Abstract
Reverse transcriptases (RTs) encoded by a wide range of mobile retroelements have had major impact on the structure and function of genomes. Among the most abundant elements in eukaryotes are the non-Long Terminal Repeat (non-LTR) retrotransposons. Here we compare the dNTP concentration requirements and error rate of the RT encoded by the non-LTR retrotransposon R2 of Bombyx mori with the well-characterized RTs of retroviruses. Surprisingly, R2 was found to have properties more similar to the lentiviral RTs, such as HIV-1, than to the oncoretroviral RTs, such as MuLV. Like HIV-1 RT, R2 RT was able to synthesize DNA at low dNTP concentrations, suggesting that R2 is able to retrotranspose in non-dividing cells. R2 RT also showed levels of misincorporation in biased dNTP pools, and replication error rates in M13 lacZα forward mutation assays, similar to HIV-1 RT. Most of the R2 base substitutions in the forward mutation assay were caused by the misincorporation of dTMP.Analogous to HIV-1, the high error rate of R2 RT appears to be a result of its ability to extend mismatches once generated. We suggest that the low fidelity of R2 RT is a by-product of the flexibility of its active site/dNTP-binding pocket required for the target-primed reverse transcription reaction used by R2 for retrotransposition. Finally, we discuss that in spite of the high R2 RT error rate, based on the frequency of R2 retrotranspositions determined in natural populations, the long term nucleotide substitution rate for R2 is not significantly above that associated with cellular DNA replication.
Keywords: reverse transcriptases, retrotransposons, dNTP dependence, fidelity, HIV
INTRODUCTION
Retroelements are mobile DNA segments that generate new DNA copies for insertion into the host genome by reverse transcribing their RNA transcripts. Retroelements have a variety of life styles, from viruses that rapidly propagate from cell to cell and organism to organism, to endogenous elements that slowly propagate in the germ line of their host over millions of years. While many of the distinctive structural and catalytic proteins involved in the propagation of retroelements have been acquired from disparate sources, the key enzyme responsible for the formation of new copies, reverse transcriptase (RT), appears to share a common origin in all retroelements.1–3
The most extensively studied RTs are derived from vertebrate retroviruses. Two basic properties of retroviral RTs differ among groups. First, RTs encoded by lentiviruses, viruses that invade the immune system of their hosts (e.g. HIV-1 and SIV), are able to synthesize DNA at lower dNTP concentrations than the RTs from oncoretroviruses, viruses that can cause cancer in their hosts (e.g. MuLV and AMV).4–6 This difference in dNTP utilization may explain why lentiviruses can infect non-dividing cells, where dNTP levels are low, while oncoretroviruses can not.7.8 Second, lentiviral RTs have error rates that are as much as 15 fold higher than the rates associated with oncoretroviral RTs.9–11 The higher mutation rates of lentiviral RTs have been attributed to their need to rapidly evade the host immune system.11
Related to the retroviruses and utilizing the same mechanism of retrotransposition are the Long-Terminal Repeat (LTR) retrotransposons.12 Best studied are those elements found in various yeast species.13–17 For example, the Ty1 element of Saccharomyces cerevisiae encodes an RT with dNTP binding affinity similar to that of HIV-1 RT,15 while its error rate is more similar to that of oncoretroviral RTs.13,14
Evolutionarily more distant to retroviruses are the non-LTR retrotransposable elements (also called Long Interspersed Nucleotide Elements or LINEs). The RTs of these elements catalyze a retrotransposition mechanism substantially different from that of retroviruses and LTR-retrotransposons.1,18 In the non-LTR retrotransposition mechanism the chromosomal target site is used to prime reverse transcription, a process called target-primed reverse transcription (TPRT).19 Retroviruses and most LTR retrotransposons use a tRNA annealed to the RNA template to prime reverse transcription.12 Non-LTR retrotransposons are the most abundant mobile insertions in the human genome, accounting for over one-third of all DNA, yet enzymatic studies of their RTs lag behind that of retroviruses and LTR-retrotransposons.18
Detailed enzymatic studies of a non-LTR retrotransposon RT have only been conducted with the protein encoded by the R2 element of Bombyx mori.20–25 R2 RT has a number of enzymatic activities that differ from the RTs encoded by retroviruses and LTR-elements. These activities include higher processivity on DNA and RNA templates, end-to-end template jumping, the ability to use the 3′ end of RNA or DNA to prime synthesis and the ability to displace an annealed RNA strand while using a DNA strand as template. In this study we analyze the dNTP concentration dependence and error rate of R2 RT. The results suggest that R2 RT is similar to HIV-1 RT in its ability to synthesize DNA at low dNTP concentrations as well as its ability to extend mismatch bases once incorporated, thus giving rise to low replication fidelity.
RESULTS
dNTP concentration dependence on RNA and DNA templates
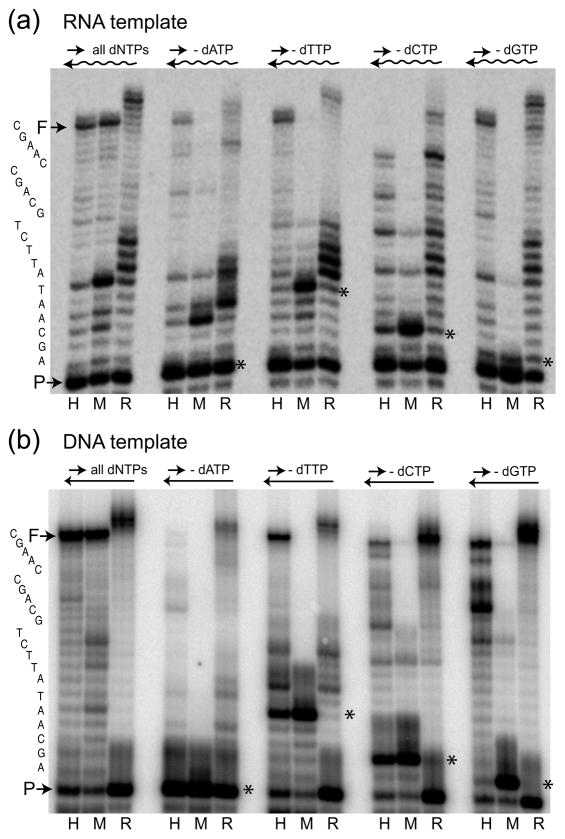
First the dNTP concentration dependence of the R2 RT was compared with that of HIV-1 and MuLV RTs. The primer extension assay employed an end-labeled 17-mer DNA primer annealed to a 38-mer RNA template. The concentration of each polymerase that generated full extension of 50% of the primer in the presence of 250μM of each dNTP was initially determined as a means to equilibrate the activity of the enzymes. The extension reactions were then repeated at this protein concentration with decreasing dNTP concentrations (250, 50, 5, 0.5, 0.05, 0.025, 0.0125 and 0.006 μM). As shown in Figure 1(a), while MuLV showed no full-length extensions at 0.05 μM (lane 5), HIV-1 and R2 RTs were both able to generate full-length extension products at a dNTP concentration as low as 0.0125 μM (lane 7).
Fig. 1.
Nucleotide concentration dependence of the RTs from HIV-1, MuLV and R2. The assay involved an end-labeled 17-mer primer (P) annealed to either a 38-mer RNA template (a) or a DNA template (b). HIV-1, MuLV and R2 RT protein concentrations were adjusted to give 50% full extensions (F) in a 15 minute incubation at 37°C in the presence of 250 μM dNTPs. The same reactions were repeated with decreasing dNTP concentrations (lane 1, 250 μM; lane 2, 50 μM; lane 3, 5 μM; lane 4, 0.5 μM: lane 5, 0.05 μM; lane 6, 0.025 μM; lane 7, 0.0125 μM; andlane 8, 0.006 μM), and the prod ucts analyzed on 14% polyacrylamide-urea gels. R2 RT extensions longer than F were the result of this polymerase adding non-templated nucleotides (terminal transferase activity).24 Products labeled J in the R2 RT reactions were generated by the polymerase jumping from the 38-mer template to the excess 17-mer primer in the incubation and continuing the polymerization.24,25
Two of the unusual properties previously noted for R2 RT can also be observed in Figure 1(a). First, at higher dNTP concentrations R2 RT exhibited terminal transferase activity by adding up to four, non-templated nucleotides when running off the RNA template. We have shown these extra nucleotides can correspond to any base; however, purines are added most efficiently.24 In contrast, the retroviral RTs added only one additional nucleotide and at lower levels. Second, R2 RT generated even longer products (band labeled J) as a result of the enzyme’s ability to jump to another (i.e. second) template and continue polymerization. The second template in this case was the 17-mer DNA primer present in the reactions. At much lower levels jumps were also made to the 38-mer RNA template (not shown). Previous studies of the mechanism used by R2 RT to jump between templates suggested the acquisition of a second template occurred by micro-homologies between the non-templated nucleotides on the synthesized DNA strand and sequences near the 3′ end of the second template.24 Consistent with this model the frequency of the template jumps corresponded to the level of non-templated additions (Figure 1A).
Differences in the ability of the three RTs to synthesize DNA at lower dNTP concentrations were more pronounced on DNA templates compared to RNA templates [Figure 1(b)]. On the DNA template, HIV-1 was unable to generate full-length products in 0.05 μM despite its ability on the RNA template (lane 5 in both panels). MuLV exhibited an even more dramatic dependence on dNTP concentration with minimal full-length products generated on the DNA template even at 5 μM (lane 3). In contrast, R2 RT exhibited less concentration dependence on the DNA template and was able to synthesize full-length DNA at the lowest dNTP levels tested (0.006 μM, lane 8). Again R2 RT readily added up to four non-template nucleotides as it ran off the DNA template but was less efficient at jumping to a second template (band labeled J).
The kinetics parameter Km for polymerases vary depending on the sequence of the template and the primer and whether the assay involves multiple or single nucleotide extensions.26–28 While the most accurate methods are single nucleotide incorporation assays, the tendency of R2 RT to add additional nucleotides as it runs off a template and to jump onto a second template complicates such assays. However, extension assays as those in Figure 1(a) and 1(b) can be used to approximate the Km values by means of standard Lineweaver-Burk plots. The Km for HIV-1 RT was determined to be 0.03 μM on the RNA template and 0.04 μM on the DNA template, in agreement with previous estimates in the range 0.03–0.13 μM and 0.03–1.9 μM on RNA and DNA templates, respectively.5, 29–31 The Km value for R2 RT on the RNA template was determined to be 0.02 μM, or somewhat lower than that of HIV-1, and 0.012 μM on the DNA template, significantly lower than that of HIV-1 RT.
Misincorporation assays on RNA and DNA templates
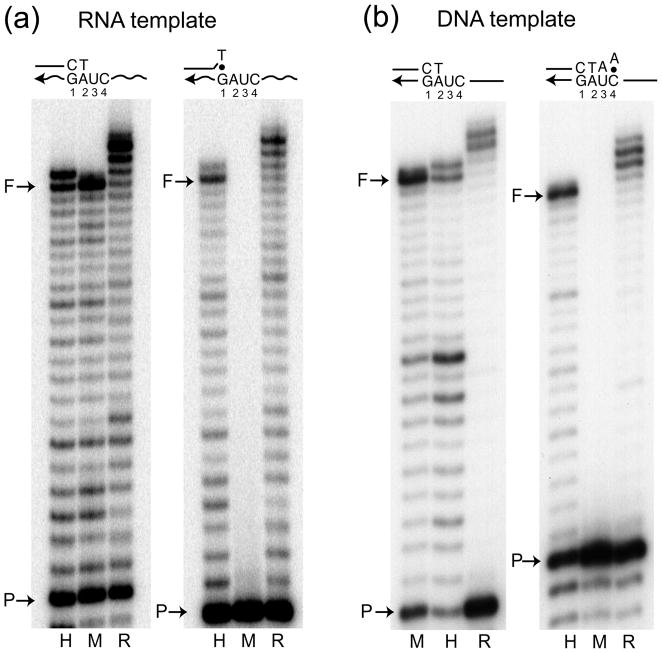
Next the fidelity of R2 RT was examined in misincorporation assays utilizing RNA and DNA templates. HIV-1 and MuLV RTs were again used for comparison, this time as examples of low and high fidelity RTs, respectively.6,32 Misincorporation assays monitor the combined influence of two steps in the generation of a mutation: misinsertion of a nucleotide and mismatch extension after misinsertion. As in the previous experiments, an end-labeled 17-mer primer was annealed to the 38-mer RNA or DNA template, and the amount of each protein needed to give equivalent levels of full extension in the presence of all four dNTPs was determined [left panel in Figures 2(a) and 2(b)]. The assays were then repeated with biased dNTP pools that contained only three dNTPs (i.e. the -dATP pool contained only dTTP, dCTP and dGTP). In the absence of one of the dNTPs, a high fidelity polymerase will stop extending one base before the template position in which the complementary dNTP was missing. These high fidelity stop sites are indicated with asterisks in Figure 2.
Fig. 2.
Nucleotide misincorporation assays in biased dNTP pools. The same labeled primer as in Figure 1 was annealed to a 38-mer RNA template (a) or DNA template (b) and extended by HIV-1 RT (lane H), MuLV RT (lane M) and R2 RT (lane R) at 37°C for 15 min. The experimental conditions included extension reactions with all four dNTPs (left set of lanes) and with biased dNTP pools containing only three dNTPs (− dATP, − dTTP, −dCTP or − dGTP sets of lanes). Reaction products were analyzed on a 14% polyacrylamide-urea denaturing gel. The terminal transferase activity of R2 RT generated products longer than full-length (F). The primary sequence of the correct extension product is shown on the far left. The sites for each biased pool where replication should have stopped unless misincorporations occurred are indicated with an *.
Consistent with previous misincorporation studies of HIV-1 and MuLV RT,6, 32 HIV-1 RT (lanes labeled H) exhibited much higher levels of extension beyond stop sites, and thus lower fidelity, compared to MuLV RT (lanes label M). MuLV showed the highest levels of misincorporation and longer products in the absence of dATP (RNA template) or dCTP (both templates), while HIV-1 showed its highest levels of longer products in the absence of dTTP and dGTP (both templates). In the case of R2 RT (lanes labeled R) similar levels of extensions past the stop sites were generated to that of HIV-1 RT on the RNA template, and even higher levels of extensions past the stop sites were observed on the DNA template.
Mismatch extension assays on RNA and DNA templates
Mismatch extension assays measure the second step in mutation synthesis: the ability of a polymerase to extend past the mismatched primer terminus generated by a misinsertion. To test the ability of the three RTs to extend mismatched primers on either RNA or DNA templates, a G/T mismatch at the 3′ end of the primer was used on the RNA template, while a C/A mismatch was used with the DNA template. The G/T and C/A mismatches were selected as they represent the replication intermediates likely involved in common base substitutions generated by HIV-1 RT.10 Consistent with previous studies,4,6 HIV-1 RT was able to extend the mismatch primers on both RNA and DNA templates almost as well as the matched primer, while MuLV RT exhibited essentially no mismatch extension [Figures 3(a) and (b)]. R2 RT was again similar to HIV-1 RT in efficiently extending the mismatched primers on both the RNA and DNA template.
Fig. 3.
Mismatch extension assays. End-labeled matched primers were annealed to the 38-mer RNA template (a) or DNA template (b) and extended by HIV-1 RT (H), MuLV RT (M) and R2 RT (R) at 37 °C for 15 min with 250 μM dNTPs. The primers corresponded to a matched 17-mer [left panels in (a) and (b)], a 16-mer with a terminal G/T mismatched annealed to an RNA template [right panel in (a)], and a 19-mer with a terminal C/A mismatched annealed to a DNA template [right panel in (b)]. Reaction products were analyzed on 14% polyacrylamide-urea denaturing gels. Again the terminal transferase activity of R2 RT generated products longer than full length.
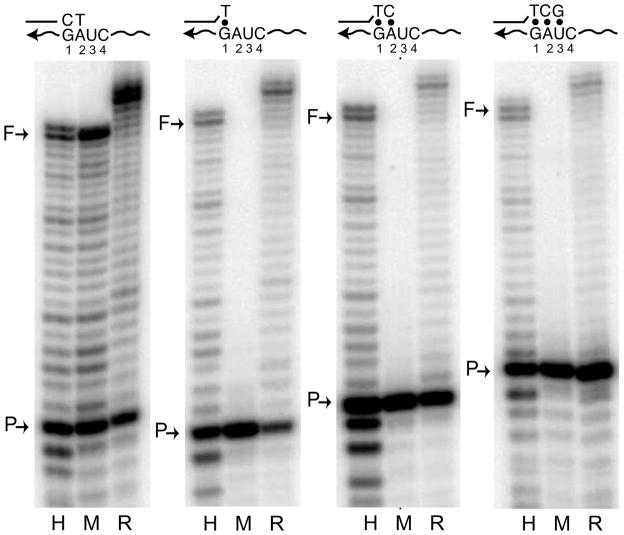
Previous studies with HIV-1 RT have shown this polymerase is capable of the elongation of mismatched primers with as many as three consecutive mismatches.33 Therefore, we compared the ability of R2 and HIV-1 RTs to extend primers with more extensive regions of mismatches on an RNA template (Figure 4). Primers were used that contain one (G/T), two (GA/TC) and three (GAU/TCG) consecutive terminal mismatches. Both R2 and HIV-1 RTs were able to initiate lower levels of synthesis even with three nucleotide mismatches. Counting the number of nucleotides added by the HIV-1 and R2 enzymes indicated that the major full-length products synthesized from the primer with three mismatches were one nucleotide shorter than those synthesized from the one and two mismatched primers. This reduction in length suggested that most products in these reactions had initiated on the RNA template one nucleotide beyond that expected for a primer with three mismatch bases.
Fig. 4.
Mismatch extension assays with one, two or three consecutive terminal mismatches. Protein activities were equilibrated with a 17-mer match primer on an RNA template as in Figure 3 (left panel). The extension reactions were repeated with a 16-mer containing one terminal mismatched nucleotide (G/T), a 17-mer with two terminal mismatches (GA/TC), or an 18-mer with three terminal mismatches (GAU/TCG). The mismatches between the primer and template are shown above the gel photographs and indicated with dots. The reactions were analyzed on 14% polyacrylamide-urea denaturing gels.
One concern in mismatch extension assays is that contaminating exonuclease activity may remove the mismatched bases at the end of the primer, and thus allow the extension of shorter primers without mismatches. Low levels of a contaminating exonuclease were probably present with the HIV-1 protein, because low levels of shorter length primers retaining their 5′ labeled ends were also generated during the reactions (bands below P). Thus the assays in Figure 4 may have over-estimated the ability of HIV-1 to extend mismatches. However, there was no evidence of the accumulation of shorter primers with either MuLV or R2 proteins suggesting these assays accurately reflected the ability of R2 RT to extend primers with extensive terminal mismatches. This mismatch extension activity of R2 RT was consistent with the ability of this enzyme to prime DNA synthesis using the ends of RNA or DNA oligonucleotides that have no sequence identity to the template.23 Indeed, the critical step of R2 retrotransposition, use of a DNA target site to prime reverse transcription, can readily occur in the absence of sequence complementarity between the DNA target and the RNA template.21
M13 lacZα forward mutation assay
Finally, we compared the replication fidelity of R2 RT with that of HIV-1 RT using a M13 lacZα forward mutation assay.34 Bacteriophage M13mp2 DNA containing a single-stranded gap of 361 nucleotides within the lacZ gene was prepared and used as the substrate for gap-filling reactions by R2 RT and HIV-1 RT. In this assay there are 113 nucleotide positions that enable detection of base substitutions and 150 positions of possible frame shifts. Two independent gap-filling reactions were performed for each RT and after each gap-filling reaction two independent transformations were performed into E. coli cells (see Materials and Methods). Phage containing mutations made by the polymerase while copying the gapped region of lacZ gene gave rise to pale blue or clear plaques while phage without mutations gave rise to blue plaques. The mutation frequency of each RT was determined as the ratio of mutant plaques to total plaques counted. As shown in Table 1, both HIV-1 and R2 RT displayed a similar mutant phage frequency of approximately 2 × 10−2. The background mutation frequency (absence of a filling reaction) was over two orders of magnitude lower (<5 × 10−5). The mutant frequency estimated here for HIV-1 RT was similar to previously published estimates obtained using the same forward mutation assays29,35 and are from 3- to 8-fold higher than the estimates derived by the same assay conducted with MuLV and AMV (0.16 × 10−2 and 0.36 × 10−2, respectively).9
Table 1.
Error rate of R2 RT and HIV-1 RT in lacZ gap-filling reactions.
| Total number of plaques | Mutant plaques | Mutant frequency | ||
|---|---|---|---|---|
| HIV-1 RT | Exp. 1 | 4499 | 102 | 2.26 × 10−2 |
| Exp. 2 | 4316 | 93 | 2.15 × 10−2 | |
| R2RT | Exp. 1 | 4062 | 70 | 1.72 × 10−2 |
| Exp. 2 | 4601 | 88 | 1.91 × 10−2 | |
Experiments 1 and 2 correspond to independent gap-filling reactions. In each experiment total plaques and mutant plaques were combined from two separate transformation reactions. Background mutation frequency (absence of gap-filling) was <5 × 10−5.
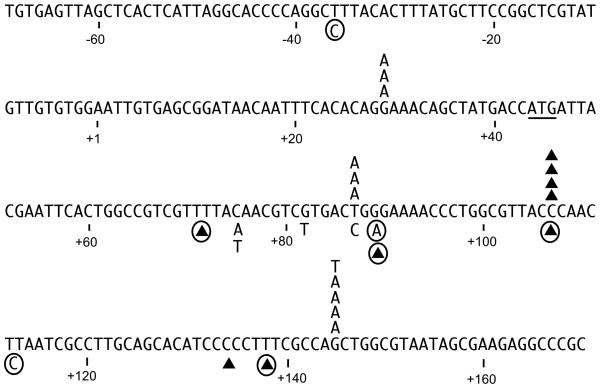
To characterize the types of mutations generated by R2 RT, independent and randomly chosen mutant phage derived from the two R2 filling reactions were isolated and the gap regions sequenced. The mutations generated by R2 RT are shown in Figure 5 above the lacZ sequence. We also sequenced mutant phage generated by HIV-1 RT to determine if the mutations obtained were similar to those of previous studies. Because the mutation spectrum for HIV-1 RT varies depending upon the strain of HIV-1 used, we have shown below the lacZ sequence in Figure 5 only the mutations generated in this study and mutation hot spots (circled mutations) seen in multiple previous studies of HIV-1. 10,35–37
Fig. 5.
Mutations generated by R2 RT in the lacZ gap-filling reactions. The wild type sequence of the M13 viral (+) template strand is shown and corresponds to the strand used as template in the RT gap-filling reactions. Position +1 is the first transcribed base while the ATG initiation codon for the lacZ open reading frame is underlined. Triangles represent one base deletions. Substitutions and deletions shown above the sequence were obtained from two independent filling and transformation experiments using R2 RT. Reaction 1 generated two T to A transversions at position 87, two G to A transitions at position 145 and a one base deletion in the run of Cs at position 106–108. All other mutations were generated by reaction 2. Substitutions and deletions shown below the primary sequence are mutations generated by HIV-1 RT. Those without circles were obtained in this report, while those that are circled are HIV-1 mutation hot spots identified in previous studies.10,36–38
The majority of the R2 RT mutations (11 of 15) corresponded to base substitutions at three hotspots. Of these substitutions seven corresponded to G to A transitions at positions 29 and 145 and three corresponded to T to A transversions at position 87. Because the strand synthesized by R2 RT in the gap-filling reaction was the complement to that shown in Fig. 5, 10 of the 11 scored misincorporations corresponded to the insertion of dTMP. In comparison, previous studies have shown that most HIV-1 RT mutations also involved misincorporations occurring at a few hot spots, with most misincorporations corresponding to the insertion of dGMP.10,36–38 In the case of the four single base deletions scored for R2 RT, all occurred in the run of Cs at positions 106 –108. This region also corresponds to a hot spot for deletions by HIV-1.10,36–38 Because the three consecutive Cs at position 106–108 did not correspond to the longest run of Cs in the template (found at positions 132–136), sequence context appeared to influence slippages. Finally, previous findings with the less error prone AMV RT in identical gap-filling assays also generated more base substitutions, which were more broadly distributed and corresponded to the misincorporation of dAMP or dGMP (not shown in the Figure).9
DISCUSSION
This study continued our comparison of the enzymatic activities of the R2 reverse transcriptase with that of retroviral reverse transcriptases. R2 RT was found similar to HIV-1 RT in being able to synthesize DNA at low dNTP concentrations on either RNA or DNA templates. The ability of HIV-1 to synthesize DNA at low dNTP concentrations has been suggested to give this virus the ability to replicate within the low dNTP concentrations of a non-dividing cell.8 We have characterized the timing of R2 retrotranspositions in the female germ line of Drosophila simulans.39 Most retrotransposition events occurred late, during the final stages of germ cell division or oocyte development; however, events were also detected earlier in development when there are only a few primordial germ cells. These early events are important to the propagation of R2 elements because one early event will be inherited by many progeny. The ability of R2 RT to efficiently synthesize DNA at low dNTP concentrations may enable R2 elements to retrotranspose even when the germ cells are not dividing.
While all RTs are expected to have low replication fidelities because of the absence of proofreading activity, the assays described here suggested that R2 RT was most similar to the RTs encoded by retroviruses, such as HIV-1, which have the lowest replication fidelity. R2 RT exhibited similar, or even lower, fidelity to HIV-1 RT in misincorporation assays and in mismatch extension assays. In the sensitive M13 lacZα forward mutation assays R2 RT and HIV-1 RT had similar error rates (Table 1), which were three to eight fold higher than the error rates associated with AMV and MuLV RT.6,9 To our knowledge this is the first time this sensitive assay has been used on a non-retroviral RT.
Error rates similar to that of retroviruses have been previously described for a number of non-retroviral RTs. Based on in vitro misinsertion assays, the RT from the LTR retrotransposon Ty1 was shown by Boutabout et al. to be less error prone than that of HIV-1 RT and more similar to oncoretroviruses.14 Similarly, based on in vitro misincorporation assays, Kirshenboim et al. estimated that the RT from the LTR retrotransposon Tf1 had error rates similar to retroviral RT.17 Using the in vivo approach of sequencing new insertions after one round of retrotransposition, Gabriel and co-workers estimated that the substitution rate for Ty1 was about 2.5 X 10−5 per nucleotide per cycle in yeast cells, 13 Gilbert and co-workers estimated that the substitution rate of the non-LTR retrotransposon L1 was about 1.5 X 10−5 per nucleotide per cycle in human cells,40 and Conlan and coworkers estimated the substitution rate for the L1.LtrB, a mobile self-splicing group II intron, was about 10−5 per nucleotide per cycle in E. coli. 41 These rates are all similar to the values determined in single cycle replication studies of retroviruses.
The high error rate associated with retroviral RT, especially HIV-1, has been suggested to generate new variants that aide the virus in escaping the mammalian immune system.11 However, like all retrotransposons, R2 elements have no apparent need for a high mutation rate. R2 elements have maintained the same structure, the same insertion strategy and have been stable components of animal genomes for hundreds of millions of years.1,42 Why then does R2 encode an RT with an error rate similar to one of the most error prone retroviruses?
The generation of mutations by a polymerase involves two steps: the incorporation of an incorrect nucleotide, which generates a template/primer intermediate with a 3′ mismatch; followed by mismatch extension, which effectively locks in the incorrect nucleotide. Previous comparative studies done between HIV-1 and MuLV RTs showed that these two RTs have similar misinsertion fidelities, and it is the higher levels of mismatch extension by HIV-1 RT that is responsible for its lower replication fidelity.4 The lower replication fidelity of HIV-1 has been attributed to greater protein flexibility in the vicinity of the active site.43 Greater flexibility, particularly in the vicinity of the dNTP binding pocket, means the enzyme will be more tolerant to distortions of the base pair geometry caused by incorrect nucleotides.
The low sequence similarity between R2 RT and retroviral RTs prevents a direct comparison of their dNTP binding pocket and active site as is possible between the low and high fidelity retroviral polymerases. However, a variety of enzymatic properties suggest the R2 RT active site is also highly flexible. First, there is remarkable flexibility in what this enzyme can use to prime synthesis. In the normal R2 retrotransposition reaction, priming of reverse transcription occurs by a nick in one strand of the DNA target site.19 In this TPRT reaction there is no requirement for the DNA primer to anneal to the 3′ end of the RNA template.20,21 Indirect evidence suggests that second strand synthesis is also primed by cleavage of the second strand of the target site.25,44 Indeed, R2 RT is able to use the 3′ end of any RNA or DNA strand to prime synthesis independent of its ability to anneal to the template.21,22 A second observation that suggest of R2 RT has a flexible active site is the enzyme’s ability to add multiple non-templated nucleotides as it runs off an RNA or DNA template (ref. 24, see also Figures 1,2) and to use microhomologies within the extra nucleotides to jump onto the 3′ end of a second template.24 This template jumping ability may also play a role in the R2 integration reactions as non-templated nucleotides are often found at the 5′ and 3′ ends of integrated R2 elements.23,24 The ability we show here of R2 RT to efficiently use primers that contain one, two or three terminal nucleotide mismatches (Figures 4) is consistent with these previously shown activities. Thus we suggest that it is the flexibility of R2 RT active site required for the TPRT method of integration that is responsible for its high error rate.
While this is the first report of the direct in vitro estimation of the error rate associated with the RT encoded by a non-LTR retrotransposon, we suggest other members of this class may also have similarly high error rates. Other non-LTR retrotransposons have been shown to prime reverse transcription from a nick at a DNA target site.19,45 In addition, experiments with the RT encoded by the L1 non-LTR retrotransposon in humans have shown that this enzyme can extend primer-template complexes with two terminally mismatched nucleotides.46
Finally, the stable inheritance of R2 with its host genome and the many species with well-resolved phylogenies in Drosophila has enabled us to previously estimate R2 nucleotide substitution rates during evolution.47,48 Using the rate of change at synonymous positions in the open reading frame of R2 as the best indicator of the baseline nucleotide substitution rate, the rate of change for R2 was within the range of that seen for typical nuclear genes.47 This finding argued against the generally held view that retrotransposons evolve rapidly because of their error prone mechanism of synthesis. The direct estimate of the error rate associated with the R2 reverse transcriptase in this study and a recently conducted study of R2 activity in populations of Drosophila simulans,49 allows a more detailed analysis of this apparent paradox of a high replication error rate yet a low rate of nucleotide substitution. The R2 retrotransposition rate determined for natural populations was estimated to be one new insertion per haploid genome every 40 generations. Because the average number of elements per genome for the population was ~50 copies, the length of time required for each elements to average one retrotransposition event was therefore about 2000 generations. Furthermore, because there are from 10–20 cell divisions in the germ line of Drosophila each generation, there are from 20,000 to 40,000 DNA replication cycles for every retrotransposition event. Assuming that the error rate for Drosophila R2 elements are similar to Bombyx R2 elements, then even though each new R2 retrotransposition generates 1,000 to 10,000 times more mutations than that associated with each round of DNA replication,50 an increase in the R2 nucleotide substitution rate resulting from retrotransposition would be difficult to detect above the background substitution rate derived from DNA replication.
MATERIALS AND METHODS
Purification of RT proteins
The R2 element of Bombyx mori encodes a single 120 kilodalton protein with RNA-directed and DNA-directed DNA polymerase activity as well as a specific DNA endonuclease that cleaves the chromosomal insertion site.18 The R2 protein was expressed in Escherichia coli JM109/pR260 and purified as described previously.18,25 The protein was stored in 0.1% Triton X-100 and 0.1 mg/ml bovine serum albumen at −20°C. HIV-1 RT (p66/p66 homodimer) and MuLV RT were also expressed and purified as described previously.51,52 Protein concentrations were determined by silver staining of SDS polyacrylamide gels using bovine serum albumen as the protein concentration standard.
[dNTP] dependent DNA polymerization
Assays were performed to compare the ability of the various RTs to synthesize DNA using either DNA or RNA as a template in the presence of different concentrations of dNTPs. A 17-mer primer (5′-CGCGCCGAATTCCCGCT-3′, Integrated DNA Technologies) was 32P-labeled at the 5′ end by T4 polynucleotide kinase (New England Biolabs). A 38-mer RNA template (5′-GCUUGGCUGCAGAAUAUUGCUAGCGGGAAUUCGGCGCG- 3′, Dharmacon Research) was annealed with the 32P-labled primer in a 1:1 ratio of template-primer (T/P). A 38-mer DNA template encoding the same sequence as the 38-mer RNA template (Integrated DNA Technologies) was used to form a DNA Template/Primer in the same manner. Assays were performed in 20 μl of 5 mM Tris-HCl pH 8.0, 100 mM KCl, 2 mM DTT, 5 mM MgCl2 and 0.1 mg/ml bovine serum albumin, containing 10 nmol T/P and sufficient RT enzyme to give 50% primer extension at 250 μM dNTP concentrations (15 nmol for HIV-1 RT, 15 nmol for MuLV RT and 6–12 nmol R2 RT). To conduct the reaction, RT and T/P were pre-incubated at 37°C for 10 min and polymerizations initiated by adding the dNTPs at 37°C for 15 min. Reactions were stopped by the addition of three volumes of 95% (v/v) ethanol containing 0.3 M sodium acetate pH 5.3 and 0.1% (w/v) SDS. After precipitation the products were resuspended and incubated at 95°C for 5 min in 10 μl 40 mM EDTA, 99% formamide loading dye. The products were separated on 14% polyacrylamide-urea gels, and quantified using a PhosphoImager (BioRad). The percent of the input primer that had undergone any level of extension (i.e. at least one nucleotide) was used to calculate the Km values using Lineweaver-Burk plots.
Misincorporation assays with DNA or RNA templates
Procedures for the misincorporation assay were modified from those of Preston and co-workers.11 The same 17-mer primer and 38-mer templates (RNA and DNA) were prepared as described in the [dNTP] dependence DNA polymerization assays. The assays were again performed in 20 μl of 5 mM Tris-HCl, pH 8.0, 100 mM KCl, 2 mM DTT, 5 mM MgCl2, 0.1mg/ml bovine serum albumin, containing 10 nmol T/P and sufficient RT proteins to give 50% primer extension in the presence of all four dNTPs at 250 μM. The biased dNTP pools contained only three dNTPs at 250 μM. The RTs were pre-incubated with the T/Ps at 37°C for 10 min, and the reactions initiated by the addition of dNTPs at 37°C for 15 min. Reactions were stopped and the products quantified on 14% polyacrylamide-urea gels as described above.
Extension of Mismatched Primers
The ability of each RT to extend 3′-end mismatches was measured using different mismatched primers annealed to either the 38-mer RNA or DNA templates. The mismatched primers were a 16-mer G/T mismatched primer (5′-CGCGCCGAATTCCCGT-3′), a 19-mer C/A mismatched primer (5′-CGCGCCGAATTCCCGCTAA-3′), a 17-mer primer with two mismatched bases (5′-CGCGCCGAATTCCCGTC-3′) and a 18-mer primer with three mismatched bases (5′-CGCGCCGAATTCCCGTCG-3′) (mismatches underlined). All primers used in this assay were 32P-labeled at their 5′ end and annealed to the 38-mer template at a 1:1 ratio of template to primer. As a control, a matched RNA or DNA template/primer was prepared by annealing the 38-mer RNA or DNA template with the 17-mer primer as described above for the misincorporation assays. Mismatch extension reactions were conducted with all four dNTPs at 250 μM in the same buffer and the same T/P and protein concentrations as the misincorporation assays.
M13 lacZα forward mutation assay
The fidelity of purified R2 RT and HIV-1 RT proteins in vitro were determined using the assays previously described by Bebenek and Kunkel.34 First, M13mp2 DNA containing a 361-nucleotide single-stranded gap was prepared. “Gapped” M13mp2 DNA (75 ng) was then incubated with 10 nmol R2 RT or 15 nmol HIV-1 RT for 20 min at 37°C under the conditions described above for the [dNTP] dependence assay with all four dNTPs (250 mM). Two independent gap-filling reactions per RT protein were performed and the extended gapped DNAs were analyzed by 0.7% agarose gel to verify the production of double-stranded M13 DNA with filled gaps. Approximately 0.15 ng of the extended gapped DNA was transformed into MC1016 cells, and the transformed cells plated onto ten M9 plates containing 5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside (X-gal) and isopropyl-1-thio-b-D-galactopyranoside (IPTG) on a lawn of CSH50 cells. From 4,000 to 4,600 plaques could be scored on the 10 plates and the mutation frequencies determined as the ratio of mutant (pale blue and clear) plaques to total plaques (mutant plus wild type plaques). Individual mutant plaques from each gap-filling assay were isolated and the gap regions in the phage DNAs sequenced (Macrogen Inc.). The background mutation frequency was estimated by transforming MC1016 cells with 1.5 ng DNA in the absence of gap-filling and observing one mutant out of a total of ~8,000 plaques.
Acknowledgments
We thank Baek Kim for help with the lacZ forward mutation assays and for his comments on the manuscript. We also thank Danna Eickbush for her comments of the manuscript, and William Burke for his help with the DNA sequencing. This work was supported by research grant GM42790 from the National Institutes of Health.
Abbreviations used
- RT
Reverse Transcriptase
- LTR
long terminal repeats
- LINE
Long Interspersed Nucleotide Elements
- HIV-1
Human Immunodeficiency Virus type 1
- MuLV
Murine Leukemia Virus
- SIV
Simian Immunodeficiency Virus
- AMV
Avian Myeloblastosis Virus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eickbush TH, Malik HS. Evolution of retrotransposons. In: Craig N, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. American Society of Microbiology Press; Washington, D.C.: 2002. pp. 1111–1144. [Google Scholar]
- 2.Arkhipova IR, Pyatkov KI, Meselson M, Evgen’ev MB. Retroelements containing introns in diverse invertebrate taxa. Nat Genet. 2003;33:123–124. doi: 10.1038/ng1074. [DOI] [PubMed] [Google Scholar]
- 3.Simon DM, Zimmerly S. A diversity of uncharacterized reverse transcriptases in bacteria. Nucleic Acids Res. 2008;36:7219–7229. doi: 10.1093/nar/gkn867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skasko M, Weiss KK, Reynolds HM, Jamburuthugoda V, Lee K, Kim B. Mechanistic differences in RNA-dependent DNA polymerization and fidelity between murine leukemia virus and HIV-1 reverse transcriptases. J Biol Chem. 2005;280:12190–12200. doi: 10.1074/jbc.M412859200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diamond TL, Roshal M, Jamburuthugoda VK, Reynolds HM, Merriam AR, Lee KY, Balakrishnan M, Bambara RA, Planelles V, Dewhurst S, Kim B. Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J Biol Chem. 2004;279:51545–51553. doi: 10.1074/jbc.M408573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Operario DJ, Reynolds HM, Kim B. Comparison of DNA polymerase activities between recombinant feline immunodeficiency and leukemia virus reverse transcriptases. Virology. 2005;335:106–121. doi: 10.1016/j.virol.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 7.Lewis PF, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol. 1994;68:510–516. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis P, Hensel M, Emerman M. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 1992;11:3053–3058. doi: 10.1002/j.1460-2075.1992.tb05376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts JD, Preston BD, Johnston LA, Soni A, Loeb LA, Kunkel TA. Fidelity of two retroviral reverse transcriptases during DNA-dependent DNA synthesis in vitro. Mol Cell Biol. 1989;9:469–476. doi: 10.1128/mcb.9.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bebenek K, Abbotts J, Roberts JD, Wilson SH, Kunkel TA. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem. 1989;264:16948–16956. [PubMed] [Google Scholar]
- 11.Preston BD, Poiesz BJ, Loeb LA. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242:1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 12.Boeke JD, Corces VG. Transcription and reverse transcription of retrotransposons. Annu Rev Microbiol. 1989;43:403–434. doi: 10.1146/annurev.mi.43.100189.002155. [DOI] [PubMed] [Google Scholar]
- 13.Gabriel A, Willems M, Mules EH, Boeke JD. Replication infidelity during a single cycle of Ty1 retrotransposition. Proc Natl Acad Sci USA. 1996;93:7767–7771. doi: 10.1073/pnas.93.15.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boutabout M, Wilhelm M, Wilhelm FX. DNA synthesis fidelity by the reverse transcriptase of the yeast retrotransposon Ty1. Nucleic Acids Res. 2001;29:2217–2222. doi: 10.1093/nar/29.11.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pandey M, Patel S, Gabriel A. Insights into the role of an active site aspartate in Ty1 reverse transcriptase polymerization. J Biol Chem. 2004;279:47840–47848. doi: 10.1074/jbc.M406019200. [DOI] [PubMed] [Google Scholar]
- 16.Bibillo A, Lener D, Klarmann GJ, Le Grice SF. Functional roles of carboxylate residues comprising the DNA polymerase active site triad of Ty3 reverse transcriptase. Nucleic Acids Res. 2005;33:171–181. doi: 10.1093/nar/gki150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirshenboim N, Hayouka Z, Friedler A, Hizi A. Expression and characterization of a novel reverse transcriptase of the LTR retrotransposon Tf1. Virology. 2007;366:263–276. doi: 10.1016/j.virol.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Research. 2008;134:221–234. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 20.Luan DD, Eickbush TH. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol Cell Biol. 1995;15:3882–3891. doi: 10.1128/mcb.15.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luan DD, Eickbush TH. Downstream 28S gene sequences on the RNA template affect the choice of primer and the accuracy of initiation by the R2 reverse transcriptase. Mol Cell Biol. 1996;16:4726–4734. doi: 10.1128/mcb.16.9.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bibillo A, Eickbush TH. High processivity of the reverse transcriptase from a non-long terminal repeat retrotransposon. J Biol Chem. 2002;277:34836–34845. doi: 10.1074/jbc.M204345200. [DOI] [PubMed] [Google Scholar]
- 23.Bibillo A, Eickbush TH. The reverse transcriptase of the R2 non-LTR retrotransposon: continuous synthesis of cDNA on non-continuous RNA templates. J Mol Biol. 2002;316:459–473. doi: 10.1006/jmbi.2001.5369. [DOI] [PubMed] [Google Scholar]
- 24.Bibillo A, Eickbush TH. End-to-end template jumping by the reverse transcriptase encoded by the R2 retrotransposon. J Biol Chem. 2004;279:14945–14953. doi: 10.1074/jbc.M310450200. [DOI] [PubMed] [Google Scholar]
- 25.Kurzynska-Kokorniak A, Jamburuthugoda VK, Bibillo A, Eickbush TH. DNA-directed DNA polymerase and strand displacement activity of the reverse transcriptase encoded by the R2 retrotransposon. J Mol Biol. 2007;374:322–333. doi: 10.1016/j.jmb.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chowdhury K, Kaushik N, Pandey VN, Modak MJ. Elucidation of the role of Arg 110 of murine leukemia virus reverse transcriptase in the catalytic mechanism: biochemical characterization of its mutant enzymes. Biochemistry. 1996;35:16610–16620. doi: 10.1021/bi961462l. [DOI] [PubMed] [Google Scholar]
- 27.Furge LL, Guengerich FP. Analysis of nucleotide insertion and extension at 8-oxo-7,8-dihydroguanine by replicative T7 polymerase exo- and human immunodeficiency virus-1 reverse transcriptase using steady-state and pre-steady-state kinetics. Biochemistry. 1997;36:6475–6487. doi: 10.1021/bi9627267. [DOI] [PubMed] [Google Scholar]
- 28.Shi Q, Singh K, Srivastava A, Kaushik N, Modak MJ. Lysine 152 of MuLV reverse transcriptase is required for the integrity of the active site. Biochemistry. 2002;41:14831–14842. doi: 10.1021/bi0258389. [DOI] [PubMed] [Google Scholar]
- 29.Matamoros T, Kim B, Menéndez-Arias L. Mechanistic insights into the role of Val75 of HIV-1 reverse transcriptase in misinsertion and mispair extension fidelity of DNA synthesis. J Mol Biol. 2008;375:1234–1248. doi: 10.1016/j.jmb.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 30.Gutiérrez-Rivas M, Menéndez-Arias L. A mutation in the primer grip region of HIV-1 reverse transcriptase that confers reduced fidelity of DNA synthesis. Nucleic Acids Res. 2001;29:4963–4972. doi: 10.1093/nar/29.24.4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandey VN, Kaushik N, Rege N, Sarafianos SG, Yadav PN, Modak MJ. Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry. 1996;35:2168–2179. doi: 10.1021/bi9516642. [DOI] [PubMed] [Google Scholar]
- 32.Kim B, Ayran JC, Sagar SG, Adman ET, Fuller SM, Tran NH, Horrigan J. New human immunodeficiency virus, type 1 reverse transcriptase (HIV-1 RT) mutants with increased fidelity of DNA synthesis. Accuracy, template binding, and processivity. J Biol Chem. 1999;274:27666–27673. doi: 10.1074/jbc.274.39.27666. [DOI] [PubMed] [Google Scholar]
- 33.Yu H, Goodman MF. Comparison of HIV-1 and avian myeloblastosis virus reverse transcriptase fidelity on RNA and DNA templates. J Biol Chem. 1992;267:10888–10896. [PubMed] [Google Scholar]
- 34.Bebenek K, Kunkel TA. Analyzing fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 35.Weiss KK, Chen R, Skasko M, Reynolds HM, Lee K, Bambara RA, Mansky LM, Kim B. A role for dNTP binding of human immunodeficiency virus type 1 reverse transcriptase in viral mutagenesis. Biochemistry. 2004;43:4490–500. doi: 10.1021/bi035258r. [DOI] [PubMed] [Google Scholar]
- 36.Rezende LF, Curr K, Ueno T, Mitsuya H, Prasad VR. The impact of multidideoxynucleoside resistance-conferring mutations in human immunodeficiency virus type 1 reverse transcriptase on polymerase fidelity and error specificity. J Virol. 1998;72:2890–2895. doi: 10.1128/jvi.72.4.2890-2895.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rezende LF, Drosopoulos WC, Prasad VR. The influence of 3TC resistance mutation M184I on the fidelity and error specificity of human immunodeficiency virus type 1 reverse transcriptase. Nucleic Acids Res. 1998;26:3066–3072. doi: 10.1093/nar/26.12.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drosopoulos WC, Prasad VR. Increased misincorporation fidelity observed for nucleoside analog resistance mutations M184V and E89G in human immunodeficiency virus type 1 reverse transcriptase does not correlate with the overall error rate measured in vitro. J Virol. 1998;72:4224–4230. doi: 10.1128/jvi.72.5.4224-4230.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Zhou J, Eickbush TH. Rapid R2 retrotransposition leads to the loss of previously inserted copies via large deletions of the rDNA locus. Mol Biol Evol. 2008;25:229–237. doi: 10.1093/molbev/msm250. [DOI] [PubMed] [Google Scholar]
- 40.Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol Cell Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conlan LH, Stanger MS, Ichiyanagi K, Belfort M. Localization, mobility and fidelity of retrotransposed Group II introns in rRNA genes. Nucleic Acids Res. 2005;33:5262–5270. doi: 10.1093/nar/gki819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 43.Harris D, Kaushik N, Pandey PK, Yadav PN, Pandey VN. Functional analysis of amino acid residues constituting the dNTP binding pocket of HIV-1 reverse transcriptase. J Biol Chem. 1998;273:33624–33634. doi: 10.1074/jbc.273.50.33624. [DOI] [PubMed] [Google Scholar]
- 44.Christensen SM, Ye J, Eickbush TH. RNA from the 5′ end of the R2 retrotransposon controls R2 protein binding to and cleavage of its DNA target site. Proc Natl Acad Sci USA. 2006;103:17602–17607. doi: 10.1073/pnas.0605476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nature Strut Mol Biol. 2006;13:655–666. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- 47.Eickbush DG, Lathe WC, III, Francino MP, Eickbush TH. R1 and R2 retrotransposable elements of Drosophila evolve at rates similar to those of nuclear genes. Genetics. 1995;139:685–695. doi: 10.1093/genetics/139.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lathe WC, III, Eickbush TH. A single lineage of R2 retrotransposable elements is an active, evolutionarily stable component of the Drosophila rDNA locus. Mol Biol Evol. 1997;14:1232–1241. doi: 10.1093/oxfordjournals.molbev.a025732. [DOI] [PubMed] [Google Scholar]
- 49.Zhou J, Eickbush TH. The pattern of R2 retrotransposon activity in natural populations of Drosophila simulans reflects the dynamic nature of the rDNA locus. PLoS Genetics. 2009;5:e1000386. doi: 10.1371/journal.pgen.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 51.Kim B. Genetic selection in Escherichia coli for active human immunodeficiency virus reverse transcriptase mutants. Methods. 1997;12:318–324. doi: 10.1006/meth.1997.0485. [DOI] [PubMed] [Google Scholar]
- 52.Weiss KK, Isaacs SJ, Tran NH, Adman ET, Kim B. Molecular architecture of the mutagenic active site of human immunodeficiency virus type 1 reverse transcriptase: roles of the beta 8-alpha E loop in fidelity, processivity, and substrate interactions. Biochemistry. 2000;39:10684–10694. doi: 10.1021/bi000788y. [DOI] [PubMed] [Google Scholar]