 1987;169(7):2967–76. doi: 10.1128/jb.169.7.2967-2976.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stohl EA, Seifert HS. Neisseria gonorrhoeae DNA recombination and repair enzymes protect against oxidative damage caused by hydrogen peroxide. J Bacteriol. 2006;188(21):7645–51. doi: 10.1128/JB.00801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeong KC, Hung KF, Baumler DJ, Byrd JJ, Kaspar CW. Acid stress damage of DNA is prevented by Dps binding in Escherichia coli O157:H7. BMC Microbiol. 2008;8:181. doi: 10.1186/1471-2180-8-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin EA, Zhang XS, Levine SM, Gill SR, Falush D, Blaser MJ. Natural transformation of helicobacter pylori involves the integration of short DNA fragments interrupted by gaps of variable size. PLoS Pathog. 2009;5(3):e1000337. doi: 10.1371/journal.ppat.1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karnholz A, Hoefler C, Odenbreit S, Fischer W, Hofreuter D, Haas R. Functional and topological characterization of novel components of the comB DNA transformation competence system in Helicobacter pylori. J Bacteriol. 2006;188(3):882–93. doi: 10.1128/JB.188.3.882-893.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stingl K, Muller S, Scheidgen-Kleyboldt G, Clausen M, Maier B. Composite system mediates two-step DNA uptake into Helicobacter pylori. Proc Natl Acad Sci U S A. 107(3):1184–9. doi: 10.1073/pnas.0909955107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang G, Wilson TJ, Jiang Q, Taylor DE. Spontaneous mutations that confer antibiotic resistance in Helicobacter pylori. Antimicrob Agents Chemother. 2001;45(3):727–33. doi: 10.1128/AAC.45.3.727-733.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.