Abstract
Purpose
To explore the activity of dasatinib alone and in combination with paclitaxel and carboplatin in ovarian cancer cells and to determine if dasatinib activity can be predicted based on evaluation of the SRC pathway.
Experimental Design
Microarray analysis was performed for IGROV1, OVCAR3, A2780 and SKOV3 ovarian cancer cells and the status of the genomic SRC signature pathway was determined. Cells were treated with carboplatin, paclitaxel and dasatinib individually and in combination. Pre- and post-treatment phospho-SRC (pSRC) and SRC protein expression was determined. Dose-response curves were constructed, and drug interaction was assessed by the Combination Index (CI) method.
Results
SRC protein expression levels reflected the SRC pathway genomic signature in the cell lines with the lowest (SKOV3) and highest (IGROV1) pathway expression, but not in those with intermediate expression (OVCAR3, A2780). Dasatinib treatment caused loss of pSRC in all cell lines, with 50% growth inhibition for IGROV1 at 70nM, OVCAR3 at 34nM, A2780 at 4.1μM and SKOV3 at 530nM. Dasatinib combined with cytotoxics yielded a synergistic effect (CI=0.46 to 0.79) in all cell lines except SKOV3.
Conclusion
Dasatinib in combination with standard chemotherapeutic agents appears to interact in a synergistic manner in some ovarian cancer cell lines. Further research is needed to evaluate tumor cell characteristics which predict response to dasatinib.
Keywords: SRC pathway, dasatinib, ovarian cancer
INTRODUCTION
Advanced stage epithelial ovarian cancer is the most lethal gynecologic malignancy with current 5-year survival of 31% [1]. Despite initial response to platinum and taxane-based chemotherapy in 70% of patients, most patients will ultimately succumb to chemorefractory disease. Hence, there is a critical need to identify novel therapies that can improve clinical outcomes for patients with this devastating illness. Attention in the past decade has been increasingly directed to the use of biologic agents to target critical cell pathways involved in carcinogenesis.
SRC is a nonreceptor tyrosine kinase that mediates multiple cell signaling pathways, including cell proliferation, growth, and survival [2]. It is aberrantly activated in a number of solid tumors, including colon, pancreas, and lung cancers [2]. SRC and its activated form, phospho-SRC (pSRC), are overexpressed in ovarian cancer cell lines and a majority of late stage ovarian cancers [2, 3, 4]. In previous studies, treatment of ovarian cancer cells in vitro or in vivo with various SRC-inhibitors resulted in decreased activation of cell growth and survival pathways, and increased the activity of standard chemotherapeutics [5, 6, 7, 8, 9]. A microarray based gene expression signature has been developed that reflects activation of the SRC pathway, and this has been associated with platinum-resistance and poor survival [5, 6]. In addition, there was a strong correlation between the level of SRC pathway activation, based on gene expression profiles, and the extent of inhibition of cell proliferation of ovarian cancer cell lines by the SRC inhibitor, SU6656 [5, 6].
Dasatinib is an oral inhibitor of SRC family kinases [2], and also inhibits at least four other protein tyrosine kinases and kinase families including BCR-ABL, c-KIT, EPHA2 and PDGFβ [10]. Dasatinib received Food and Drug Administration (FDA) approval in 2006 for the treatment of patients with Philadelphia chromosome positive acute lymphocytic leukemia (ALL) and chronic myelogenous leukemia (CML) [11], and is currently being studied in a variety of solid tumors. We hypothesize that SRC inhibitors, such as dasatinib, may have utility in the treatment of patients with ovarian cancer. The aim of our study was to evaluate the antiproliferative activity of dasatinib alone and in combination with paclitaxel and carboplatin in ovarian cancer cell lines. In addition, we sought to determine if the genomic SRC pathway signature or SRC protein levels could predict the activity of dasatinib as a single-agent, as well as the drug interaction effect of dasatinib in combination with cytotoxic agents.
MATERIALS AND METHODS
Drugs
Dasatinib (BMS-354825) was provided by Bristol-Myers-Squibb (Princeton, NJ). Carboplatin and paclitaxel were purchased from Sigma (St. Louis, MO). Paclitaxel and dasatinib were dissolved in dimethylsulfoxide (DMSO), and carboplatin was dissolved in distilled water. Concentrated stock solutions of all drugs were stored at −25°C.
Ovarian Cancer Cell Lines and RNA extraction
SKOV3 and OVCAR3 human ovarian cancer cell lines were obtained from the American Type Culture Collection (Manassas, VA). The A2780 and IGROV1 human ovarian cancer cell lines were gifts from Drs. Robert Brown and Johnathan Lancaster, respectively. Cell line authentication was performed by DNA genotyping at the University of Colorado DNA Sequencing and Analysis Core (Denver, CO). Analysis of the IGROV1 cell line showed more than two alleles at some autosomal loci, suggesting the presence of cells from more than one individual.
The cells were maintained in monolayer culture in RPMI 1640 (Gibco) media supplemented with 10% fetal bovine serum (Hyclone, Logan UT), 1% sodium pyruvate, 1% nonessential amino acids in a humidified chamber containing 5% CO2. All cell lines were mycoplasma-free. RNA from the cell lines was extracted using the Qiagen RNeasy Mini Kit after homogenization of the cell pellets using the Qiashredder (Qiagen, Valencia, CA). RNA quality and integrity was assayed using an Agilent 2100 Bioanalyzer [6].
Gene Expression Microarrays and Statistical Analysis
The SRC pathway signature was developed by Bild et al as described previously [6]. RNA was prepared per the NCI protocol, available at http://nciarray.nci.nih.gov/reference/ under “Alternative Methods and Protocols.” Gene expression values were obtained using Affymetrix Human Genome U133A 2.0 Plus array chips at the Duke DNA Microarray Facility. The SRC pathway signature was applied to the ovarian cancer cell lines as described in Gatza et al [12]. Briefly, expression estimates were generated by robust multi-array average (RMA) [13], log2 transformed, and standardized at the feature level to have the mean and standard deviation as in SRC/green fluorescent protein (GFP) expressing cells. Data were filtered to the 85 genes in the signature and decomposed into three factors based on a singular value decomposition (SVD) of the SRC/GFP expressing cells. Predictive probabilities of SRC pathway deregulation in the ovarian cell lines were obtained from the Bayesian probit regression model using a Monte Carlo Markov Chain (MCMC) algorithm with 1,000 burn-in and 5,000 iterations.
Immunocytochemistry
Cells were seeded 2,000 cells/well in 96-well plates, grown to 40% confluence and fixed with a 1:1 ratio of acetone and alcohol. Immunostaining was performed using IgG (negative control) and a rabbit monoclonal SRC antibody (36D10, Cell Signaling). Assessment of SRC membrane and cytoplasmic staining was subjectively scored as absent (0), weak (1+), moderate (2+), strong (3+) and very strong (4+).
Western Blot Analysis
Cells were seeded at 3×106 cells per plate, and allowed to reach confluence over 24 hours. Cells were incubated at 37°C for 72 hours with the IC50 dose of each drug or drug combination. Controls were treated with DMSO. Protein extraction was performed using Invitrogen’s Cell Extraction Buffer with the addition of protease and phosphatase inhibitors (Roche Complete Mini Protease inhibitor cocktail 11–836–153–001, Sigma cocktail 1 P2850 and cocktail 2 P5726). Insoluble proteins were pelleted by centrifugation, and the protein in the supernatants quantified (Pierce 660 nm Protein Assay 22660). Forty micrograms of total protein was resolved on 10% SDS-PAGE gels (BioRad Criterion 3450009 ).Western transfer and blocking was performed according to established protocols. Primary antibodies obtained from Cell Signaling Technology included: Src36D10 (#2109; used at 1:2000) and pSrc (Tyr416) (#2101; 1:1000). Actin antibody was obtained from Sigma (A4700; 1:3000). Protein expression was detected using horseradish peroxidase-conjugated IgG secondary antibody (Jackson Labs), followed by Western Lightning chemiluminescence (Perkin Elmer) and exposure to film (Kodak BioMax Light Film). SRC, pSRC, and actin expression were quantified by densitometric scanning using the Scion Image software (Scion Corporation, Frederick, MD). Results were normalized to the corresponding actin protein levels in each lane.
Cell proliferation assay
Tumor cells were seeded at 2,000 cells/well in a 96-well plate, and allowed to reach 40% confluence over 24 hours. Cells were incubated with each drug at 37°C for 72 hours: paclitaxel (range 0.5 – 25nM); carboplatin (20–1000μM); dasatinib (25 −2000nM). Control wells contained RPMI media only. Cell proliferation was measured using the CellTiter 96 AQueous One Solution Cell Proliferation Assay according to manufacturer’s recommendations (Promega, Madison, WI). All experiments were performed in triplicate. Single-agent dose response curves were constructed and the IC50 computed from the best fitting transition functions (determined by F-statistic) using GraphPad Prism software, version 4.03 (San Diego, CA). The cells were subsequently treated with combinations of paclitaxel and carboplatin; dasatinib and paclitaxel; dasatinib and carboplatin; and the three-drug combination of paclitaxel, carboplatin, and dasatinib at fixed-ratio molar concentrations ranging from 0.125 to 4 multiples of the single-drug IC50 doses. The cell proliferation assay and IC50 computations were performed as above.
In vitro drug interaction studies and statistical methodologies
In order to evaluate the growth inhibition of the agents in combination, we used the median effect method, which takes into account the potency of each drug combination and the shape of the dose-response curve [14, 15]. Composite dose response curves were obtained from three independent experiments and the median effective dose, Dm (equivalent to the IC50) was computed using CalcuSyn software (Biosoft, Cambridge, UK). Drug interaction was assessed by the combination index (CI) method of Chou and Talalay [14, 15]:
where D is the dose that yields x% growth inhibition and α=0 for mutually exclusive drugs (the drugs have similar sites of action). A synergistic, additive, or antagonistic drug interaction is defined as CI < 1, CI = 1, CI >1, respectively.
RESULTS
SRC pathway expression
Evaluation of the SRC pathway was based on a previously defined SRC pathway gene signature [6]. The relative expression of the SRC pathway signature on a scale from 0–100% was: SKOV3 (38.3%), OVCAR3 (42.3%), A2780 (55.1%), and IGROV1 (60.4%) [Table 1].
Table 1.
Expression of the SRC pathway and SRC protein in comparison to 1) the potency of single agent paclitaxel, carboplatin and dasatinib based on concentration required for 50% inhibition of cell proliferation; and 2) the combination index of dasatinib in combination with paclitaxel and carboplatin
| SKOV3 | OVCAR3 | A2780 | IGROV1 | |
|---|---|---|---|---|
| SRC pathway activity# | 38.3% | 42.3% | 55.1% | 60.4% |
| SRC protein expression | 1.713 | 2.049 | 0.404 | 2.27 |
| Dasatinib IC50 | 530 nM | 34 nM | 4100 nM | 70 nM |
| Paclitaxel IC50* | N/A | 3 nM | 7 nM | 2 nM |
| Carboplatin IC50 | 341 nM | 32.6 nM | 84.6 nM | 29.1 nM |
| Dasatinib-cytotoxic CI§ | 1.25 | 0.496 | 0.462 | 0.791 |
scale of activity ranges from 0 to 100%
Paclitaxel IC50 could not be reached despite increasing concentrations of drug
Three-drug CI reported except in the SKOV3 cell line, where only a dasatinib + carboplatin CI could be calculated.
SRC protein expression
Immunohistochemical assessment of SRC protein expression in the untreated cell lines was lowest in the SKOV3 cell line (1+), while the A2780 cell line had heterogeneous 2+ immunostaining, OVCAR3 demonstrated heterogeneous 3+ staining, and IGROV1 exhibited diffuse 4+ membrane and cytoplasmic staining [Figure 1].
Figure 1.

Representative SRC protein expression relative to IgG in two ovarian cancer cell lines. SRC expression was high (3+) for OVCAR3 with intense membrane staining and low (1+) for SKOV3.
Western blot analysis revealed low relative SRC protein expression in the A2780 cell line (0.404) and increasing SRC protein expression in the SKOV3 (1.713), OVCAR3 (2.049) and IGROV1 (2.27) cell lines [Figure 2]. SRC protein expression reflected the SRC pathway genomic signature in the cell lines with the lowest (SKOV3) and highest (IGROV1) pathway expression, but not in those with intermediate expression (OVCAR3, A2780) [Table 1]. Relative SRC protein expression did not change after treatment with single-agent paclitaxel, carboplatin, or dasatinib, or with the various combinations of the agents. Similarly, relative pSRC protein expression did not change after treatment with single-agent paclitaxel in any of the cell lines. In contrast, after exposure to carboplatin, relative pSRC expression was completely abrogated in the A2780 cells and was reduced 74.8% in the IGROV cell line. After treatment with either dasatinib alone or the three-drug combination, relative pSRC expression was absent in all cell lines.
Figure 2.

Immunoblot analysis of SRC and pSRC pre- and post-treatment. A. Baseline relative SRC expression is highest in IGROV1 and OVCAR3, and lowest in SKOV3 and A2780. B. Carboplatin results in a 74.8% decrease in pSRC expression in the IGROV1 cell line, and complete absence in the A2780 cell line. Treatment with dasatinib resulted in an absence of pSRC expression despite stable SRC expression in all cell lines, indicating inhibition of the SRC pathway.
Antiproliferative activity of single agent dasatinib, paclitaxel and carboplatin
The IC50 results for single agent dasatinib, paclitaxel and carboplatin are detailed in Table 1. The degree of dasatinib-inhibited cell growth was greatest in the IGROV1 and OVCAR3 cell lines (IC50 70nM and 34nM) and lower in A2780 (4100nM) and SKOV3 (530nM) [Table 1, Figure 3]. Carboplatin and paclitaxel had similar IC50 values in the IGROV1, OVCAR3 and A2780 cell lines. In the SKOV3 cell line, 50% cell inhibition could not be achieved with increasing doses of paclitaxel. Likewise, SKOV3 was the most resistant to carboplatin, with an IC50 that was ten-times higher than the most platinum-sensitive cell lines, IGROV1 and OVCAR3.
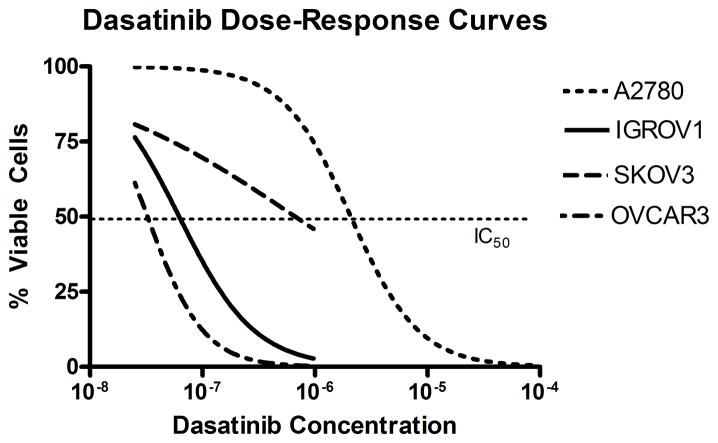
Figure 3.
Dasatinib dose-response curves for A2780 (IC50 4100nM), IGROV1 (70nM), OVCAR3 (40nM) and SKOV3 (530nM).
Drug interaction assessment for combination dasatinib and cytotoxic chemotherapy
The median effect analysis for combining dasatinib with the cytotoxic agents revealed significant synergy in all the cell lines except SKOV3 [Table 2, Figure 4]. The addition of dasatinib to either paclitaxel or carboplatin had a synergistic effect in OVCAR3 and A2780 cell lines. In the IGROV1 line, the addition of dasatinib to carboplatin was synergistic (CI 0.752), while the addition of dasatinib to paclitaxel was antagonistic (CI 1.381). When the three-drug combination was assessed, the addition of dasatinib to combination paclitaxel and carboplatin was strongly synergistic (CI 0.462–0.791) in cell lines with either SRC deregulation and/or high protein expression [Table 2, Figure 4]. Conversely, the addition of dasatinib to carboplatin in SKOV3 produced an antagonistic interaction (CI 1.25) [Table 2, Figure 4].
Table 2.
The combination index (CI) at fixed molar ratios in ovarian cancer cell lines. CI <1 is synergistic, CI=1 is additive, CI>1 is antagonistic
| Cell Line | Drugs | Molar Ratio | Combination Index (CI) |
|---|---|---|---|
| IGROV1 | |||
| Paclitaxel:Carboplatin | 1:15900 | 1.052 | |
| Paclitaxel:Dasatinib | 1:38.5 | 1.381 | |
| Carboplatin:Dasatinib | 414:1 | 0.752 | |
| Paclitaxel:Carboplatin:Dasatinib | 1:15900:38.5 | 0.791 | |
| A2780 | |||
| Paclitaxel:Carboplatin | 1:5833 | 1.194 | |
| Paclitaxel:Dasatinib | 1:8.775 | 0.876 | |
| Carboplatin:Dasatinib | 1:665 | 0.447 | |
| Paclitaxel:Carboplatin:Dasatinib | 1:5833:8.775 | 0.462 | |
| OVCAR3 | |||
| Paclitaxel:Carboplatin | 1:9429 | 1.63 | |
| Paclitaxel:Dasatinib | 1:9.68 | 0.207 | |
| Carboplatin:Dasatinib | 1:974 | 0.568 | |
| Paclitaxel:Carboplatin:Dasatinib | 1:9429:9.68 | 0.496 | |
| SKOV31 | |||
| Paclitaxel:Carboplatin | 1:12 | NA | |
| Paclitaxel:Dasatinib | 1:52.8 | NA | |
| Carboplatin:Dasatinib | 1:641.5 | 1.25 | |
| Paclitaxel:Carboplatin:Dasatinib | 1:52.8:641.5 | NA | |
The combination index for paclitaxel combinations could not be calculated in the SKOV3 cell line due to high paclitaxel resistance.
Figure 4.

Combination index plots (CI) of Chou-Talalay in the IGROV1 (A), OVCAR3 (B), A2780 (C) and SKOV3 (D) cell lines. In the cell lines SRC pathway deregulation (Panels A, C) or high SRC protein expression (Panel B), dasatinib combinations are synergistic, indicated by a combination index (CI) <1. In the SKOV3 cell line (D), which has a regulated SRC pathway and low SRC protein expression, the addition of dasatinib to carboplatin does not yield a synergistic interaction, indicated by a CI >1 (Panel D). P=paclitaxel; C=carboplatin; D=dasatinib.
DISCUSSION
Our data demonstrates that dasatinib inhibits the SRC pathway, as manifested by the absence of pSRC in ovarian cancer cell lines after treatment. Dasatinib demonstrated cytotoxic activity in ovarian cancer cell lines and treatment in combination with both paclitaxel and carboplatin yielded synergistic activity. Given our preclinical data as well as the findings reported by others [5, 6, 7, 8, 9], the SRC pathway is an appealing therapeutic target in ovarian cancer.
Several authors have reported enhanced activity of chemotherapeutic agents with the addition of anti-SRC therapies in ovarian cancer cell lines [5, 7, 8, 9]. SRC inhibition with PP2 or SU6656 in combination with paclitaxel or cisplatin resulted in decreased viability of ID8 murine ovarian cancer cells as well as SKOV3 and CaOV3 human ovarian cancer cells as compared to treatment with a SRC-inhibitor or either cytotoxic drug alone [7, 8]. Moreover, SRC inhibition by PP2, SU6656, or a SRC-dominant negative fusion construct restored paclitaxel and cisplatin sensitivity in the resistant ID8TaxR cell line. SRC inhibition in combination with paclitaxel yielded a 3–4-fold increase of caspase-9 independent caspase-3 cleavage compared to single-agent paclitaxel in the ovarian cancer cells. Furthermore, treatment with combination paclitaxel and PP2 resulted in accumulation of cleaved caspase-3 in resistant ID8TaxR and CaOV3TaxR cells but not in cells treated with paclitaxel alone [8].
Han and colleagues reported that the combination of docetaxel and the SRC inhibitor, AP23846, in the human ovarian cancer cell line HeyA8 resulted in 50-fold enhancement of growth inhibition compared to docetaxel alone [9]. Similar experiments in the engineered HeyA8-multidrug resistant (MDR) cell line demonstrated a 250-fold enhancement in growth inhibition with the combination compared to docetaxel alone. Measurement of caspase-3 cleavage products after treatment with single-agent docetaxel or AP23846 compared to combination treatment in HeyA8 or SKOV3ip1 cell lines suggested an additive effect, with doubling of caspase-3 cleavage products after combination therapy compared to treatment with either drug alone. In an orthotopic murine model utilizing HeyA8 or SKOV3ip1 cell lines, single-agent treatment with either AP23994 (an oral analogue of AP23846) or docetaxel produced a 70% decrease in tumor burden regardless of tumor cell origin. However, combination treatment with AP23994 and docetaxel further reduced the tumor burden by 95–98%. Further experiments in the HeyA8-MDR cell line model revealed significant reductions in tumor weight after treatment with either AP23994 alone (75%) or in combination with docetaxel (88%), but not with single-agent docetaxel [9]. Recently Konecny and colleagues reported that 71% of ovarian cancer cell lines in a panel of 34 were highly sensitive to dasatinib, with either additive or synergistic activity observed with the addition of carboplatin or paclitaxel [16]. These findings suggest that SRC inhibition may have an apoptosis-priming role, which may account for the synergy seen with the addition of SRC inhibitors to cytotoxics. Therefore, the efficacy of dasatinib combinations may be optimized by sequential administration of dasatinib followed by the cytotoxic agent, especially when combined with mitotic inhibitors such as paclitaxel.
Of particular interest is the identification of biomarkers to predict response and direct targeted therapy. In this paper, we examined SRC protein expression and SRC pathway activation as potential predictive markers. The small number of cell lines examined in this study precluded any definitive conclusions regarding the utility of these markers. Other biomarkers that have been reported to be potentially predictive of dasatinib activity include Yes, Lyn and EphA2, as well as caveolin-1 and -2, moesin, annexin-1 and uPA [16].
Response to dasatinib in the A2780 cell line was not consistent with what was expected based on SRC pathway activity. While the microarray analysis suggested significant SRC pathway activity, the highest IC50 value (4100 nM) for dasatinib was observed in these cells. Furthermore, dasatinib as a single agent had greater anti-proliferative activity in cell lines predicted to have a lower SRC pathway activity. Konecny and colleagues similarly reported stronger SRC expression in the A2780 cell line compared to either OVCAR3 or SKOV3 but evaluated individual gene expression rather than pathway activity. Yet the IC50 for dasatinib was greater (2829 nM) in the A2780 cell lines than the values found for either the SKOV3 (5nM) or OVCAR3 (7nM) [16]. Moreover, in our study, the A2780 cells revealed paradoxical low immunocytochemical and immunoblot SRC protein expression given the presence of significant SRC pathway activity, perhaps indicating that genomic SRC pathway deregulation does not ensure that subsequent gene translation and appropriate post-translational modification will transpire, or that an alternative mechanism for SRC pathway deregulation exists in these cells that does not involve high levels of SRC or pSRC. Another potential and more plausible explanation is that there are multiple deregulated pathways in the A2780 cell line [5, 17]. Both high MYC and PI3K pathway deregulation have been reported in A2780 cells. Additionally, Konecny et al reported that the A2780 cells exhibited relatively high expression of other genes not involved in the SRC signaling pathway, such as MSN and CDH2 [16]. The varying sources of this cell line as well as spontaneous transformation of cell lines may account for the conflicting results regarding SRC pathway activation reported in these studies. Nevertheless, the findings do suggest that additional pathways, such as MYC and PI3K, may also play a prominent role in A2780 cell proliferation. Merely inhibiting the SRC pathway may not be sufficient to inhibit proliferation in cells such as those from A2780 that exhibit deregulation of multiple pathways. In cancers with aberrant function of multiple pathways, a cocktail of targeted biologic agents may be required to successfully abrogate proliferation.
While we acknowledge the limitations of our study, which include the small number of cell lines evaluated, limited range of SRC pathway activity between the cell lines, and the use of in vitro models, the striking synergy seen when dasatinib was combined with either carboplatin and/or paclitaxel is intriguing. In vivo and clinical studies are necessary to further evaluate the synergistic activity and toxicity of the doublet and triplet regimens. The first stage of a phase II study evaluating single-agent dasatinib in women with recurrent or persistent ovarian cancer (GOG protocol #170M) has been completed, and the interim analysis is ongoing. Additionally, multiple phase I and II trials in other solid tumors have shown stabilization of disease or tumor response after treatment with single-agent dasatinib, and trials of dasatinib combinations are ongoing [18, 19, 20, 21]. Given the breadth of preclinical data demonstrating significant synergistic or additive effects of dasatinib in combination with paclitaxel and platinum-analogues, and promising results of these early clinical trials, further evaluation of dasatinib in combination with cytotoxic agents for the treatment of ovarian cancer is warranted.
Research Highlights.
Dasatinib has activity in ovarian cancer cells.
Dasatinib acts synergistically with carboplatin and paclitaxel.
Acknowledgments
We thank Dr. Monique Spillman for her assistance with cell line authentication, which was performed at the University of Colorado DNA Sequencing and Analysis Core, and Dr. Johnathan Lancaster for his thoughtful advice and discussions.
This study was supported by the National Cancer Institute grant supporting the Duke Clinical Oncology Research Career Development Award (K12) along with a research grant from Bristol-Myers-Squibb.
Footnotes
Conflict of Interest Statement: The authors have no financial interests or other competing interests to disclose. This study was an investigator initiated study supported by Bristol-Myers-Squibb, Princeton, New Jersey. It was designed by the faculty at Duke University Medical Center. The final responsibility for the manuscript and the decision to submit for publication was made by the principal investigator.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Luo F, Barrett YC, Yang Z, et al. Identification and validation of phospho-Src, a novel and potential pharmacodynamic biomarker for dasatinib, a multi-targeted kinase inhibitor. Cancer Chemother Pharmacol. 2008;62:1065–74. doi: 10.1007/s00280-008-0699-5. [DOI] [PubMed] [Google Scholar]
- 3.Budde RJ, Ke S, Levin VA. Activity of pp60c-Src in 60 different cell lines derived from human tumors. Cancer Biochem Biophys. 1994;14:171–5. [PubMed] [Google Scholar]
- 4.Wiener JR, Windham TC, Estrella VC, et al. Activated Src protein tyrosine kinase is overexpressed in late-stage human ovarian cancers. Gynecol Oncol. 2003;88:73–9. doi: 10.1006/gyno.2002.6851. [DOI] [PubMed] [Google Scholar]
- 5.Dressman HK, Berchuck A, Chan G, et al. An integrated genomic-based approach to individualized treatment of patients with advanced-stage ovarian cancer. J Clin Oncol. 2007;25:517–25. doi: 10.1200/JCO.2006.06.3743. [DOI] [PubMed] [Google Scholar]
- 6.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 7.Pengetnze Y, Steed M, Roby KF, et al. Src tyrosine kinase promotes survival and resistance to chemotherapeutics in a mouse ovarian cancer cell line. Biochem Biophys Res Commun. 2003;309:377–83. doi: 10.1016/j.bbrc.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 8.Chen T, Pengetnze Y, Taylor CC. Src inhibition enhances paclitaxel cytotoxicity in ovarian cancer cells by caspase-9-independent activation of caspase-3. Mol Cancer Ther. 2005;4:217–24. [PubMed] [Google Scholar]
- 9.Han L, Landen CN, Trevino JG, et al. Antiangiogenic and antitumor effects of Src inhibition in ovarian carcinoma. Cancer Res. 2006;66:8633–39. doi: 10.1158/0008-5472.CAN-06-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Reeves K, Luo FR, et al. Identification of candidate predictive and surrogate molecular markers for dasatinib in prostate cancer: rationale for patient selection and efficacy monitoring. Genome Biol. 2007;8:R255. doi: 10.1186/gb-2007-8-11-r255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinberg M. Dasatinib: a tyrosine kinase inhibitor for the treatment of chronic myelogenous leukemia and philadelphia chromosome-positive acute lymphoblastic leukemia. Clin Ther. 2007;29:2289–308. doi: 10.1016/j.clinthera.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Gatza ML, Lucas JE, Barry WT, et al. A Pathway-based classification of human breast cancer. Proc Natl Acad Sci USA. 2010;107:6994–9. doi: 10.1073/pnas.0912708107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 14.Chou T, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 15.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–81. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 16.Konecny GE, Glas R, Dering J, et al. Activity of the multikinase inhibitor dasatinib against ovarian cancer cells. British J of Cancer. 2009;101:1699–708. doi: 10.1038/sj.bjc.6605381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potti A, Dressman HK, Bild A, et al. Genomic signatures to guide the use of chemotherapeutics. Nature Medicine. 2006;12:1294–300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- 18.Demetri GD, Russo PL, MacPhersson IRJ, et al. Phase I dose-escalation and pharmacokinetic studies of dasatinib in patients with advanced solid tumors. Clin Cancer Res. 2009;15:6232–40. doi: 10.1158/1078-0432.CCR-09-0224. [DOI] [PubMed] [Google Scholar]
- 19.Mayer EL, Krop IE. Advances in targeting Src in the treatment of breast cancer and other solid malignancies. Clin Cancer Res. 1020;16:3526–32. doi: 10.1158/1078-0432.CCR-09-1834. [DOI] [PubMed] [Google Scholar]
- 20.Yu EY, Wilding G, Posadas E, et al. Phase II study of dasatinib in patients with metastatic castration-resistant prostate cancer. Clin Cancer Res. 2009;15:7421–28. doi: 10.1158/1078-0432.CCR-09-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.http://www.cancer.gov/search/ResultsClinicalTrials.aspx?protocolsearchid=8245829