Abstract
BCAR3 binds to the carboxy-terminus of p130Cas, a focal adhesion adapter protein. Both BCAR3 and p130Cas have been linked to resistance to anti-estrogens in breast cancer, Rac activation and cell motility. Using R743A BCAR3, a point mutant that has lost the ability to bind p130Cas, we find that BCAR3-p130Cas complex formation is not required for BCAR3-mediated anti-estrogen resistance, Rac activation or discohesion of epithelial breast cancer cells. Complex formation was also not required for BCAR3-induced lamellipodia formation in BALB/c-3T3 fibroblasts but was required for optimal BCAR3-induced motility. Although both wildtype and R743A BCAR3 induced phosphorylation of p130Cas and the related adapter protein HEF1/NEDD9, chimeric NSP3:BCAR3 experiments demonstrate that such phosphorylation does not correlate with BCAR3-induced anti-estrogen resistance or lamellipodia formation. Wildtype but not R743A BCAR3 induced lamellipodia formation and augmented cell motility in p130Cas-/- murine embryonic fibroblasts (MEFs), suggesting that while p130Cas itself is not strictly required for these endpoints, complex formation with other CAS family members is, at least in cells lacking p130Cas. Overall, our work suggests that many, but not all, BCAR3-mediated signaling events in epithelial and mesenchymal cells are independent of p130Cas association. These studies also indicate that disruption of the BCAR3-p130Cas complex is unlikely to reverse BCAR3-mediated anti-estrogen resistance.
Keywords: BCAR3, p130Cas, breast cancer, anti-estrogen resistance, Rac activation, cell motility
1. Introduction
The interaction of integrin receptors with extracellular matrix proteins regulates both the ability of breast cancer cells to grow in the presence of anti-estrogens and their invasive properties. Breast tumors that develop resistance to anti-estrogen therapy often simultaneously acquire a more aggressive and metastatic phenotype. In a cell-line model of anti-estrogen resistance, Dorssers and colleagues identified BCAR1 (p130Cas) and BCAR3 as genes whose over-expression induce estrogen independent growth in normally estrogen-dependent cell lines [1, 2]. p130Cas is a focal adhesion adapter protein best known for its ability to convert integrin receptor signals to Src family kinase-mediated tyrosine phosphorylation of the p130Cas “substrate domain” [3, 4]. Such p130Cas phosphorylation in turn leads to recruitment of Crk, and the atypical Rac GDP exchange factor DOCK180, with resultant Rac activation and augmented cell motility [5, 6]. While elimination of p130Cas expression in mice by homologous recombination results in embryonic lethality due to a cardiac phenotype, p130Cas-/- murine embryonic fibroblasts (MEFs) are notable both for defects in cell motility and Src-mediated transformation [7]. In keeping with such cell line studies, Dorssers and colleagues have reported that high-level expression of p130Cas in primary breast tumors confers an increased likelihood of lack of response to hormonal therapy, early disease progression and shorter overall survival [8-10].
Although BCAR3 and p130Cas were identified quite independently as potential “anti-estrogen resistance” genes in the studies cited above, contemporaneous studies of the murine homologue of BCAR3, AND-34, demonstrated that these two proteins normally can be found in cells as part of a complex, with the carboxy-terminus of AND-34 bound to the carboxy-terminus of p130Cas [11, 12]. AND-34 (both human and murine proteins will henceforth be referred to as BCAR3) is a 95 kDa protein with an amino-terminal SH2 domain and a carboxy-terminal domain with sequence homology to the cdc25 Ras subfamily GDP exchange factor catalytic domain. Both BCAR3 and p130Cas are members of gene families. In humans, there are three BCAR3-like proteins: NSP1, NSP2/BCAR3 and NSP3/SHEP1/CHAT and four p130Cas-like proteins: p130Cas, HEF1/NEDD9/Cas-L, Efs/Sin and HEPL [4]. BCAR3 also forms a complex in lymphoid cells with endogenous HEF1 [13]. Among NSP family members, over-expression of only BCAR3 induces significant anti-estrogen resistance [14].
Consistent with the fact that BCAR3 binds to an adapter protein known to facilitate integrin-mediated signaling and motility, BCAR3 levels regulate breast cancer cell-line morphology and cell motility. Despite the Ras subfamily GEF-like domain, initial studies in murine fibroblasts stably transfected with BCAR3 revealed lamellipodia and ruffling characteristic of Rac GTPase signaling [15]. Subsequent “pulldown” studies confirmed that BCAR3 expression resulted in robust activation of both Rac and Cdc42 GTPases, most likely as an indirect result of PI3K activation [13, 15, 16]. Over-expression of BCAR3 in ERα-positive epithelial breast cancer cell lines that normally have quite low basal levels of BCAR3 expression results in conversion of cells to a mesenchymal-like growth pattern, with a loss of cell cohesiveness and a loss of cadherin-mediated homotypic adhesion at cell borders [14]. In contrast, siRNA-mediated down-regulation of BCAR3 in mesenchymal ERα-negative breast cancer cell lines that normally express substantial levels of BCAR3 results in conversion to an epithelioid growth pattern, with cohesive apposition of cells to each other [17]. Concordantly, studies by Bouton and colleagues have shown that the over-expression of BCAR3 in epithelial breast cancer cell lines induces cell motility, while the down-regulation of BCAR3 in mesenchymal breast cancer cell lines attenuates migration and invasion [18].
Given that BCAR3 binds to p130Cas and that p130Cas is an important integrator of adhesion-related signaling, it seems reasonable to hypothesize that BCAR3 expression regulates cell morphology and motility as a direct result of its ability to modulate p130Cas-mediated signaling. Consistent with such a hypothesis, prior experiments with BCAR3 deletion constructs lacking the GEF-like domain (BCAR3 ΔGEF) required for association with p130Cas have demonstrated both a loss of BCAR3-mediated Rac activation and BCAR3-induced cell motility [15, 18]. Interestingly, however, not all BCAR3-associated signaling requires formation of a BCAR3-p130Cas complex. Recent phosphoamino-acid analysis studies have shown that BCAR3 expression induces serine phosphorylation of p130Cas, characterized by a reduction in p130Cas migration on low bis-acrylamide PAGE gels [17]. Among three sites of p130Cas serine phosphorylation identified by mass spectrometry, mutation of serine 437 (rat p130Cas) to alanine reduced such a BCAR3-induced shift in p130Cas PAGE migration. Such BCAR3-mediated p130Cas phosphorylation is adhesion and actin microfilament-dependent and temporally distinct from previously well-characterized rapid FAK and Src-kinase-mediated p130Cas tyrosine phosphorylation. BCAR3-mediated p130Cas phosphorylation requires the amino-terminal BCAR3 SH2 domain but occurs in the absence of the GEF-like domain and thus is independent of BCAR3-p130Cas complex formation.
The association of both BCAR3 and p130Cas over-expression with breast cancer anti-estrogen resistance has suggested that disrupting the BCAR3-p130Cas complex might be an important novel approach to restoring sensitivity to anti-estrogens. Small-angle X-ray scattering studies of the complex of BCAR3 with the CAS family member HEF1 revealed that the GEF-like domain of BCAR3, which folds similarly to a Cdc25-like GEF domain, binds tightly to helix 2 of a four-helix bundle carboxy-terminal binding domain in HEF1 [19]. As predicted by these structural data, mutation of arginine 743 of BCAR3 to alanine eliminated BCAR3 association with HEF1 and, as shown in the current study, p130Cas. As deletion of BCAR3’s GEF-like domain could have effects on BCAR3-associated signaling quite independent of BCAR3’s ability to form a complex with p130Cas, we here have utilized the R743A BCAR3 mutant to more directly examine the importance of BCAR3- p130Cas complex formation in BCAR3-associated anti-estrogen resistance, cytoskeletal changes, Rac activation and cell motility. Finally, we have addressed the role of p130Cas itself in BCAR3-associated lamellipodia formation and motility using p130Cas-/- MEFs.
2. Materials and Methods
2.1 Cell lines
MCF-7 human breast cancer cell lines were obtained from ATCC. The HA-BCAR3 stably transfected MCF-7 cell line II-6 has been previously described [14]. BALB/c-3T3 murine fibroblasts were a gift of Dr. Douglas Faller (Cancer Center, BUSM). MCF-7 and BALB/c-3T3 lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Mediatech, Inc.) supplemented with 10% heat-inactivated fetal calf serum (Biomeda), 2.2 mM L-glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin. p130Cas-/- mouse embryonic fibroblasts (MEFs), a gift from Dr. Kathrin Kirsch (Biochemistry, BUSM), were originally created by Honda et al [7]. p130Cas-/- MEFs were cultured as above except for the use of 5% calf serum (Thermo Scientific HyClone).
2.2 Lentiviral expression constructs
The majority of the HA-tagged BCAR3 and NSP3 constructs utilized in these studies have been previously described as pcDNA3 plasmids [14]. Inserts from these constructs were subcloned into the the bicistronic lentivirus expression plasmid pHAGE6-wtCMV-DsRedExpress-BrCr1-ZsGreen-W using the NotI and BamHI sites in the phage [20]. PCR (Ultra Pfu Fusion, Stratagene) was used to generate inserts with 5′ NotI and 3′ BclI. These clones lose the DsRed fluor, but retain ZsGreen and, therefore, infected cells can be sorted. The HA-tagged BCAR3 neomycin-selectable pCDNA3 expression vector was mutated to encode alanine rather than arginine at codon 743 by site-directed mutagenesis using the QuikChange II kit (Stratagene) and was cloned by the same strategy into the phage 6 vector [14]. Chimeric NSP3:BCAR3, containing the AND-34 GEF-like domain, was generated by an overlapping PCR strategy. The initial amino acid position of the GEF-like domain in AND-34 is 538 and in NSP3 424. A previously described plasmid-based HA-tagged rat p130Cas construct was subcloned by the same PCR strategy (using NotI-BamHI) into the lentiviral vector pHAGE2-FullEF1a-ZsGreen-IRES-dTomatoW [12]. This results in a construct that loses ZsGreen but retains dTomato fluor and therefore cells transduced with this construct may be sorted.
2.3 Lentiviral Transduction
HEK-293T cells in 10 cm plates at 50-70% confluency were transfected with lentiviral vector constructs plus packaging vectors using Fugene 6 as described by the manufacturer (Roche). After two days culture, media containing packaged phage particles was harvested and replenished with new media every 12 hours for three cycles. Virion particles were concentrated by centrifugation of the total collected filtered supernatants using an SW28 Beckman rotor at 1.5 hrs at 16.5k rpm. For lentiviral transduction, 50 ml of the concentrated lentiviral supernatants was added with polybrene (final concentration 8 mg/mL) to MCF-7, BALB/c-3T3 or p130Cas-/- MEFs cultured in 6-well plates, followed by centrifugation of the plates at 800g for 45 minutes. After at least 2 days of tissue culture, the transduced cells were trypsinized and resuspended in DMEM with 3% FCS for MoFlo sorting. Typically between 100,000 to 200,000 cells were collected. Expression was confirmed by western analysis as assayed for the HA epitope tag.
2.4 Transient Transfection, Cell Lysis, Immunoprecipitation and Phosphatase Treatment
MCF-7 cells were transiently co-transfected with plasmids encoding HEF1 and HA-tagged NSP proteins (NSP1, BCAR3 and NSP3) using Lipofectamine 2000 (Invitrogen) as previously described [13, 14]. Adherent cells were plated on 10 cm2 tissue culture dishes, grown to approximately 80-90% confluence and washed with 10 ml of phosphate buffered saline (PBS). Cell monolayers in each dish were lysed in 0.5 ml of NP40 lysis buffer (1% NP40, 1X PBS, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, and protease inhibitors). Cell lysates were centrifuged in a microcentrifuge for 30 minutes at 14,000 rpm (relative centrifugal force 16,000) at 4°C and the supernatants were transferred to clean tubes, and protein concentrations of the lysates were determined using the BCA protein assay (Bio-Rad). Whole cell lysate samples were prepared by denaturing the protein in Laemlli SDS-PAGE sample buffer at 100°C for 5 minutes. For immunoprecipitations, cell lysates were adjusted for equivalent protein and volume. Portions of the adjusted cell lysates were saved as pre-immunoprecipitation references. 500 μg portions of the remaining cell lysates were used for immunoprecipitation by adding 10-20 μl bead volumes of protein G-Sepharose beads (GE Healthcare) and a previously described peptide affinity-purified polyclonal rabbit anti-BCAR3 antisera or a mouse monoclonal anti-p130Cas antibody (BD) [15]. Immuno-complexes bound to protein G-Sepharose beads were recovered by brief centrifugation after mixing cell lysate slurries by end-over-end rotation for 3 hours at 4°C. Pelleted beads were washed three times in lysis buffer. Whole cell lysates and immunoprecipitations were suspended in Laemlli SDS-PAGE sample buffer and heat-denatured for 5 minutes at 100 degrees. Lambda protein phosphatase (40 U/mL, New England Biolabs) treatment of HEF1 immunoprecipitates was carried out as previously described [17].
2.5 SDS-PAGE and Western Analysis
Whole cell lysates and immunoprecipitates in Laemlli SDS-PAGE sample buffer were electrophoretically separated in SDS low-bis polyacrylamide gels (30% T, 0.67% C). Western transfer of proteins from gels onto polyvinylidene fluoride (PVDF; Millipore) was performed in transfer buffer containing 10% methanol. Dried membranes were briefly pre-wetted with methanol, equilibrated with Tris-buffered saline with TWEEN (TBST; 25 mM Tris-HCl pH 7.6, 130 mM sodium chloride, 2.5 mM potassium chloride, 0.1% TWEEN-20), and blocked with TBST containing 5% non-fat milk for 30 minutes prior to immunoblotting with primary antibodies: mouse monoclonal anti-HA (Covance), mouse monoclonal anti-p130Cas (BD), mouse monoclonal anti-HEF1 2G9 (Cell Signaling), anti-tubulin (Abcam). Primary antibodies diluted with either TBST containing 5% milk. Membranes were washed with TBST and immunoblotted for 2 hours with either goat anti-mouse or goat anti-rabbit secondary horseradish peroxidase (HRP)-conjugated antibodies (Santa Cruz) diluted in TBST containing 5% milk. After secondary antibody blotting, membranes were washed with several changes of TBST. Peroxidase activities on washed membranes were developed with reagents supplied in the SuperSignal West Pico Chemiluminescent Substrate kit (Pierce), and HRP activities on membranes were digitized using a model 4000 MM Kodak Image Station.
2.6 Light and immunofluorescent microscopy
Light microscopy was performed using an Olympus CKX41 microscope equipped with a 4X, 10X, 20X and 40X objectives. For immunofluorescent studies, cell lines were cultured plated on fibronectin-coated coverslips (BD). Once the cells had reached a satisfactory degree of confluency, they were rinsed with PBS, fixed with 3.7% paraformaldehyde on ice for 10 minutes, rinsed with tris-buffered saline (TBS), and permeabilized with 0.05% Triton-X100 in TBS for 10 minutes prior to being blocked overnight with 2% BSA in TBS. Blocked cells were rinsed with TBS and incubated with primary or directly conjugated antibodies including mouse monoclonal anti-HA conjugated to Alexa Fluor 594 (Invitrogen), mouse monoclonal (mAb) anti-E-cadherin (BD), mAb anti-HA (Covance) and mAb anti-p130Cas (BD). After incubating the fixed cells with 1:100 dilutions of the primary or directly conjugated antibodies in 2% BSA for 2 hours at room temperature, the cells were rinsed several times with TBS. Secondary antibodies were added for an additional 2 hours and included goat anti-mouse IgG conjugated to Alexa Fluor 647 (Invitrogen) diluted 1:1500 in 2% BSA and goat anti-mouse IgG conjugated to Alexa Fluor 594 (Invitrogen) diluted 1:100 in 2% BSA. In some cases the fixed cells were also stained with phalloidin conjugate Alexa Fluor 594 (Invitrogen). After incubation with the antibodies, all cells were rinsed several times in TBS and briefly with distilled water prior to being mounted on slides with either Prolong Gold anti-fade reagent with DAPI or Slow Fade Gold without DAPI (Invitrogen) and allowed to cure overnight. Imaging was performed on a Nikon TE2000-E scope equipped with filters to detect DAPI, Alexa Fluor 594, and Alexa Fluor 647. Images were captured using a Photometrics CoolSnap HQ2 camera and NIS Elements (Nikon) software and processed using ImageJ (NIH) and Photoshop (Adobe).
2.7 Rac Activation Assay
The levels of activated Rac were determined by pulldown analysis as previously described [13, 21]. In brief, MCF-7 cells were transiently transfected with either pCDNA3 vector, HA-BCAR3(WT), HA-BCAR3(R743A) or HA-BCAR3(ΔGEF) using Fugene 6 (Roche). After incubation for 48 hours, the transfected cells were washed with PBS and lysed in pulldown lysis buffer (50 mmol/L Tris-HCl (pH 7.4), 200 mmol/L NaCl, 5 mmol/L, MgCl2, 1% NP40, 15% glycerol, and protease inhibitors). Protein concentration was determined using a BCA protein assay (Bio-Rad) and 1 mg of whole cell lysate was incubated for 1 hour at 4 degrees with 10 μl glutathione-sepharose 4B beads that had been pre-incubated with GST-PAK-RBD. Following incubation, the beads were rinsed three times with ice-cold lysis buffer before being boiled with Laemlli SDS-PAGE sample buffer. Western analysis using 12% low-bis formulation polyacrylamide gels was conducted. Transfers of the pulldowns and of whole cell lysates were probed with mouse monoclonal anti-Rac (BD).
2.8 Quantification of cells exhibiting lamellipodia
Lamellipodia formation was quantified by counting the number of cells exhibiting lamellipodia in images, captured at 10x magnification, of BALB/c-3T3 and p130Cas-/- MEF cell populations at mid-confluency. The percentage of cells exhibiting lamellipodia was determined by finding the mean of three sets of three captures for each cell line. The propagated error of the mean was calculated and significance was determined by one-way ANOVA with Tukey’s post hoc test.
2.9 Scratch Motility Assay
Confluent cell monolayers were scratched with a pipette tip and images of the wounds were captured at 10X magnification using an Olympus CKX41 microscope. Following the scratch, BALB/c-3T3 and p130Cas-/- MEFs cell lines were cultured, as described above, for 16 hours and 24 hours, respectively, prior to being imaged again. The rate of migration was determined by measuring the total distance that the cells had migrated from the initial wound margin in the given time. The total distance of cellular migration was determined to be the mean of 13 measurements made from each wound margin. The propagated error of the mean was calculated and significance was determined by one-way ANOVA with Tukey’s post hoc test.
3. Results
3.1 BCAR3-mediated anti-estrogen resistance is independent of p130Cas complex formation and BCAR3-mediated p130Cas phosphorylation
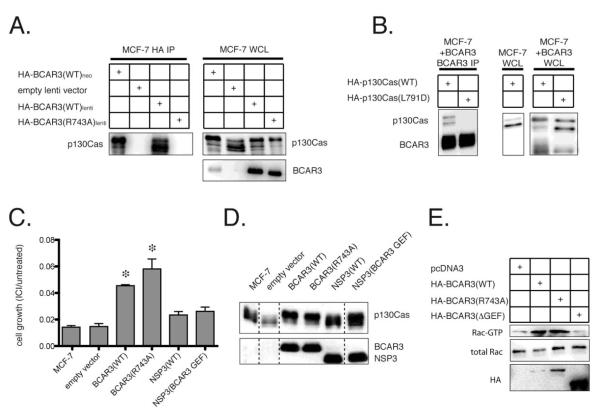
The ERα-positive human epithelial breast cancer cell line MCF-7 is known to express only low basal levels of BCAR3 and over-expression of BCAR3 in MCF-7 cells has been shown to induce both anti-estrogen resistance and a mesenchymal-like morphologic shift in growth pattern [14, 15]. To assess the role of BCAR3-p130Cas complex formation in these processes, we transduced MCF-7 cells with a control ZsGreen-expressing lentiviral vector or the same bicistronic vector also expressing either hemagglutinin (HA) epitope-tagged wildtype or R743A BCAR3. Polyclonal stably transduced ZsGreen-positive cells were isolated by flow cytometry and assessed by Western analysis. Consistent with our prior study reporting that R743A BCAR3 failed to associate with the CAS family member HEF1, anti-HA immunoprecipitation studies showed a dramatic reduction in the association of R743A BCAR3 with endogenous MCF-7 p130Cas (Fig 1A, left panel).
Figure 1. BCAR3-mediated anti-estrogen resistance and Rac activation are independent of BCAR3-p130Cas complex formation.
(A) A clonal MCF-7 cell line stably transfected with HA-tagged BCAR3 (II-6 cells) or polyclonal MCF-7 cell populations stably transduced with either a control lentivirus or wildtype or R743A HA-tagged BCAR3-expressing lentivirus were immunoprecipitated with HA antibody and probed with p130Cas antibody (left panel). Whole cell lysates (WCL) were probed with HA antibody to demonstrate expression of the exogenous BCAR3 and with p130Cas antibody to assess phosphorylation of endogenous p130Cas as determined by a shift in p130Cas gel mobility (right panel). (B) MCF-7 cells stably transfected with HA-tagged BCAR3 were transiently transfected with wildtype or L791D p130Cas, followed by immunoprecipitation with BCAR3 antibody and probing with an HA antibody. WCL from these two cell types as well as MCF-7 cells transiently transfected with wildtype p130Cas were probed with HA antibody to examine the expression and electrophoretic mobility of exogenous BCAR3 and p130Cas. (C) Following treatment with 100nM fulvestrant (ICI 182,780), parental MCF-7 cells or polyclonal populations stably transduced with empty, wildtype BCAR3, R743A BCAR3, NSP3 or NSP3/BCAR3 GEF-expressing lentivirus were assessed for cell growth. Experiments were performed three times. Asterisks indicate significant differences in cell growth (p < 0.05). (D) The PAGE migration of p130Cas was examined in WCL of either wildtype MCF-7 cells or polyclonal MCF-7 cells stably transduced with control, wildtype or R743A BCAR3 or wildtype or chimeric NSP3/BCAR3 GEF lentivirus. Expression of the NSP family members was confirmed by probing with HA antibody. (E) GST-PAK1 pulldown assay to detect GTP-bound Rac in MCF-7 cells transiently transfected with empty vector, HA-BCAR3(WT), HA-BCAR3(R743A) or HA-BCAR3(ΔGEF). WCL were probed for total Rac and HA. The pulldown assay was performed in triplicate.
As previously reported for MCF-7 cell lines stably transfected with a neomycin-selectable BCAR3 expression plasmid, stable polyclonal lentiviral over-expression of BCAR3 induced phosphorylation of p130Cas detectable as a reduction in p130Cas migration on low-bis PAGE (Fig 1A, right panel). Despite the lack of physical association with p130Cas, whole cell lysates of the polyclonal MCF-7 cell population stably transduced with R743A BCAR3 showed equivalent BCAR3-mediated p130Cas phosphorylation to the cells transduced with wildtype BCAR3. As previously reported using another cell line model, we found that mutation of p130Cas leucine 791 to aspartate (L791D) also blocked formation of a BCAR3-p130Cas complex in MCF-7 cells (Fig 1B, left panel). Once again, disruption of the BCAR3-p130Cas complex did not eliminate BCAR3’s ability to induce phosphorylation of L791D p130Cas, although in this case phosphorylation was reduced relative to that observed with wildtype p130Cas (Fig 1B, right panels).
When polyclonal lentivirus-transduced MCF-7 cells were tested for growth in the presence of the pure ER antagonist fulvestrant (ICI 182,780; 100 nM), cells transduced with wildtype BCAR3 grew significantly better than either wildtype MCF-7 cells or MCF-7 cells transduced with a lentiviral control vector (p < 0.05, Fig 1C). Entirely similar results were observed with MCF-7 cells transduced with R743A BCAR3, indicating that BCAR3-p130Cas complex formation is not required for BCAR3-mediated anti-estrogen resistance. In prior studies utilizing single stably transduced MCF-7 clones, we found that BCAR3 was unique among NSP family members both in its ability to induce anti-estrogen resistance and in its ability to induce p130Cas phosphorylation [14, 17]. Consistent with these studies, polyclonal NSP3-transduced MCF-7 cells treated with fulvestrant did not grow significantly faster than vector-only transduced cells, although a non-significant trend towards greater growth was observed in such experiments (Fig 1C). Also as expected, such lines did not demonstrate phosphorylation of endogenous p130Cas as judged by reduced PAGE migration (Fig 1D). To establish whether replacing the carboxy-terminal NSP3 sequence with the corresponding region of BCAR3 rendered the resulting chimeric proteins capable of inducing anti-estrogen resistance, we generated polyclonal MCF-7 cell populations stably transduced with such a chimeric construct. While this NSP3 chimeric construct, NSP3 (BCAR3 GEF), induced p130Cas phosphorylation as judged by the presence of a more slowly migrating p130Cas species, it failed to induce anti-estrogen resistance. These studies demonstrate that p130Cas phosphorylation, at least as judged by this assay, is not sufficient for BCAR3-mediated anti-estrogen resistance (Figs 1C, D).
3.2 Complex formation with p130Cas is not required for BCAR3 membrane localization, BCAR3-induced cellular discohesion or Rac activation in epithelial breast cancer cells
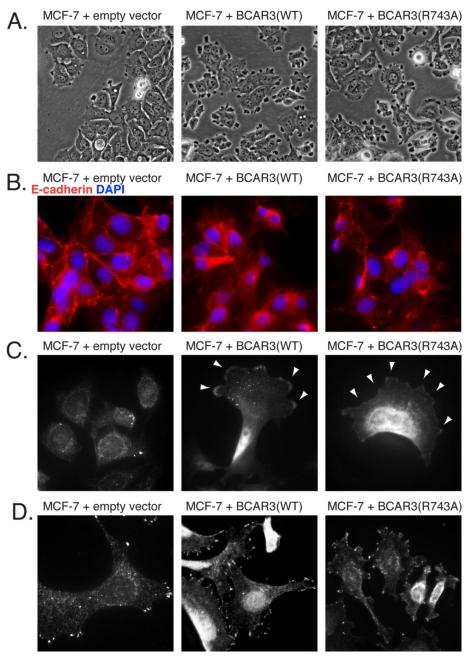
The polyclonal wildtype BCAR3-transduced MCF-7 cells demonstrated both a loss of cohesive epithelial growth pattern and the acquisition of “tufted” surface protrusions relative to control lentivirus transduced MCF-7 cells, a finding in keeping with our prior studies using single cloned lines of MCF-7 cells stably transfected with BCAR3 plasmid expression vectors. Entirely similar morphologic changes were observed in MCF-7 cells stably transduced with R743A BCAR3 indicating that the association of BCAR3 and p130Cas is not necessary for these alterations (Fig 2A). While intercellular cohesion is maintained in MCF-7 cells stably transduced with an empty lentiviral vector, as demonstrated by the presence of E-cadherin between apposing cells, MCF-7 cells transduced with either wildtype or R743A BCAR3 exhibit decreased E-cadherin localization at points of intercellular contact, indicating a loss of cohesion (Fig 2B). Immunofluorescent studies demonstrated that both wildtype and R743A BCAR3 localized similarly to the membrane at the leading edge of polarized MCF-7 cells (Fig 2C). When the intracellular distribution of p130Cas was examined in the same three stably transduced polyclonal MCF-7 cell lines, expression of wildtype or R743A BCAR3 did not alter the localization of p130Cas. In all cell lines examined, p130Cas localized focally in a punctate manner within the cell membrane, fully consistent with its known association with focal adhesions (Fig 2D). Of note, this concentrated localization of p130Cas was distinctly different from the more diffuse membrane distribution observed for either wildtype or R743A BCAR3.
Figure 2. BCAR3-induced cellular dissociation and BCAR3 membrane localization are independent of BCAR3-p130Cas complex formation.
(A) Polyclonal populations of MCF-7 cells stably transduced with an empty lentiviral vector, HA-BCAR3(WT) or HA-BCAR3(R743A) were photographed in culture at 40X magnification. (B) E-cadherin localization was assessed by immunofluorescent analysis of the same cell populations. Cells were counter-stained with DAPI and visualized at 60X magnification. (C) Stably transduced MCF-7 cells were probed with HA antibody to assess HA-tagged BCAR3 localization at 60X magnification. (D) Endogenous p130Cas localization in stably transduced MCF-7 cells was assessed with a p130Cas antibody at 60X magnification.
Our prior studies suggested that BCAR3 over-expression induces Rac activation by an indirect PI3K-dependent signaling pathway in a manner that was dependent upon BCAR3-p130Cas complex formation, as deletion of the BCAR3 GEF-like domain responsible for interaction with p130Cas abrogated BCAR3-mediated PI3K and Rac activation [16]. Given that the BCAR3 morphologic changes observed in MCF-7 cells are likely to be driven by Rac activation, we considered the possibility that the deletion of the GEF-like domain led to alterations in BCAR3-mediated signaling that were independent of the role of this domain in BCAR3-p130Cas complex formation. We therefore next sought to establish whether BCAR3-mediated Rac activation was also independent of the association of BCAR3 and p130Cas using the more specific R743A point mutation. As expected, transient transfection of MCF-7 cells with wildtype BCAR3 led to an increase of GTP-bound Rac relative to MCF-7 control cells. While, as previously observed, deletion of BCAR3’s GEF-like domain eliminated such BCAR3-mediated Rac activation, transfection with R743A BCAR3 induced Rac activation equivalently to wildtype BCAR3 (Fig 1E).
3.3 BCAR3 over-expression induces HEF1 phosphorylation in fibroblasts
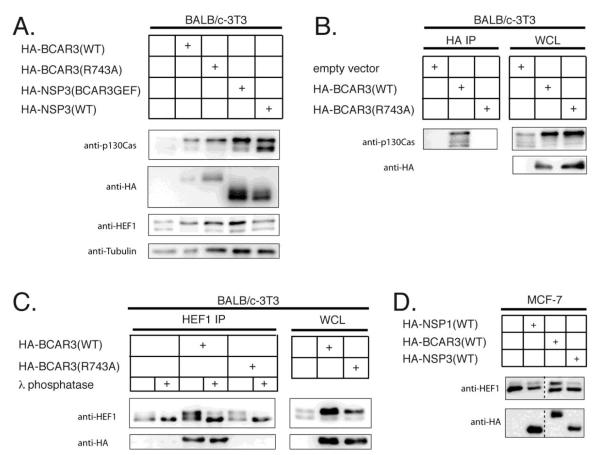
Over-expression of BCAR3 in single clones of NIH-3T3 cells previously has been shown to induce lamellipodia formation and ruffling, a phenotype that is consistent with Rac activation [15]. To assess the role of p130Cas association in BCAR3-mediated lamellipodia formation in fibroblasts, wildtype and R743A BCAR3 were expressed in the murine fibroblast cell line BALB/c-3T3 by the polyclonal lentiviral transduction approach. Both wildtype and R743A BCAR3 were expressed at relatively equal levels and both induced comparable phosphorylation of p130Cas in BALB/c-3T3 cells (Fig 3A). As in MCF-7 cells, immunoprecipitation studies demonstrated that wildtype, but not R743A BCAR3, associated with endogenous p130Cas in BALB/c-3T3 cells (Fig 3B).
Figure 3. BCAR3-mediated p130Cas and HEF1 phosphoryation is independent of complex formation in stable polyclonal BALB/c-3T3 cells.
(A) Polyclonal populations of stably transduced BALB/c-3T3 cells expressing wildtype BCAR3, R743A BCAR3, wildtype NSP3 or chimeric NSP3/BCAR3 GEF were assessed for endogenous p130Cas, over-expressed HA-tagged proteins, endogenous Hef1 and tubulin by Western analysis. (B) HA immunoprecipitation of wildtype and R743A HA-BCAR3 from stably transduced polyclonal populations of BALB/c-3T3 cells. Immunoprecipitated lysates were probed with p130Cas antibody while WCL were probed with p130Cas and HA antibodies to detect endogenous p130Cas and over-expressed HA-tagged BCAR3, respectively. (C) λ phosphatase-treated HEF1 immunoprecipitates and whole cell lysates from BALB/c-3T3 cells were probed with HEF1 and HA antibodies to determine if the observed BCAR3-induced gel mobility shift represents a phosphorylated species. (D) NSP-mediated gel mobility shift of HEF1 was assessed by probing whole cell lysates from MCF-7 cells transiently co-transfected with plasmids encoding HEF1 and HA-tagged NSP family proteins with HEF1 antibody. The expression of exogenous NSP1, BCAR3 and NSP3 was confirmed by reactivity to anti-HA.
Both wildtype and R743A BCAR3 also induced a reduction in the migration of HEF1, a CAS family member previously shown to form complexes with BCAR3 in murine and human B lymphocytes (Fig 3A) [13]. In order to establish whether such a BCAR3-induced reduction in HEF1 PAGE migration was due to phosphorylation, HEF1 was immunoprecipitated from the stably transduced cell lines and treated with lambda phosphatase. Phosphatase treatment led to depletion of the more slowly migrating HEF1 isoform and concomitant accumulation of the faster migrating isoform (Fig 3C). To examine the specificity of this process, MCF-7 cells were transiently transfected with plasmids encoding HEF1 and either NSP1, BCAR3 or the non-hematopoietic isoform of NSP3. Only BCAR3 induced HEF1 phosphorylation as judged by augmentation of the levels of the more slowly migrating form of HEF1 (Fig 3D). In aggregate, these studies suggest that among the three NSP family isoforms tested, BCAR3 is unique in its ability to induce HEF1 phosphorylation and that it does so in a manner that does not require the formation of a BCAR3-HEF1 complex.
3.4 BCAR3-p130Cas complex formation is dispensable for BCAR3-mediated lamellipodia formation but not BCAR3-induced cell motility
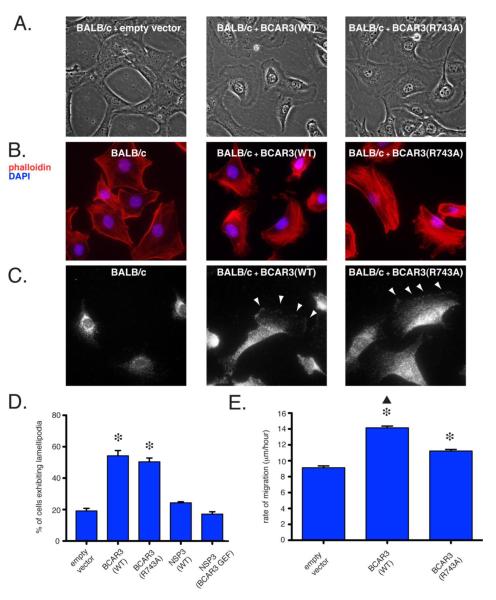
Polyclonal BALB/c-3T3 cell populations stably transduced with either wildtype or R743A BCAR3 exhibit increased lamellipodia formation relative to vector-only transduced counterparts (Fig 4A). Further, a large subset of BALB/c-3T3 cells over-expressing either wildtype or R743A BCAR3 exhibit a redistribution of actin stress fibers, consistent with the induction of lamellipodia and suggestive of a motile phenotype (Fig 4B). As had been observed in the epithelial MCF-7 cells, both wildtype and R743A BCAR3 were detected at the membrane on the leading edge of polarized BALB/c-3T3 cells, indicating that stable association with p130Cas is not required for such localization (Fig 4C). Quantitatively, whereas 19% of BALB/c-3T3 cells transduced with a control lentiviral vector exhibited lamellipodia, 54% of BALB/c-3T3 stably over-expressing wildtype BCAR3 and 50% of BALB/c-3T3 stably over-expressing R743A BCAR3 displayed lamellipodia (Fig 4D; p < 0.05 for both forms of BCAR3 relative to vector-only transduced cells). To determine whether serine phosphorylation played a pivotal role in BCAR3-mediated lamellipodia formation, we generated polyclonal stably transduced BALB/c-3T3 populations over-expressing NSP3 or NSP3(BCAR3 GEF). As previously observed in MCF-7 cells, NSP3(BCAR3 GEF) but not NSP3 induced p130Cas phosphorylation in BALB/c-3T3 cells. Similarly, the phosphorylation of HEF1 was induced by NSP3(BCAR3 GEF) but not by wildtype NSP3 (Fig 3A). Neither cell population showed a significant increase in lamellipodia formation relative to control BALB/c-3T3 cells, demonstrating that p130Cas phosphorylation is not sufficient for BCAR3-mediated lamellipodia formation (Fig 4D).
Figure 4. BCAR3-induced membrane lamellipodia formation is independent of association with p130Cas in BALB/c-3T3 cells, while BCAR3-induced motility is not.
(A) Phase contrast images of polyclonal populations of BALB/c-3T3 cells stably transduced with either an empty lentiviral vector, wildtype BCAR3 or R743A BCAR3 at 40X magnification. (B) BALB/c-3T3 cells and polyclonal populations of BALB/c-3T3 cells stably transduced with wildtype BCAR3 or R743A BCAR3 were stained with phalloidin (red) and DAPI (blue) to visualize actin stress fibers and nuclei, respectively (40x magnification). (C) BALB/c-3T3 cells and BALB/c-3T3 cells stably transduced with either wildtype or R743A BCAR3 were probed with anti-HA primary and Alexa Fluor 647 secondary antibodies to determine BCAR3 localization at 60X magnification. (D) Quantification of BCAR3-induced lamellipodia formation in BALB/c-3T3 and stably transduced populations of BALB/c-3T3 cells. Error bars depict the propagated error of the mean and significance was determined by one-way ANOVA (asterisk, p < 0.05 relative to BALB/c-3T3). (E) Mean rates of migration of BALB/c-3T3 cells were determined by measuring the distance traveled by cells over a 16 hour period following the scratching of a confluent cell monolayer. Significance was determined by one-way ANOVA (p < 0.05).
Expression of BCAR3 has previously been linked to augmented cell motility and BCAR3-induced migration has been shown to increase synergistically when p130Cas is co-expressed [22]. As assessed by a “scratch” assay, both wildtype and R743A BCAR3 induced significantly greater migration of polyclonal BALB/c 3T3 transductants than vector-only controls. Notably, however, the increase in migration induced by wildtype BCAR3 was significantly greater than that induced by R743A BCAR3 (p < 0.05; Fig. 4E).
3.5 p130Cas is not required for BCAR3-induced lamellipodia formation or motility
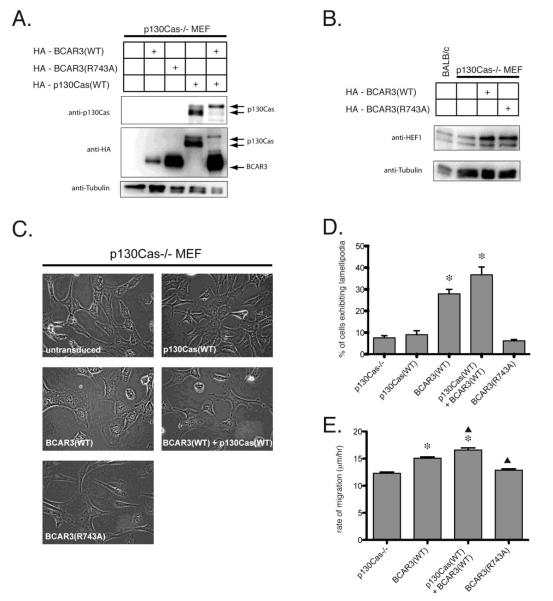
The experiments described above suggest that association with p130Cas is not required for BCAR3-mediated morphologic changes or lamellipodia formation but does influence BCAR3-induced cell motility. As these experiments did not directly assess a requirement for p130Cas itself in these processes, we next sought to examine these endpoints using MEFs derived from a strain of mice in which homologous recombination was used to eliminate functional p130Cas gene expression [7]. Of note, BCAR3 is not endogenously expressed at high levels in p130Cas-/- MEFs and could not be detected by BCAR3 immunoprecipitation in p130Cas-/- MEFs or by p130Cas immunoprecipitation in p130Cas-/- MEFs reconstituted with wildtype p130Cas (unpublished data). p130Cas-/- MEFs were stably transduced with HA-tagged forms of BCAR3 and/or p130Cas and Western analysis confirmed expression of these proteins in the transduced lines (Fig 5A). The established polyclonal cell lines allowed us to distinguish phenotypes induced by p130Cas expression, BCAR3 expression, and by expression of the combination of these two proteins.
Figure 5. BCAR3-induced lamellipodia formation and motility in the absence of p130Cas.
(A) Western analysis of polyclonal populations of p130Cas-/- MEFs stably transduced with combinations of HA-BCAR3(WT), HA-BCAR3(R743A) and HA-p130Cas(WT). WCL were probed with antibodies to detect endogenous p130Cas, over-expressed HA-tagged proteins and tubulin. (B) As for panel A, a Western analysis of endogenous HEF1 expression and phosphorylation. (C) Phase contrast images of p130Cas-/- MEFs and polyclonal populations of stably transduced p130Cas-/- MEFs (40X magnification). p130Cas-/- MEFs and p130Cas-/- MEFs expressing either wildtype or R743A BCAR3 are shown in the left column and p130Cas-/- MEFs reconstituted with wildtype p130Cas and also expressing wildtype or R743A BCAR3 are shown in the right column. (D) Quantification of BCAR3-induced lamellipodia formation in p130Cas-/- MEFs and stably transduced polyclonal populations of p130Cas-/- MEFs. (E) Mean rates of migration of p130Cas-/- MEFs were determined by measuring the distance traveled by cells over a 24 hour period following the scratching of a confluent cell monolayer. Significance was determined by one-way ANOVA (asterisk p < 0.05 relative to p130Cas-/-; triangle p < 0.05 relative to p130Cas-/- + BCAR3(WT)).
The morphologic phenotype of p130Cas-/- MEFs expressing wildtype BCAR3 differed from both p130Cas-/- MEFs and p130Cas MEFs stably reconstituted with p130Cas (Fig 5C). p130Cas-/- MEFs had a characteristic elongated morphology and only a small percentage of such cells (8%) demonstrated lamellipodia (Fig 5D). Reconstitution with p130Cas resulted in cells with a rounder morphology as well as a greater degree of cellular branching, but no increase in the number exhibiting lamellipodia (9%). Over-expression of wildtype BCAR3 in p130Cas-/- MEFs significantly increased the formation of lamellipodia (28%, p < 0.05) but failed to induce the branched cell phenotype. Over-expression of wildtype BCAR3 in p130Cas reconstituted p130Cas-/- MEFs led to both the formation of lamellipodia (37%) as well as increased cellular branching. Although reproducibly observed, the BCAR-induced lamellipodia exhibited by cells lacking p130Cas expression were not as large as those induced by BCAR3 in BALB/c-3T3 cells. In some contrast to the results previously obtained with BALB/c-3T3 cells, p130Cas-/- MEFs over-expressing R743A BCAR3 did not exhibit abundant lamellipodia (7%). As expected, BCAR3 induced phosphorylation of p130Cas in the reconstituted cells (Fig 5A). BCAR3-induced HEF1 phosphorylation as judged by reduced PAGE migration in p130Cas-/- MEFs over-expressing both wildtype and R743A BCAR3 (Fig 5B).
The motility of p130Cas-/- MEFs has been previously reported to be impaired and reconstitution of p130Cas expression restores the migratory capacity of these cells [7]. As judged by the scratch assay, over-expression of wildtype BCAR3 also induced a significant increase in the rate of migration of p130Cas-/- MEFs (Fig 5E). Combined expression of p130Cas and BCAR3 induced more rapid migration than when BCAR3 was expressed alone. In concordance with the results obtained in BALB/c-3T3 cells, over-expression of R743A BCAR3 did not increase the rate of migration of p130Cas-/- MEFs (Fig 5E). These results indicate that while BCAR3 may induce migration in a p130Cas-independent manner, the ability of BCAR3 to form a complex with other CAS family members appears to play an important role in this process.
4. Discussion
In the studies described in this report, we have shown that BCAR3 complex formation with p130Cas (or the related family member HEF1) is not required for BCAR3-induced ERα-positive breast cancer cell-line anti-estrogen resistance, cell discohesion, Rac activation, or fibroblast lamellipodia formation. These results are surprising in that both BCAR3 and p130Cas (BCAR1) were identified in a screen for genes capable of inducing anti-estrogen resistance and the endogenous proteins were subsequently found to form a stable complex with each other in several cell types. Further, a variety of studies have implicated both BCAR3 and p130Cas in signaling pathways that positively regulate Rac activation and cell motility. Not surprisingly, therefore, such earlier studies had suggested that BCAR3 was likely to act by facilitating p130Cas-mediated signaling in a manner that was dependent upon formation of a complex between the two proteins. Consistent with this model, elimination of the carboxy-terminal GEF-like domain of BCAR3 that binds to p130Cas resulted in loss of both BCAR3-induced anti-estrogen resistance and Rac activation [16]. The current studies have addressed the role of BCAR3-p130Cas association more specifically by taking advantage of a recent structural study that identified a BCAR3 point mutation, R743A, that abrogates complex formation [19]. In contrast to prior studies with carboxy-terminal BCAR3 deletion mutants, experiments with this point mutant now demonstrate that BCAR3 association with p130Cas is dispensable for BCAR3-induced growth of MCF-7 cells in the presence of fulvestrant (ICI 182,780), a clinically efficacious estrogen antagonist that induces proteosomal degradation of the estrogen receptor. BCAR3’s amino-terminal SH2 domain has previously been found to be required for anti-estrogen resistance [16]. In aggregate, these studies suggest that fulvestrant resistance requires binding of the BCAR3 SH2 domain to a currently unknown tyrosine phosphorylated ligand and that formation of a BCAR3-p130Cas complex is not obligatory for the BCAR3-induced signaling that leads to such anti-estrogen resistance.
Despite the fact that p130Cas signaling has been linked to activation of the atypical Rac GDP exchange factor DOCK180, BCAR3-p130Cas complex formation was also found to be unnecessary for BCAR3-induced Rac activation and cellular discohesion in breast cancer epithelial cells. This result is in marked contrast to results obtained previously with the carboxy-terminal deleted form of BCAR3 which failed to induce Rac activation, suggesting that this region of BCAR3 plays a role in BCAR3-induced Rac activation independent of its ability to allow complex formation with CAS family members. Notably, immunofluorescent studies indicate that the localization of the R743A mutant form of BCAR3 to the leading edge membrane was indistinguishable from that of wildtype BCAR3 in both epithelial breast cancer and fibroblast cell lines. This finding is in keeping with a prior report by Schrecengost et al indicating that both wildtype and carboxy-terminal-deleted BCAR3 localized to the membrane of breast cancer cell lines [18]. BCAR3-induced lamellipodia formation in fibroblasts, a process characteristic of Rac-mediated signaling, also occurred equivalently with wildtype or R743A BCAR3 over-expression. Dail et al have reported that when modified to constitutively localize to the plasma membrane, over-expression of NSP3 induces ruffling in NIH-3T3 fibroblasts in a manner that is dependent on p130Cas association [23]. In the current study, BCAR3-induced lamellipodia formation in fibroblasts was not mimicked by over-expression of either wildtype non-hematopoietic NSP3, which does not constitutively localize to the membrane, or by a chimeric form of NSP3 in which the carboxy-terminal sequences responsible for binding to p130Cas were replaced with the analogous portion of BCAR3. These studies demonstrate that a recently identified complex-independent BCAR3-mediated signaling pathway that results in serine phosphorylation of p130Cas also does not correlate with either BCAR3-induced anti-estrogen resistance or lamellipodia formation.
Consistent with the concept of p130Cas-independent BCAR3-mediated signaling, it is notable that the morphologic effects of transduction of p130Cas knockout MEFs with a lentiviral BCAR3 expression construct are quite distinct from those observed following reconstitution with p130Cas. Upon reconstitution of p130Cas-/- MEFs with p130Cas, cells acquired a more rounded and branched phenotype but failed to alter their basal level of lamellipodia. BCAR3-transduced MEFs, in contrast, had augmented levels of lamellipodia, but failed to recapitulate the branched phenotype characteristic of p130Cas reconstitution. In cells transduced with both lentiviral constructs, fibroblasts exhibited cell rounding, branching and an increase in lamellipodia. These studies support the hypothesis that BCAR3 does not exert its morphologic effects in fibroblasts by way of a linear signaling pathway in which BCAR3 augments or facilitates p130Cas-mediated signaling.
Despite the negative results reported above, complex formation between BCAR3 and p130Cas is nonetheless likely to have important consequences in the physiology of cells that express both of these proteins. Bouton and colleagues have previously shown that over-expression of BCAR3 induces cell motility. In an initial study, while transient transfection of a plasmid encoding BCAR3 alone did not augment motility in C3H10T1/2-5H murine fibroblasts, the same BCAR3 construct enhanced migration induced by transient transfection of a plasmid encoding p130Cas [22]. Deletion of BCAR3’s carboxy-terminal GEF domain abrogated the ability of BCAR3 to enhance such p130Cas over-expression-induced migration. The authors suggested that BCAR3 may have enhanced motility by recruiting p130Cas from focal adhesions to lamellipodial membranes. In a second study in epithelial breast cancer cell lines, BCAR3 expression induced motility even when used alone, although the role of BCAR3 association with p130Cas in this process was not specifically addressed [18].
In some contrast to the prior work of Bouton and colleagues in fibroblast cell lines, we find that stable polyclonal lentiviral expression of BCAR3 in BALB/c-3T3 cells is sufficient to induce motility. In contrast to the hypothesis that BCAR3 induces motility solely as a result of its ability to associate with and recruit p130Cas to lamellipodial membranes, we observed BCAR3-induced migration in the absence of BCAR3-p130Cas complex formation in BALB/c-3T3 cells. Further, BCAR3-induced migration is observed in MEFs completely lacking p130Cas expression. However, our studies with the R743A mutant form of BCAR3 suggest that BCAR3 complex formation with CAS family members contributes to BCAR3-induced cell motility, as while R743A BCAR3 does induce motility in the BALB/c-3T3 cells relative to a vector-only control, the degree of motility induced was less than that observed with wildtype BCAR3. Consistent with this finding, in p130Cas-/- MEFs, the R743A BCAR3 mutant’s loss of ability to bind to other CAS family members such as HEF1 led to a complete abrogation of induced motility. These observations parallel a report in which complex formation with p130Cas was shown to be required for NSP3’s ability to augment COS-7 cell migration to EGF [23]. Similarly, Chat-H, a hematopoietic-restricted NSP3 isoform, has been reported to induce T lymphocyte migration in a manner that requires complex formation with HEF1 [24, 25].
Prior studies have demonstrated that BCAR3 binds endogenous HEF1 in murine B lymphocytes [13]. In the current study, over-expression of BCAR3 induced a reduction in PAGE migration in endogenous HEF1 in both BALB/c-3T3 cells and p130Cas-/- MEFs. This BCAR3-induced mobility shift was identified to be λ phosphatase-sensitive, confirming that as previously documented for p130Cas, BCAR3 over-expression induces phosphorylation of HEF1. However under the conditions utilized in these experiments, we failed to observe a coincident BCAR3-mediated reduction in HEF1 stability, suggesting that BCAR3-induced HEF1 phosphorylation is distinct from the previously reported adhesion-mediated phosphorylation of HEF1 that leads to proteasomal degradation [26]. Whereas the non-hematopoietic isoform of NSP3 utilized in our study does not induce HEF1 phosphorylation, Chat-H, which constitutively localizes to the plasma membrane, has been reported to induce HEF1 phosphorylation [24]. Our studies do however suggest that BCAR3 binding to p130Cas, or in the absence of p130Cas, HEF1, plays an important role in BCAR3’s ability to augment cell motility.
In conclusion, the studies reported here suggest that an important subset of the functional effects observed following over-expression of BCAR3 are independent of binding to CAS family members or, in certain instances, the presence of p130Cas. Should BCAR3 ultimately be found to play a meaningful role in clinical breast cancer resistance to tamoxifen or fulvestrant, these findings are of particular importance in that they suggest that small molecules that disrupt the BCAR3-p130Cas complex would be unlikely to reverse this process. Instead, the focus of future efforts in this field should center on identifying the signaling pathway activated by binding of the BCAR3 SH2 domain to its relevant ligand.
Acknowledgements
The authors gratefully acknowledge excellent technical assistance from Alex Bloom, as well as advice and reagents from Dr’s Douglas Faller (Cancer Center, BUSM), Kathrin Kirsch (Biochemistry, BUSM), and Maria Kukuruzinska (Molecular and Cell Biology, BU Goldman School of Dental Medicine). This work was supported by NIH RO1 CA114094, the Logica Foundation and by the Breast Cancer Alliance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].van Agthoven T, van Agthoven TL, Dekker A, van der Spek PJ, Vreede L, Dorssers LC. EMBO J. 1998;17:2799–2808. doi: 10.1093/emboj/17.10.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Brinkman A, van der Flier S, Kok EM, Dorssers LC. J Natl Cancer Inst. 2000;92:112–120. doi: 10.1093/jnci/92.2.112. [DOI] [PubMed] [Google Scholar]
- [3].Ruest PJ, Shin NY, Polte TR, Zhang X, Hanks SK. Mol Cell Biol. 2001;21:7641–7652. doi: 10.1128/MCB.21.22.7641-7652.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cabodi S, Del Pilar Camacho-Leal M, Di Stefano P, Defilippi P. Nat Rev Cancer. 10:858–870. doi: 10.1038/nrc2967. [DOI] [PubMed] [Google Scholar]
- [5].Klemke RL, Leng J, Molander R, Brooks PC, Vuori K, Cheresh DA. J Cell Biol. 1998;140:961–972. doi: 10.1083/jcb.140.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kiyokawa E, Hashimoto Y, Kurata T, Sugimura H, Matsuda M. J Biol Chem. 1998;273:24479–24484. doi: 10.1074/jbc.273.38.24479. [DOI] [PubMed] [Google Scholar]
- [7].Honda H, Oda H, Nakamoto T, Honda Z, Sakai R, Suzuki T, Saito T, Nakamura K, Nakao K, Ishikawa T, Katsuki M, Yazaki Y, Hirai H. Nat Genet. 1998;19:361–365. doi: 10.1038/1246. [DOI] [PubMed] [Google Scholar]
- [8].van der Flier S, Chan CM, Brinkman A, Smid M, Johnston SR, Dorssers LC, Dowsett M. Int J Cancer. 2000;89:465–468. doi: 10.1002/1097-0215(20000920)89:5<465::aid-ijc11>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- [9].Dorssers LC, Grebenchtchikov N, Brinkman A, Look MP, van Broekhoven SP, de Jong D, Peters HA, Portengen H, Meijer-van Gelder ME, Klijn JG, van Tienoven DT, Geurts-Moespot A, Span PN, Foekens JA, Sweep FC. Clin Cancer Res. 2004;10:6194–6202. doi: 10.1158/1078-0432.CCR-04-0444. [DOI] [PubMed] [Google Scholar]
- [10].van Agthoven T, Sieuwerts AM, Meijer-van Gelder ME, Look MP, Smid M, Veldscholte J, Sleijfer S, Foekens JA, Dorssers LC. J Clin Oncol. 2009;27:542–549. doi: 10.1200/JCO.2008.17.1462. [DOI] [PubMed] [Google Scholar]
- [11].Cai D, Clayton LK, Smolyar A, Lerner A. J Immunol. 1999;163:2104–2112. [PubMed] [Google Scholar]
- [12].Gotoh T, Cai D, Tian X, Feig L, Lerner A. J Biol Chem. 2000;275:30118–30123. doi: 10.1074/jbc.M003074200. [DOI] [PubMed] [Google Scholar]
- [13].Cai D, Felekkis KN, Near RI, O’Neill GM, van Seventer JM, Golemis EA, Lerner A. J Immunol. 2003;170:969–978. doi: 10.4049/jimmunol.170.2.969. [DOI] [PubMed] [Google Scholar]
- [14].Near RI, Zhang Y, Makkinje A, Vanden Borre P, Lerner A. J Cell Physiol. 2007;212:655–665. doi: 10.1002/jcp.21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cai D, Iyer A, Felekkis K, Near R, Luo Z, Chernoff J, Albanese C, Pestell RG, Lerner A. Cancer Research. 2003;63:6802–6808. [PubMed] [Google Scholar]
- [16].Felekkis KN, Narsimhan RP, Near R, Castro AF, Zheng Y, Quilliam LA, Lerner A. Mol Cancer Res. 2005;3:32–41. [PubMed] [Google Scholar]
- [17].Makkinje A, Near RI, Infusini G, Vanden Borre P, Bloom A, Cai D, Costello CE, Lerner A. Cellular Signaling. 2009;21:1423–1435. doi: 10.1016/j.cellsig.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Schrecengost RS, Riggins RB, Thomas KS, Guerrero MS, Bouton AH. Cancer Res. 2007;67:6174–6182. doi: 10.1158/0008-5472.CAN-06-3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Garron ML, Arsenieva D, Zhong J, Bloom AB, Lerner A, O’Neill GM, Arold ST. J Mol Biol. 2009;386:190–203. doi: 10.1016/j.jmb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- [20].Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Felekkis K, Quilliam LA, Lerner A. Methods Enzymol. 2005;407:55–63. doi: 10.1016/S0076-6879(05)07006-0. [DOI] [PubMed] [Google Scholar]
- [22].Riggins RB, Quilliam LA, Bouton AH. J Biol Chem. 2003;278:28264–28273. doi: 10.1074/jbc.M303535200. [DOI] [PubMed] [Google Scholar]
- [23].Dail M, Kalo MS, Seddon JA, Cote JF, Vuori K, Pasquale EB. J Biol Chem. 2004 doi: 10.1074/jbc.M402929200. [DOI] [PubMed] [Google Scholar]
- [24].Regelmann AG, Danzl NM, Wanjalla C, Alexandropoulos K. Immunity. 2006;25:907–918. doi: 10.1016/j.immuni.2006.09.014. [DOI] [PubMed] [Google Scholar]
- [25].Alexandropoulos K, Regelmann AG. Immunol Rev. 2009;232:160–174. doi: 10.1111/j.1600-065X.2009.00831.x. [DOI] [PubMed] [Google Scholar]
- [26].Zheng M, McKeown-Longo PJ. J Cell Sci. 2006;119:96–103. doi: 10.1242/jcs.02712. [DOI] [PubMed] [Google Scholar]