Abstract
Dehydroquinate dehydratase (DHQD) catalyzes the third step in the biosynthetic shikimate pathway. We present three crystal structures of the Salmonella enterica type I DHQD which address the functionality of a surface loop that is observed to close over the active site following substrate binding. Two wild type structures with differing loop conformations and kinetic and structural studies of a mutant provide evidence of both direct and indirect mechanisms of loop involvement in substrate binding. In addition to allowing amino acid side chains to establish a direct interaction with the substrate, loop closure necessitates a conformational change of a key active site arginine, which in turn positions the substrate productively. The absence of DHQD in humans and its essentiality in many pathogenic bacteria makes the enzyme a target for the development of nontoxic antimicrobials. The structures and ligand binding insights presented here may inform the design of novel type I DHQD inhibiting molecules.
The seven enzymes of the shikimate pathway catalyze sequential reactions to generate chorismate – a crucial branch point in the synthesis of the aromatic amino acids (1). Due to the essentiality of the shikimate pathway enzymes in a number of organisms and the absence of mammalian shikimate homologs, the shikimate pathway has been regarded as a viable target for the development of novel non-toxic antimicrobials, anti-fungals, and herbicides (2–5).
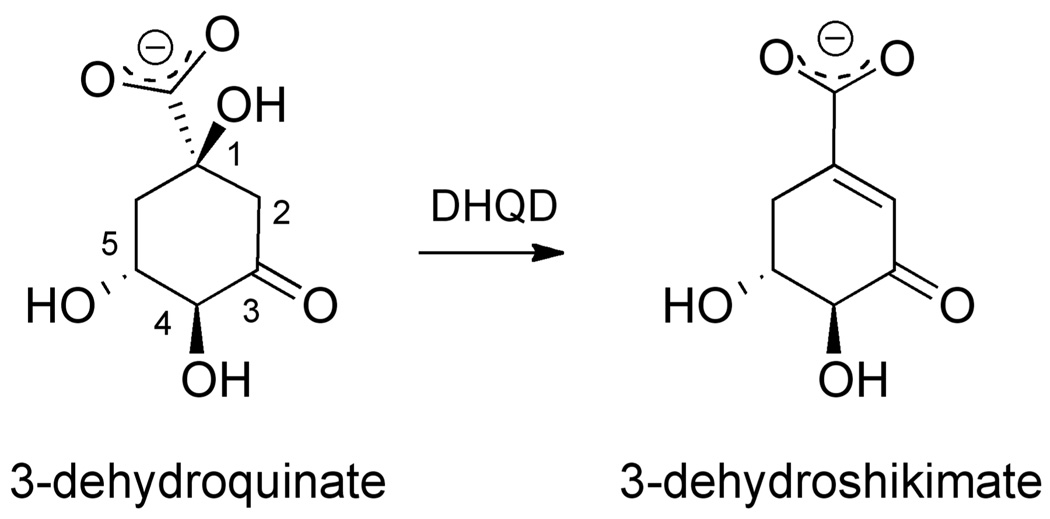
The dehydration of dehydroquinate to dehydroshikimate represents the third step in the shikimate pathway (Figure 1). Biochemical and genetic studies have shown this reaction can be catalyzed by two phylogenetically unrelated enzyme families (6). Type I dehydroquinate dehydratases (DHQDs) are found in plants, fungi, and some bacterial species (7, 8). In bacteria, these ~29 kDa enzymes assemble into homo-dimers and catalyze a reaction which proceeds via a covalent Schiff base intermediate to result in a cis-elimination (9–11). Distinct from type I enzymes, type II DHQDs are found exclusively within bacteria. These ~17 kDa enzymes assemble into homo-dodecamers and catalyze a reaction that proceeds via a non-covalent enolate intermediate to result in a trans-elimination (12–14).
Figure 1.
Reaction catalyzed by DHQD.
We recently reported crystal structures of the Samonella enterica type I DHQD in binary complex with the substrate 3-dehydroquinate in both a non-covalent pre-Schiff base state and a covalent Schiff base bound reaction intermediate state (15). In addition, we analyzed the Clostridium difficile type I DHQD in covalent Schiff base bound complex with the product 3-dehydroshikimate (15). These crystal structures provided crucial insight into the reaction mechanism, suggesting that a single active site histidine cycles through multiple protonation events to catalyze the reaction and the formation/hydrolysis of the Schiff base intermediate (15). Notably, these DHQD complexes consistently display a surface loop in a “closed” conformation that allows it to interact with the substrate/product. Here we report three substrate/product-free structures of S. enterica DHQD that reveal an “open” conformation of this surface loop. Along with kinetic study of loop mutants, these new crystal structures establish two distinct functions for the surface loop – substrate binding and positioning an active site arginine residue – and demonstrate that appropriate loop function is vital to the DHQD catalyzed reaction.
EXPERIMENTAL PROCEDURES
Cloning, expression, and purification of DHQD
Standard Center for the Structural Genomics of Infectious Diseases protocols were used for cloning, over-expression, and purification of S. enterica DHQD (16). In short, S. enterica DHQD was cloned into the MCSG7 expression vector as previously described (15). Following transformation into the BL21 (DE3) E.coli strain, cells were grown for 4 hours at 37° C. At which point, the temperature was reduced to 25° C and protein over-expression induced by the addition of isopropyl-1-thio-D-galactopyranoside to a final concentration of 0.5 mM. After overnight growth, cells were harvested by centrifugation, resuspended in a buffer containing 10 mM Tris-HCl pH 8.3, 500 mM NaCl, 10% glycerol, and 5 mM β-mercaptoethanol and lysed by sonication. DHQD was purified by Ni-NTA affinity chromatography and eluted in a buffer containing 10 mM Tris-HCl pH 8.3, 500 mM NaCl, 5 mM β-mercaptoethanol. Immediately following purification, DHQD was concentrated to 7.5 mg/ml and stored in a buffer containing 0.5 M NaCl and 10 mM Tris-HCl, pH 8.3.
Site-directed mutagenesis
Following the supplier’s technical manual, the QuikChange XL Site-Direct Mutagenesis Kit (Agilent) was used to generate S232A and Q236A variants; successful incorporation of mutations was confirmed by DNA sequencing.
DHQD activity assay
As previously described (15), the DHQD bioactivity assay measured the increase in absorbance at λ = 234 nm to follow the formation of the conjugated enone carboxylate within dehydroshikimate (ε = 12 mM−1cm−1) (17).
Protein crystallization and data collection
Sitting drop crystallization was set up at room temperature using a 1:1 ratio between DHQD to reservoir. Further details regarding conditions of crystal growth is outlined in Table 1. Harvested crystals where transferred to mother liquor before being frozen in liquid nitrogen. Diffraction data were collected at 100° K at the Life Sciences Collaborative Access Team at the Advance Photon Source, Argonne, Illinois.
Table 1.
Crystallization conditions, data collection, and refinement statistics
| Variant | open loop- P212121 | closed loop- P21 | Q236A |
| PDB code | 3L2I | 3OEX | 3O1N |
| Displayed color in figures |
green | yellow | magenta |
| Active Site ligand | none | chloride | chloride |
| Protein buffer solution |
500 mM NaCl 10 mM Tris-HCl pH 8.3 |
500 mM NaCl 10 mM Tris-HCl pH 8.3 |
500 mM NaCl 10 mM Tris-HCl pH 8.3 |
| Crystallization condition |
200 mM MgCl2 20% PEG 3350 |
170 mM NH4OAc pH 4.6 25.5% PEG 4000 15% glycerol |
200 mM MgCl2 100 mM Tris pH 8.5 20% PEG 3350 |
| Space group | P212121 | P21 | P1 |
| Unit Cell dimensions |
|||
| a,b,c (Å) | a=36.92 b=76.91 c=171.85 |
a=69.26 b=75.76 c=99.90 |
a=45.43 b=46.48 c=55.79 |
| α, β, γ (°) | α=90.00 β=90.00 γ=90.00 |
α=90.00 β=102.65 γ=90.00 |
α=104.23 β=97.50 γ=98.70 |
| Resolution (Å) | 28.64-1.85 (1.90-1.85) |
30.00-1.90 (1.93-1.90) |
26.62-1.03 (1.05-1.03) |
| No. reflections | 42787 (3010) | 69261 (3428) | 196964 (8952) |
| Completeness (%) | 99.9 (98.6) | 99.9 (99.7) | 92.7 (84.9) |
| Redundancy | 6.5 (4.5) | 3.3 (2.7) | 4.3 (3.4) |
| Rmerge I | 0.077 (0.355) | 0.085 (0.398) | 0.068 (0.333) |
| I/σ(I) | 21.3 (4.1) | 12.6 (2.8) | 15.0 (3.7) |
| molecules per au | 2 | 4 | 2 |
| No. of atoms | 4572 | 8540 | 4951 |
| Rwork/Rfree | 0.168/0.214 | 0.152/0.204 | 0.140/0.161 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.016 | 0.014 | 0.012 |
| Bond angles (°) | 1.526 | 1.431 | 1.464 |
| Average B-factors | |||
| Protein | 23.1 | 23.2 | 14.2 |
| Waters | 33.3 | 33.1 | 26.8 |
| Ligand | - | 23.5 | 11.6 |
Highest resolution shell in parenthesis
Structure determination and refinement
All structural work was performed by the Center for Structural Genomics of Infectious Diseases (18). Data were processed using HKL-3000 for indexing, integration, and scaling (19). Structures were solved by molecular replacement using Phaser (20). The Salmonella typhi apo DHQD structure (PDB code 1GQN) was used as the starting model for the open loop-P212121 DHQD structure and the open loop-P212121 structure as a starting model for subsequently determined closed loop-P21 and Q236A variant structures. Structures were refined with Refmac (21). Models were displayed in Coot (22) and manually corrected based on electron density maps. All structure figures were prepared using PyMOL Molecular Graphics System, Version 1.3 (Schrödinger, LLC).
RESULTS AND DISCUSSION
Identification of a surface loop that closes over the active site
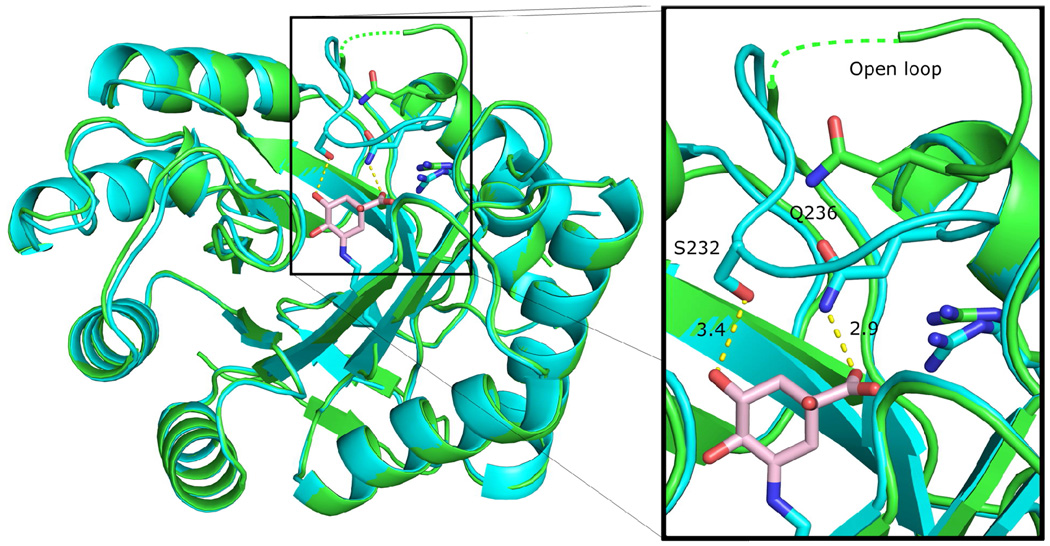
We recently reported crystal structures of the type I DHQD in which the substrate 3-dehydroquinate or the product 3-dehydroshikimate are covalently linked to Lys170 in pre- and post-dehydration Schiff base bound intermediate states (15). To characterize additional functional states of the enzyme we solved an apo state structure of the S. enterica DHQD in the P212121 space group (Table 1, open loop-P212121). A superposition of the apo structure with the previously reported reaction intermediate bound crystal structure shows a mostly identical structural core (Figure 2). However, one significant difference is observed in residues spanning Val228 to Gln236. In the apo structure, these residues are distant from the active site and residues Lys230 to Ser232 could not be modeled because they display poor electron density (Figure S1a). In contrast, in the intermediate bound structure all residues in the region are well ordered (Figure S1b) and are displaced to a position in which they form a side of the active site (Figure 2, inset).
Figure 2.
Dynamic loop behaviour in DHQD. Overlay of open loop-P212121 (green) and pre-dehydration intermediate bound (cyan, PDB code 3M7W) DHQD structures (RMSD = 0.28 Å over 198 Cα atoms, calculated with Val228 to Gln236 residues omitted). A dotted green line traces where the disordered loop residues may lie. Inset highlights differential loop behaviour between the two structures and shows interaction of closed loop residues with the reaction intermediate (shown in pink).
Closure of this loop is likely due to the potential for formation of favorable hydrogen bonding interactions when substrate/product is bound. The reaction intermediate bound structures reveal that in its closed conformation, side chains of loop residues Ser232 and Gln236 are within hydrogen bonding distance of the reaction intermediate’s 5-hydroxyl and 1-carboxyl groups, respectively (Figure 2, inset). The ligand-responsive property of the surface loop observed in these crystal structures is consistent with previously reported type I DHQD structures from Clostridium difficile (15), Salmonella typhi (10, 23), Staph aureus (24), Aquifex aeolicus (PDB code 2YSW), Geobacillus kaustophilus (PDB code 2YR1), and Streptococcus pyogenes (PDB code 2OCZ), which display a similar open and/or disordered apo state and closed and ordered ligand bound loop behavior. Similar behavior of this loop in multiple type I DHQDs suggests that inducible loop closure may be a general feature of substrate binding in these enzymes.
Crystal structure with non-standard loop behavior
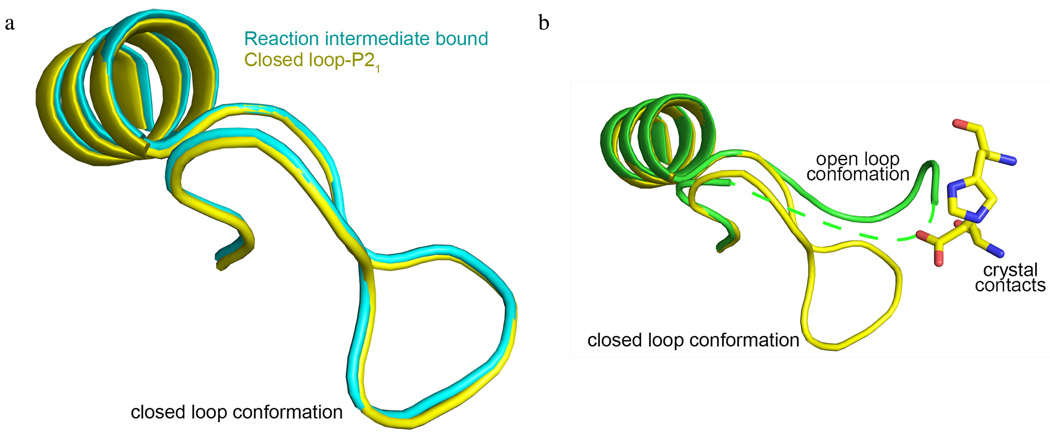
We solved a second DHQD structure in the absence of biological ligand. Contrary to the open loop-P212121 crystal, this crystal adopted the P21 space group (Table 1, closed loop-P21). Surprisingly, the behavior of the surface loop differs between these two crystal forms. Similar to the reaction intermediate bound structures, all four molecules within the P21 asymmetric unit display a closed loop conformation (Figures S1c & 3a). An overlay of these two structures reveals that in the P21 space group the open loop conformation would clash with residues from a crystallographic symmetry mate (Figure 3b). This suggests that adoption of the closed loop conformation may reflect constraints imposed by crystal packing rather than an accurate representation of solution state loop behavior in the absence of ligand.
Figure 3.
The closed loop conformation in the closed loop-P21 structure likely results from crystal packing. (a) Superposition of pre-dehydration intermediate bound (cyan) and closed loop-P21 (yellow) structures. The reaction intermediate is shown in pink. (b) Superposition of the closed loop-P212121 (yellow) and open loop-P212121 (green) structures. Residues His51 and Glu89 of a symmetry related molecule are proximal to the closed loop and would clash with an open loop conformation. A dotted green line traces where the disordered residues may lie.
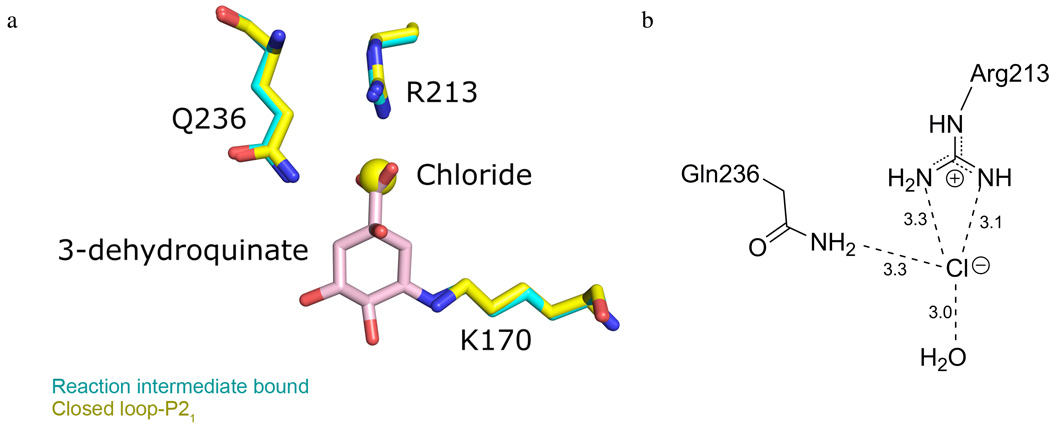
Interestingly, within the closed loop-P21 structure a chloride from the crystallization buffer is observed at the enzyme’s active site (Figure 4). An overlay of the closed loop-P21 structure with the previously reported pre-dehydration reaction intermediate bound structure reveals that the chloride is bound in precisely the same position as the reaction intermediate’s 1-carboxyl group (Figure 4). This observation provides a structural rationale for the previous report of this anion acting as a competitive inhibitor of type I DHQDs, (17) which was also found to be true of the S. enterica DHQD (data not shown). The absence of ordered chloride in other DHQD crystal structures, including our open loop-P212121 structure, despite crystallization conditions containing similarly high concentrations of the anion, suggests that chloride binding may be loop conformation dependent – i.e., the anion may preferentially bind the closed loop state of the protein. This apparent requirement of loop closure for chloride binding, in combination with the similar localization of chloride and 1-carboxyl group, suggests that a functional role of loop closure may be in coordinating the substrate’s anionic 1-carboxyl group to bind the substrate in a conformation conducive to catalysis.
Figure 4.
Chloride binding within the closed loop-P21 active site. (a) Superposition of closed loop- P21 (yellow) and pre-dehydration reaction intermediate bound (cyan) structures. The chloride and 1-carboxyl group localize to the same position within the active site. The reaction intermediate is shown in pink. (b) A schematic representation of the closed loop-P21 active site shows key chloride interactions.
Catalytic deficiencies of S232A and Q236A loop mutants
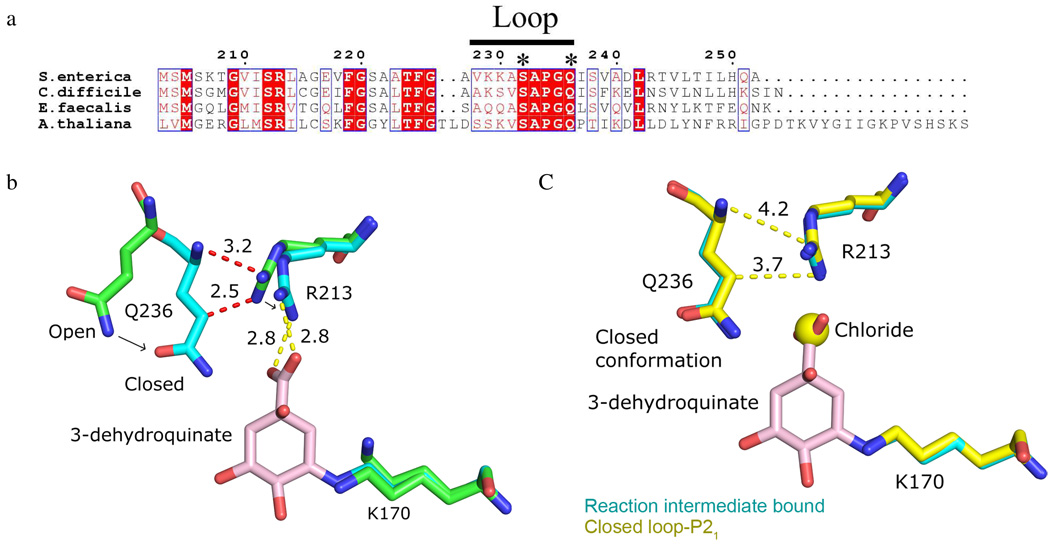
Alignment of multiple DHQD surface loop sequences from distantly related organisms reveals a five residue stretch of complete conservation (Figure 5a), implying a functionally significant role to this part of the loop. Notably, the two residues within this conserved stretch, Ser232 and Gln236, directly interact with the substrate/product (Figure 2, inset). To probe the function of these loop residues we generated the Ser232 → Ala (S232A) and Gln236 → Ala (Q236A) mutants. The dehydration activity of wild type (WT) and mutant enzymes was assayed and Michaelis-Menten kinetics determined (Table 2). Compared to WT enzyme both the S232A and Q236A variants have attenuated activity, which is reflected in both a decreased kcat and an increased Km (Table 2). While the S232A mutation results in a moderate 50-fold decrease in kcat/Km, the Q236A mutation results in a more severe 1000-fold decrease (Table 2).
Figure 5.
Loop closure results in a conformational change of Arg213. (a) Sequence alignment of the type I DHQDs from S. enterica, C. difficile, E. faecalis, and A. thaliana done in ClustalW2 version 2 using ESPript version 2.2. Asterisks denote S. enterica Ser232 and Gln236, which are observed to hydrogen bond with the reaction intermediate. (b) Superposition of open loop-P212121 (green) and pre-dehydration reaction intermediate bound (cyan) active sites. The reaction intermediate bound conformation of Arg213 positions the residue to form a bidentate salt bridge (yellow dashes) with the reaction intermediate’s (shown in pink) 1-carboxyl group. Distances that would be unusually short between the open loop-P212121 conformation of Arg213 and reaction intermediate bound conformation of Gln236 (red dashes) are shown in angstroms. Arrows denote the direction of Gln236 and Arg213 movement as the loop transitions from open to closed states. (c) Superposition of pre-dehydration reaction intermediate (cyan) and closed loop- P21 (yellow) structures. Arg213 adopts identical conformations in each structure.
Table 2.
Kinetic characterization of wild type, S232A, and Q236A DHQD
| kcat (sec−1) | Km (µM) | kcat/Km (sec−1µM−1) | |
| Wild type | 210 ± 5.1 | 21 ± 2.9 | 10 |
| S232A | 107 ± 7.4 | 548 ± 85 | 0.20 |
| Q236A | 6.9 ± 0.35 | 682 ± 204 | 0.01 |
The crystallographically-observed interactions between Ser232 and Gln236 with the substrate/product indicate that both residues have a direct role in substrate binding. Since both Ser232 and Gln236 side chains make a single interaction with the substrate, the residues might be expected to be of similar functional importance. However, the 20-fold lower efficiency of the Q236A mutant versus the S232A mutant implies a more critical role for Gln236 than Ser232 in the reaction and suggests that Gln236 may have a more extensive role than merely binding the substrate.
Loop closure induces Arg213 conformational change
Having identified a differential effect of the S232A and Q236A mutations on the enzyme’s kinetics we set out to identify the source of Ser232 and Gln236’s differing functionalities. A comparison of apo and ligand bound structures reveals that the transition from apo to reaction intermediate bound states results in a conformational change of the Arg213 side chain (Figure 5b). This change is likely to be functionally significant because the residue moves from a position in the apo structure where its guanidinium group points away from the active site to a position in the reaction intermediate bound structure where the guanidinium group forms a bidentate salt bridge with the reaction intermediate's 1-carboxyl group (Figure 5b).
Two features of Arg213’s local environment differ between the apo and reaction intermediate bound states and thus one or both are likely responsible for inducing the change in Arg213 conformation. First, the presence of the reaction intermediate differs between the two structures. As such, substrate binding may bias Arg213 to a conformation in which the favorable interaction between its guanidinium and the substrate’s 1-carboxyl group can form. Second, the loop conformation differs within the two structures. With the loop adopting its closed conformation, the apo conformation of Arg213 would be unusually close to the loop residue Gln236 (Figure 5b). Thus, loop closure could produce an unfavorable Gln236/Arg213 clash if Arg213 did not change conformations. This latter possibility is particularly intriguing because, with Gln236 involvement, it could explain the unexpectedly large effect of the Q236A mutation on the enzyme’s kinetic parameters.
Because it isolates the effect of loop closure, the closed loop-P21 structure allows for deconvolution of the relative influence of the two forces, substrate binding and loop closure, acting on Arg213. An analysis of this structure reveals that despite the absence of biological ligand, when the loop is closed, Arg213 adopts its reaction intermediate bound conformation (Figure 5c). Thus, even when lacking the potential for formation of the guanidinium-carboxyl interaction, loop closure is sufficient to cause the Arg213 conformational change. This finding suggests that loop closure, and specifically the side chain of Gln236, may be critical for inducing the change in Arg213 conformation.
Loop mutant fails to induce Arg213 conformational change
A limitation in basing interpretations of the effect of loop closure on Arg213 conformation on the closed loop-P21 structure results from the presence of chloride within this structure’s active site. It is possible that the chloride has a compensatory effect for the absence of the 1-carboxyl group. In which case, it may be the presence of this anion rather than loop closure which is directly responsible for the change in Arg213 conformation. To address this possibility and to further probe the functional role of Gln236, the Q236A variant enzyme was subjected to crystallographic study.
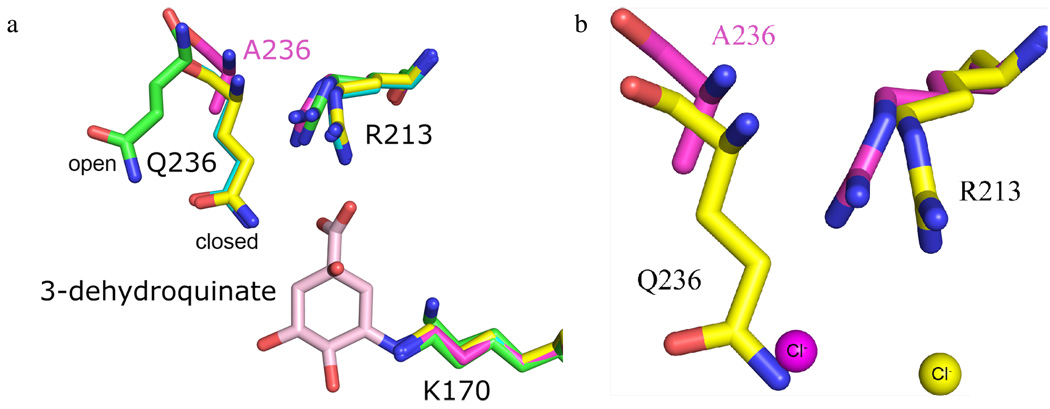
Interestingly, the Q236A enzyme crystallized in a new crystal form which diffracted to a significantly higher resolution than any of the WT crystals (Table 1, Q236A variant). Analysis of this structure reveals that it behaves like a hybrid of the two WT structures. One molecule in this structure’s asymmetric unit is similar to the WT open loop-P212121 structure and displays an open and partially disordered loop conformation (Figure S1d). The other molecule within the asymmetric unit is similar to the WT closed loop-P21 structure and, despite lacking the presence of biological ligand, adopts a closed loop conformation (Figure S1e). An examination of the mutant protein’s crystal packing reveals that the open conformation of the closed loop is disallowed by precisely the same crystal packing as the WT closed loop-P21 structure (Figure S2). Also similar to behavior of the WT enzyme, the presence of chloride is observed within the asymmetric unit’s closed-loop but not open-loop molecule (Figure 6a). This correlation between loop conformation and active site chloride occupancy within a single crystal provides compelling evidence that chloride binding is loop conformation dependent, providing further support for loop closure playing a key role in anion coordination. Despite co-crystallization and crystal soaking attempts, no biological ligand was observed in the Q236A variant active site, a result consistent with the mutant enzyme’s increased Km (Table 2).
Figure 6.
The Q236A variant enzyme fails to induce a conformational change of Arg213 and has altered anion coordinating properties. (a) Superposition of open loop-P212121 (green), pre-dehydration reaction intermediate bound (cyan), closed loop-P21 (yellow), and closed loop-Q236A (magenta) active sites. The reaction intermediate is shown in pink. In both closed loop WT structures Arg213 adopts its ligand bound conformation. In the Q236A variant structure Arg213 remains in its apo conformation. (b) Superposition of the closed loop-P21 (yellow) and closed loop-Q236A (magenta) active sites. Corresponding to the altered Arg213 conformation, the chloride is displaced 3.0 Å within the Q236A active site relative to its position in the WT enzyme.
An overlay of the closed loop-Q236A molecule with WT structures reveals that, despite loop closure, Arg213 remains in its apo conformation in the Q236A variant enzyme (Figure 6b). This finding allows the impact of Gln236 on Arg213 conformation to be deconvoluted from that of chloride. Because the Q236A variant enzyme is capable of binding chloride in its apo conformation, the possibility that the Arg213 conformational change observed in the WT closed loop-P21 structure is a consequence of the chloride binding can be ruled out. Having eliminated this confounding variable, it can be concluded that the differing Arg213 conformation between WT open loop-P212121 and closed loop-P21 structures is the direct result of loop closure.
Furthermore, the difference in Arg213 conformation between WT closed loop-P21 and closed loop-Q236A structures demonstrates that Gln236 is the critical determinant responsible for loop closure’s induction of the Arg213 conformational change. Without the side chain of Gln236, loop closure no longer results in a conformational change of Arg213. Thus, analysis of this structure establishes that: 1) loop closure results in Arg213 adopting its substrate binding conformation and, 2) that Gln236 is the sole residue responsible for this conformational change.
The functional significance of the Q236A variant enzyme’s inability to induce a conformational change of Arg213 is suggested by the behavior of the bound chloride anion. In the WT enzyme the anion is coordinated by Arg213 and is positioned precisely where the substrate’s 1-carboxyl group is found in the reaction intermediate bound structures (Figure 4b). The similar localization of 1-carboxyl group and chloride argues that Arg213 is likely to be functionally relevant for coordinating the 1-carboxyl group in the substrate binding event. In the closed loop-Q236A structure, chloride is still coordinated by Arg213 but with the residue adopting its apo conformation the anion is displaced ~3 Å (Figure 6c). Thus, without Gln236 inducing a change in Arg213 conformation the position of anion coordination shifts from the position of 1-carboxyl group binding to a position removed from the enzyme’s catalytic center. Notably, if the 1-carboxyl group were to bind in the observed chloride position, then the substrate’s 3-carbonyl carbon would be too far removed from the Schiff base forming Lys170 for formation of the covalent reaction intermediate to occur. As such, an inappropriate coordination of the carboxyl group to this position should greatly hinder enzyme activity.
In light of the evidence presented here, we propose that loop closure has two functions. First, as evidenced by the ligand bound crystal structures and kinetic study of the S232A variant enzyme, loop residues directly interact with the substrate in manner which facilitates substrate binding. Second, as demonstrated by structural and kinetic studies of the Q236A variant enzyme, loop closure and specifically Gln236 facilitate a conformational change of Arg213 which positions the residue to form a key interaction with the substrate’s 1-carboxyl group. Thus, loop closure acts by both a direct and an indirect mechanism to facilitate substrate binding.
CONCLUSIONS
We present three crystal structures of the type I S. enterica DHQD that address the function of a surface loop which is observed to close over the active site. An exception to the typical open and disordered apo state loop behavior, the first structure in which the loop adopts a closed conformation in the absence of substrate/product is presented. This structure demonstrates that loop closure results in a conformational change in the substrate binding residue Arg213. Kinetic studies of two mutants reveal that the loop is functionally important for catalysis. A crystal structure of the Q236A mutant enzyme demonstrates that Gln236 is required for loop closure to induce the Arg213 conformational change. A consequence of Arg213 forgoing its conformational change in this mutant is a ~3 Å shift in the position of chloride localization. The fact that the bound anion moves from the position of substrate carboxyl group binding to a position removed from the enzyme’s catalytic center argues that Arg213’s conformational change is critical for binding the 1-carboxyl group so as to position the substrate in the catalytically favored orientation. Based on the data presented here we propose that the loop residues influence substrate binding by two separate mechanisms. Closure of the loop facilitates substrate binding by directly interacting with the substrate and by inducing a conformational change of Arg213 to its substrate binding position.
As a requisite step in aromatic amino acid biosynthesis DHQD is a target for antimicrobial enzyme inhibitors. The findings presented here provide insight that may aid in targeted design of novel DHQD inhibitors. The attenuation in enzymatic activity resulting from compromised loop function validate an inhibition strategy based upon the prevention of loop closure, suggesting that a molecule capable of stabilizing the open state of the loop or destabilizing the closed state may be an effective allosteric DHQD inhibitor.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Elisabetta Sabini for facilitating communication between groups involved in this work.
Abbreviations
- DHQD
dehydroquinate dehydratase
- Q236A
glutamine-236 → alanine DHQD variant
- S232A
serine-232 → alanine DHQD variant
- PDB
Protein Data Bank
- WT
wild type
Footnotes
Accession numbers: Protein Data Bank codes 3L2I (open loop-P212121), 3OEX (closed loop-P21), 3O1N (Q236A variant)
The Center for Structural Genomics of Infectious Diseases has been funded in whole or in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract no. HHSN272200700058C (to W.F.A.). Use of the Advanced Photon Source at Argonne National Laboratory was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported in part by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817).
SUPPORTING INFORMATION AVAILABLE
The model and associated electron density for the DHQD loop region from the structures discussed is shown in Supplemental Figure 1. Superposition of the open loop and closed loop apo structures is shown in Supplemental Figure 2. This material is available free of charge via the Internet at http://pubs.acs.org.”
REFERENCES
- 1.Bentley R. The shikimate pathway--a metabolic tree with many branches. Crit Rev Biochem Mol Biol. 1990;25:307–384. doi: 10.3109/10409239009090615. [DOI] [PubMed] [Google Scholar]
- 2.Benowitz AB, Hoover Jennifer L, Payne David J. Antibacterial Drug Discovery in the Age of Resistance. Microbe. 2010;5:390–396. [Google Scholar]
- 3.Microbe GM, Shah DM. Amino-Acid Biosynthesis Inhibitors as Herbicides. Annu Rev Biochem. 1988;57:627–663. doi: 10.1146/annurev.bi.57.070188.003211. [DOI] [PubMed] [Google Scholar]
- 4.Marques MR, Pereira JH, Oliveira JS, Basso LA, de Azevedo WF, Santos DS, Palma MS. The inhibition of 5-enolpyruvylshikimate-3-phosphate synthase as a model for development of novel antimicrobials. Curr Drug Targets. 2007;8:445–457. doi: 10.2174/138945007780058951. [DOI] [PubMed] [Google Scholar]
- 5.Noble M, Sinha Y, Kolupaev A, Demin O, Earnshaw D, Tobin F, West J, Martin JD, Qiu CY, Liu WS, DeWolf WE, Tew D, Goryanin II. The kinetic model of the shikimate pathway as a tool to optimize enzyme assays for high-throughput screening. Biotechnol Bioeng. 2006;95:560–573. doi: 10.1002/bit.20772. [DOI] [PubMed] [Google Scholar]
- 6.Kleanthous C, Deka R, Davis K, Kelly SM, Cooper A, Harding SE, Price NC, Hawkins AR, Coggins JR. A comparison of the enzymological and biophysical properties of two distinct classes of dehydroquinase enzymes. Biochem J. 1992;282(Pt 3):687–695. doi: 10.1042/bj2820687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lumsden J, Coggins JR. Subunit Structure of Arom Multienzyme Complex of Neurospora-Crassa - Possible Pentafunctional Polypeptide-Chain. Biochemical Journal. 1977;161:599–607. doi: 10.1042/bj1610599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polley LD. Purification and Characterization of 3-Dehydroquinate Hydrolase and Shikimate Oxidoreductase - Evidence for a Bifunctional Enzyme. Biochim Biophys Acta. 1978;526:259–266. doi: 10.1016/0005-2744(78)90310-8. [DOI] [PubMed] [Google Scholar]
- 9.Butler JR, Alworth WL, Nugent MJ. Mechanism of Dehydroquinase Catalyzed Dehydration .1. Formation of a Schiff-Base Intermediate. J Am Chem Soc. 1974;96:1617–1618. [Google Scholar]
- 10.Gourley DG, Shrive AK, Polikarpov I, Krell T, Coggins JR, Hawkins AR, Isaacs NW, Sawyer L. The two types of 3-dehydroquinase have distinct structures but catalyze the same overall reaction. Nat Struct Biol. 1999;6:521–525. doi: 10.1038/9287. [DOI] [PubMed] [Google Scholar]
- 11.Hanson KR, Rose IA. Absolute Stereochemical Course of Citric Acid Biosynthesis. P Natl Acad Sci USA. 1963;50:981–988. doi: 10.1073/pnas.50.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price NC, Boam DJ, Kelly SM, Duncan D, Krell T, Gourley DG, Coggins JR, Virden R, Hawkins AR. The folding and assembly of the dodecameric type II dehydroquinases. Biochemical Journal. 1999;338:195–202. [PMC free article] [PubMed] [Google Scholar]
- 13.Harris JM, Gonzalez-Bello C, Kleanthous C, Hawkins AR, Coggins JR, Abell C. Evidence from kinetic isotope studies for an enolate intermediate in the mechanism of type II dehydroquinases. Biochem J. 1996;319(Pt 2):333–336. doi: 10.1042/bj3190333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker EJ, Bello CG, Coggins JR, Hawkins AR, Abell C. Mechanistic studies on type I and type II dehydroquinase with (6R)- and (6S)-6-fluoro-3-dehydroquinic acids. Bioorg Med Chem Lett. 2000;10:231–234. doi: 10.1016/s0960-894x(99)00660-5. [DOI] [PubMed] [Google Scholar]
- 15.Light SH, Minasov G, Shuvalova L, Duban M, Caffrey M, Anderson WF, Lavie A. Insights into the mechanism of type I dehydroquinate dehydratases from structures of reaction intermediates. J Biol Chem. 2011;286:3531–3539. doi: 10.1074/jbc.M110.192831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim Y, Bigelow L, Borovilos M, Dementieva I, Duggan E, Eschenfeldt W, Hatzos C, Joachimiak G, Li H, Maltseva N, Mulligan R, Quartey P, Sather A, Stols L, Volkart L, Wu RY, Zhou M, Joachimiak A. High-Throughput Protein Purification for X-Ray Crystallography and Nmr. Adv Protein Chem. 2008;75:85–105. doi: 10.1016/S0065-3233(07)75003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaudhuri S, Lambert JM, Mccoll LA, Coggins JR. Purification and Characterization of 3-Dehydroquinase from Escherichia-Coli. Biochemical Journal. 1986;239:699–704. doi: 10.1042/bj2390699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson WF. Structural genomics and drug discovery for infectious diseases. Infect Disord Drug Targets. 2009;9:507–517. doi: 10.2174/187152609789105713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 21.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Lee WH, Perles LA, Nagem RA, Shrive AK, Hawkins A, Sawyer L, Polikarpov I. Comparison of different crystal forms of 3-dehydroquinase from Salmonella typhi and its implication for the enzyme activity. Acta Crystallogr D Biol Crystallogr. 2002;58:798–804. doi: 10.1107/s0907444902003918. [DOI] [PubMed] [Google Scholar]
- 24.Nichols CE, Lockyer M, Hawkins AR, Stammers DK. Crystal structures of Staphylococcus aureus type I dehydroquinase from enzyme turnover experiments. Proteins. 2004;56:625–628. doi: 10.1002/prot.20165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.