Abstract
The high affinity receptor for IgE, FcɛRI on mast cells and basophils plays an essential role in immunological defense. Upon multivalent antigen binding, FcɛRI becomes phoshorylated by the protein-tyrosine kinase Lyn, as a result of receptor clustering in lipid rafts. FcɛRI has been shown to be ubiquitinated. Ubiquitination can lead to degradation by proteasomes, but it can also act as a sorting signal to internalize proteins destined to the endosomal/lysosomal pathway. We have analyzed whether FcɛRI ubiquitination takes place within rafts. We report biochemical and imaging evidence in rat basoleukemia cells for the presence of ubiquitinated FcɛRI in clustered rafts upon receptor activation. Moreover, we demonstrated that the ubiquitin ligases Cbl and Nedd4 colocalize with FcɛRI patches and showed that both ligases become associated with lipid rafts after activation of IgE signaling. Because Cbl is known to interact with the FcɛRI signaling complex, ubiquitination is likely to be an important parameter regulating IgE-triggered signaling occurring in rafts.
Like other multichain immune recognition receptors, the IgE receptor (FcɛRI) interacts with protein-tyrosine kinases to activate the signaling pathway (1). Upon antigen cross-linking of IgE-bound FcɛRI, contact between the receptor and protein-tyrosine kinases, such as Lyn and Syk, occurs within specialized lipid microdomains called rafts (1). These are constituted of cholesterol and sphingolipid assemblies resistant to extraction by selective detergent (2). After contact, immunoreceptor tyrosine-based activation motifs present on the β and the γ chains of the FcɛRI become phosphorylated by Lyn, allowing Syk to bind phosphorylated immunoreceptor tyrosine-based activation motifs. Subsequently, lysine residues in the cytoplasmic region of β and γ subunits of the receptors become ubiquitinated (3).
A good candidate to mediate FcɛRI ubiquitination could be the protooncogene Cbl. Cbl was shown to participate in cis- and trans-ubiquitination (4) and is phosphorylated upon FcɛRI engagement (5). Moreover, Cbl has been shown to negatively regulate the Syk kinase (6) and is involved in IgE-triggered signaling. We recently demonstrated that the ubiquitin ligase Nedd4 (7) could be recruited into rafts in epithelial cells (8). Nedd4 contains ubiquitin conjugating hect, WW, and C2 domains and was shown to ubiquitinate membrane receptors (7). This enzyme could be a potential candidate for mediating ubiquitination of proteins in FcɛRI patches.
Here, we have examined the role of lipid rafts in IgE-triggered activation of FcɛRI ubiquitination. We could show that ubiquitin ligases (Cbl and Nedd4) were recruited into rafts upon IgE triggering possibly to regulate the dynamics of raft-associated proteins involved in cell-signaling responses.
Materials and Methods
Reagents.
Rat basophilic leukemia (RBL) 2H3 cells and the pMT123 plasmid coding for the hemagglutinin (HA)-tagged ubiquitin were kindly provided by P. Chavrier (Institut Curie, Paris) and D. Bohmann (European Molecular Biology Laboratory, Heidelberg), respectively. Anti-Nedd4 antibody was a generous gift of D. Rotin (Hospital for Sick Children, Toronto). Anti-HA and anti-Lyn antibodies were from Santa Cruz. Dinitrophenol (DNP)-BSA and FITC-conjugated anti-DNP were from Molecular Probes. Anti-DNP IgE was purchased from Calbiochem. Fluorochrome-conjugated secondary antibodies were from Jackson ImmunoResearch.
Electroporation, Sensitization, Activation, and Cyclodextrin Treatment.
Cells were grown in RPMI, 10% FCS (GIBCO/BRL). Cells were trypsinized and washed, and 2 × 107 RBL cells were placed in a 1-ml electroporation gene pulser cuvette. A Bio-Rad microporator was used for loading the pMT123 plasmid (≈10 μg/ml). RBL cells were electroporated at a field strength of 330 V, 960 μF applied for 2 s. After loading, cells were kept 10 min on ice and washed with 10 ml of culture medium.
Cells were sensitized by overnight incubation with 100 ng/ml DNP-specific IgE in culture medium containing 0.1% FCS. After cells had been washed with serum-free medium, IgE receptors were activated by addition of 1 μg/ml DNP-BSA at room temperature for less that 5 min.
Cells were treated in lipid-free medium with 5 mM Me-β-cyclodextrin (MeCD, Sigma) for 30 min at 37°C. (We observed some cell loss when using a higher concentration, i.e., 10 mM.) We checked that this treatment led to 60% extraction of cellular cholesterol by quantitative measurement of cholesterol analyzed on TLC after CuAc/H3PO4 staining. We verified that the cells were still alive after such a treatment by EtBr exclusion, analysis of actin cytoskeleton organization and transferrin uptake. As reported previously, we noticed lower transferrin uptake as compared with untreated cells in agreement with previous reports (9–11).
Detergent Extraction and Floatation.
When necessary, RBL cells were transfected with the pMT123 plasmid coding for a HA-tagged ubiquitin chimera, and ubiquitinated proteins were detected by using an anti-HA antibody. Detergent extraction and floatation were performed as described in refs.12 and 13. Briefly, scraped cells were incubated with 0.05% Triton X-100 on ice for 30 min in TNE (25 mM Tris⋅HCl, pH 7.4/150 mM NaCl/5 mM EDTA/1 mM DTT/protease inhibitor mixture). Samples were made 40% OptiPrep (Nycomed) and overlaid with 30% and 0% OptiPrep step gradients. Samples were centrifuged at 55,000 rpm in TLS 55 rotor for 2 h at 4°C, and fractions collected from the top were analyzed by Western blotting. Blots were developed with chemoluminescent probes.
Immunofluorescence.
Sensitized cyclodextrin-treated and control cells exposed or not to DNP-BSA were fixed in 4% paraformaldehyde and processed for immunolabeling as described in ref. 13. Confocal (x, y) sections acquired on a Zeiss 410 confocal microscope are presented.
Results and Discussion
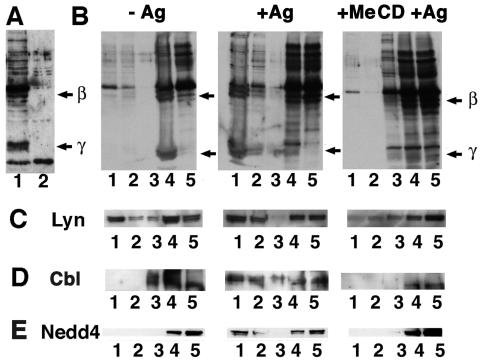
Sensitized RBL cells, coated with anti-DNP IgE, were stimulated with multivalent DNP-BSA, leading to receptor clustering and activation. As a result, dramatic ubiquitination of both β and γ chains of the FcɛRI was observed, consistent with previous results (Fig. 1 A and B) (3). After treating RBL cells with MeCD, a cholesterol-removing agent that disturbs raft function, the pool of activated FcɛRI that was ubiquitinated decreased dramatically, although the extent of ubiquitination of other proteins remained unaffected as judged by Western blotting (Fig. 1A). This observation suggested that rafts play a role in the FcɛRI ubiquitination process. To further investigate this issue, an HA-tagged version of ubiquitin was transiently expressed in cells subsequently sensitized and stimulated or not with DNP-BSA, and we analyzed raft association of ubiquitinated FcɛRI by floatation experiments in Optiprep gradients after Triton X-100 extraction in the cold. We could detect ubiquitinated forms of the FcɛRI in the lowest buoyant fractions corresponding to detergent-resistant membranes (DRMs; see ref. 2; Fig. 1B). This floatation pattern was abolished after treating cells with MeCD before clustering (Fig. 1B). Such a distribution pattern is typical of raft components as exemplified by the behavior of Lyn (13).
Figure 1.
Ubiquitinated FcɛRI and Nedd4 are present within DRMs in RBL cells. RBL cells were transfected with an HA-tagged ubiquitin chimera. (A) Cellular extracts from sensitized RBL cells activated with DNP-BSA for less than 5 min at room temperature and treated (lane 1) or not (lane 2) with MeCD were analyzed by Western blotting. Anti-HA antibody was used to detect HA-tagged ubiquitinated proteins. Arrows indicate where β and γ subunits of the FcɛRI migrate. (B) Sensitized RBL cells were activated (+Ag) or not (−Ag) with DNP-BSA for less than 5 min at room temperature before lysis and analysis by SDS/PAGE and Western blotting. After floatation of Triton X-100 cell extracts, proteins were precipitated by using MeOH/CHCl3 followed by an acetone wash. The analysis was carried out by SDS/PAGE and Western blotting using an anti-HA antibody. Arrows indicate where β and γ subunits of the FcɛRI migrate. (C) Filters presented in B were reprobed with anti-Lyn (C), anti-Cbl (D), and anti-Nedd4 (E) antibodies.
We also followed the distribution of the ubiquitinated FcɛRI in immunofluorescence experiments (Fig. 2A). Upon FcɛRI clustering, DNP-BSA colocalized at the cell surface in patches with ubiquitinated proteins as revealed by using an anti-HA antibody (and an anti-ubiquitin antibody; unpublished results). We could also observe a partial colocalization of ubiquitinated activated FcɛRI and Lyn (Fig. 2B). This copatching was inhibited by MeCD treatment. At this point, we concluded that ubiquitinated pools of the FcɛRI were present in raft patches on the cell surface that mostly likely correspond to DRMs.
Figure 2.
Distribution of ubiquitinated proteins and Lyn upon activation with multivalent antigen. (A) RBL cells were transfected with a HA-tagged ubiquitin chimera. Sensitized RBL cells were activated (+Ag) or not (−Ag) with DNP-BSA for less than 5 min at room temperature and then fixed and processed for immunolabeling with FITC-conjugated anti-DNP and polyclonal anti-HA antibodies plus tetramethyl isothiocyanate (TRITC)-conjugated anti-rabbit antibodies. Two magnifications are presented for activated cells. The red channel (Left), the green channel (Center), and the merged channels (Right) are shown. (B) RBL cells were activated, fixed, and immunolabeled with FITC-conjugated anti-DNP and polyclonal anti-Lyn antibodies plus TRITC-conjugated anti-rabbit antibodies (with or without MeCD treatment) or with polyclonal anti-HA antibodies plus FITC-conjugated anti-rabbit antibodies and monoclonal anti-Lyn antibodies plus TRITC-conjugated anti-rat antibodies. Only merged pictures are shown. (Bars = 10 μm.)
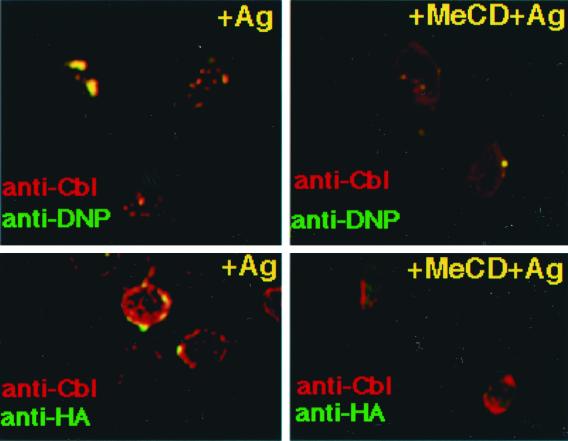
After FcɛRI engagement, a dramatic change in Cbl association with DRMs was observed. If cholesterol was extracted by using MeCD before clustering, little Cbl was found in DRMs (Fig. 1D). Furthermore, as determined by immunofluorescence microscopy, Cbl colocalized with FcɛRI patches after IgE binding as judged by the copatching with DNP-BSA (Fig. 3). Partial copatching of Cbl was also observed with ubiquitinated proteins and with Lyn (Fig. 3 and unpublished results).
Figure 3.
Distribution of ubiquitinated proteins and Cbl upon activation with multivalent antigen with or without MeCD treatment. Activated RBL cells were immunolabeled with polyclonal anti-Cbl antibodies plus TRITC-conjugated anti-rabbit antibodies. Costaining was performed by using either FITC-conjugated anti-DNP antibodies or monoclonal anti-HA primary plus FITC-conjugated secondary antibodies. Merged pictures are shown. (Bars = 10 μm.)
Nedd4 is endogenously expressed in RBL cells and was found to be associated with DRMs in a cholesterol-dependent manner (Fig. 1E). Moreover, the immunolabeling patterns also suggested that both FcɛRI and Nedd4 colocalize upon FcɛRI clustering (Fig. 4).
Figure 4.
Distribution of ubiquitinated proteins and Nedd4 upon activation with multivalent antigen with or without MeCD treatment. Activated RBL cells were immunolabeled with FITC-conjugated anti-DNP and polyclonal anti-Nedd4 plus TRITC-conjugated anti-rabbit antibodies. The red channel (Left), the green channel (Center), and the merged channels (Right) are shown. (Bars = 10 μm.)
What could be the role of ubiquitination occurring within rafts? Ubiquitination could act as a dynamic signal triggering lateral removal of proteins from rafts, thereby limiting the duration of the signaling response. According to such an hypothesis, the epidermal growth factor receptor that has been shown associated with rafts was recently proposed to be transiently ubiquitinated before internalization (14). Alternatively, ubiquitin has been proposed to operate as an endocytic signal (15). The ubiquitinated γ chain of the FcɛRI has been shown to consist of species carrying one or two ubiquitins (3). Interestingly, monoubiquitin seems to contain the endocytosis signal, whereas tetraubiquitin constitutes the minimal proteasomal targeting signal (16, 17). Cross-linked FcɛRI undergoes rapid endocytosis transporting the FcɛRI-IgE complex to proteolytic degradation in lysosomes (18). Based on our observations, therefore, one may speculate that phosphorylation-dependent ubiquitination regulates the half-life of clustered cell surface FcɛRI, and possibly of other partners implicated in the signaling pathway. The ligase candidate for mediating FcɛRI ubiquitination could be Cbl via a binding involving its COOH terminus to the SH2 domain present in the FcɛRIβ subunit. This ubiquitin ligase has been demonstrated to be present in the complex because it binds Syk that is associated with immunoreceptor tyrosine-based activation motifs (6). Neither the β chain nor the γ chain of the rat FcɛRI carries PPXY or PxxP motifs that could interact with the WW domain of Nedd4, although a C2 domain-mediated FcɛRI-Nedd4 binding cannot be excluded. Nedd4 could be required to ubiquitinate proteins involved in intercellular adhesion or in downstream events.
Ubiquitin-dependent regulation of cell signaling within rafts might well be generalized to other receptors. Among the immunoreceptors, the T cell receptor undergoes phosphorylation-dependent ubiquitination (19), possibly also involving Cbl. Interestingly, expression of a constitutively active form of p56lck causes internalization and lysosomal degradation of the assembled cell surface T cell receptors (20). It is worth noting that protein-tyrosine kinases have been shown to interact with ubiquitin-binding proteins, e.g., the p56lck binding protein p62 (21). Ubiquitination might also play a role in the context of CD40-mediated B-cell/T-cell interaction. Indeed, the protein adaptor TRAF2 which triggers association of CD40 with rafts (22) contains zinc RING domain which in other proteins, e.g., Cbl, participate in cis- and trans-ubiquitination (5). Other classes of raft-associated receptors could be targets of ubiquitin-based regulatory mechanisms as it has recently been reported that Cbl regulates insulin-stimulated glucose transport (23). When insulin binds to its kinase receptor, Cbl is recruited by the adaptor protein CAR. Upon phosphorylation of Cbl, the CAR-Cbl complex dissociates from the receptor and moves to DRMs where the complex binds the raft-associated protein flotillin. Altogether, these studies and results presented here emphasize the role of ubiquitination as a regulator of the activation of raft-associated receptors.
How ubiquitin exactly operates in the maintenance of the signaling response within rafts remains to be clarified. Would inhibition of ubiquitination lead to sustained activation of the signaling response? Is ubiquitination a general way to remove raft proteins from the plasma membrane for degradation in lysosomes? Studies of these issues should give new clues to how down-regulation of raft signal responses is achieved.
Acknowledgments
F.L. thanks F. G. van der Goot for her support and J.-M. Matter for sharing equipment. N. Fivaz, T. Harder, D. Rotin, and F. G. van der Goot are acknowledged for critical reading of the manuscript. This work was supported by a European Union network grant and a grant from the Deutsche Forschungsgemainschaft (SFB352).
Abbreviations
- DNP
dinitrophenol
- DRM
detergent-resistant membrane
- HA
hemagglutinin
- MeCD
Me-β-cyclodextrin
- RBL
rat basophilic leukemia
- TRITC
tetramethyl isothiocyanate
References
- 1.Sheets E D, Holowka D, Baird B. Curr Opin Chem Biol. 1999;3:95–99. doi: 10.1016/s1367-5931(99)80017-9. [DOI] [PubMed] [Google Scholar]
- 2.Simons K, Toomre D. Nat Rev. 2000;1:31–41. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 3.Paolini R, Kinet J-P. EMBO J. 1993;12:779–786. doi: 10.1002/j.1460-2075.1993.tb05712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freemont P S. Curr Biol. 2000;10:R84–R87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- 5.Ota Y, Beitz L O, Scharenberg A M, Donovan J A, Kinet J P, Samelson L E. J Exp Med. 1996;184:1713–1723. doi: 10.1084/jem.184.5.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ota Y, Samelson L E. Science. 1997;276:418–420. doi: 10.1126/science.276.5311.418. [DOI] [PubMed] [Google Scholar]
- 7.Harvey K F, Kumar S. Trends Cell Biol. 1999;9:166–169. doi: 10.1016/s0962-8924(99)01541-x. [DOI] [PubMed] [Google Scholar]
- 8.Plant P, Lafont F, Lecat S, Verkade P, Simons K, Rotin D. J Cell Biol. 2000;149:1473–1484. doi: 10.1083/jcb.149.7.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodel S V, Skretting G, Garred O, Vilhardt F, van Deurs B, Sandvig K. Mol Biol Cell. 1999;10:961–974. doi: 10.1091/mbc.10.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Subtil A, Gaidarov I, Kobylarz K, Lampson M A, Keen J H, McGraw T E. Proc Natl Acad Sci USA. 1999;96:6775–6780. doi: 10.1073/pnas.96.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verkade P, Harder T, Lafont F, Simons K. J Cell Biol. 2000;148:727–739. doi: 10.1083/jcb.148.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lafont F, Verkade P, Galli T, Wimmer C, Louvard D, Simons K. Proc Natl Acad Sci USA. 1999;96:3734–3738. doi: 10.1073/pnas.96.7.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheets E D, Holowka D, Baird B. J Cell Biol. 1999;145:877–887. doi: 10.1083/jcb.145.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stang E, Johannessen L E, Knardal S L, Madshus I H. J Biol Chem. 2000;275:13940–13947. doi: 10.1074/jbc.275.18.13940. [DOI] [PubMed] [Google Scholar]
- 15.Hicke L. FASEB J. 1997;11:1215–1225. doi: 10.1096/fasebj.11.14.9409540. [DOI] [PubMed] [Google Scholar]
- 16.Shih S C, Sloper-Mould K E, Hicke L. EMBO J. 2000;19:187–198. doi: 10.1093/emboj/19.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thrower J S, Hoffman L, Rechsteiner M, Pickart C M. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isersky C, Rivera J, Segal D M, Triche T. J Immunol. 1983;131:388–396. [PubMed] [Google Scholar]
- 19.Cenciarelli C, Wilhelm K G, Jr, Guo A, Weissman A M. J Biol Chem. 1996;271:8709–8713. doi: 10.1074/jbc.271.15.8709. [DOI] [PubMed] [Google Scholar]
- 20.D'Oro U, Vacchio M S, Weissman A M, Ashwell J D. Immunity. 1997;7:619–628. doi: 10.1016/s1074-7613(00)80383-0. [DOI] [PubMed] [Google Scholar]
- 21.Vadlamudi R K, Joung I, Strominger J L, Shin J. J Biol Chem. 1996;271:20235–20237. doi: 10.1074/jbc.271.34.20235. [DOI] [PubMed] [Google Scholar]
- 22.Hostager B S, Catlett I M, Bishop G A. J Biol Chem. 2000;275:15392–15398. doi: 10.1074/jbc.M909520199. [DOI] [PubMed] [Google Scholar]
- 23.Baumann C A, Ribon V, Kanzaki M, Thurmond D C, Mora S, Shigematsu S, Bickel P E, Pessin J E, Saltiel A R. Nature (London) 2000;407:202–207. doi: 10.1038/35025089. [DOI] [PubMed] [Google Scholar]