Abstract
Histone methylation recognition is accomplished by a number of evolutionarily conserved protein domains, including those belonging to the methylated lysine-binding Royal family of structural folds. One well-known member of the Royal family, the chromodomain, is found in the HP1/Chromobox and CHD subfamilies of proteins, in addition to a small number of other proteins that are involved in chromatin remodeling and gene transcriptional silencing. Here we discuss the structure and function of the chromodomain within these proteins as histone methylated lysine binders, and how the functions of these chromodomains can be modulated by additional post-translational modifications or binding to nucleic acids.
At the heart of epigenetic transcriptional regulation is the recognition of chromatin in active or repressed states by proteins that directly or indirectly affect changes on transcription. Whether chromatin is in an active or repressed state is reflected by the covalent modifications found on the tails and cores of the DNA-packing histones. The deposition or removal of these modifications coincide with gene transcriptional activation or silencing. In recent years, a number of evolutionarily conserved domains have been identified that interact with specific histone modifications associated with a particular transcriptional state. Furthermore, many of these protein domains are capable of recognizing multiple levels of valency (e.g. unmodified, mono-, di- and trimethylation). Proteins often contain tandem repeat domains or multiple domains that have distinct, specialized histone recognition modes. Two adjacent domains in combination may be required to recognize a single histone modification, typically with one domain recognizing the modification and the other binding neighboring residues in the histone sequence. Single domains have also been shown to recognize two proximal modifications on the same histone tail. Although structural studies have yet to show tandem domain recognition of multiple modifications, it is believed that not only could this be possible, but that these modifications may occur on trans histone tails. Therefore, recognition of combinatorial modifications offers a mechanism by which gene transcription can be controlled in a subtly complex manner (1).
Chromodomains (2) are histone methylated-lysine recognition modules that belong to a larger, structurally related family of protein domains referred to as the Royal family (3). The Royal family includes Tudor, chromo-, Malignant brain tumor (MBT), PWWP and Agenet domains and is descended from a common ancestral fold with an evolutionarily conserved ability to recognize methylated ligands. The basic fold of these Royal domains includes a curved three-stranded β-sheet and an adjacent helix; for the non-chromodomain Royal folds, additional strands help form a β-barrel structure sometimes referred to as a Tudor barrel. The methylated ligand is coordinated by well-conserved two, three or four conserved aromatic residues that form a “cage” around the moiety. A detailed summary of the different folds involved in methylated histone lysine (and other histone modification) recognition has been covered by a recent structure-based review (1). Here, we will describe recent advancements in our understanding of the structure and function of chromodomains in transcriptional regulation.
Chromodomains are found in a relatively small and specific group of proteins
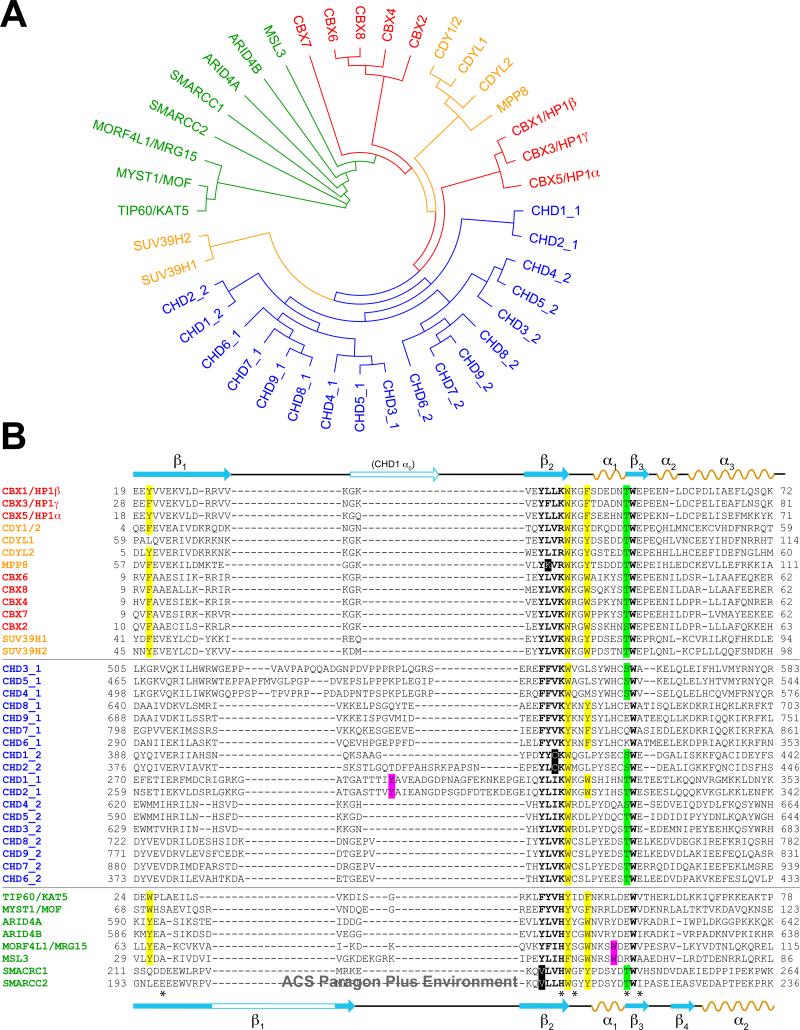
The number of proteins that contain chromodomains within the human genome is surprisingly low given the complexity of site and state-specific histone lysine methylation in gene transcription, and as compared to that of the acetyl-lysine binding bromodomain family in humans (4). A phylogenetic tree (Figure 1A) generated by a sequence-based alignment of all known chromodomains in the human genome and a minor structural variant, the chromo barrel domain (Figure 1B), suggests that subfamily classification may give clues to the functions of these domains within encompassing proteins. We propose three separate subfamilies: The Heterochromatin protein 1 (HP1)/Chromobox (CBX) subfamily; the Chromodomain helicase-DNA-binding (CHD) subfamily; and the chromo barrel domain family. The chromodomains of HP1 and the CBX homologs of Drosophila Polycomb (Pc) are highly related and thus are believed to have arisen from a common ancestor, with its initial branch shared by the chromodomain-Y-linked (CDY) proteins and the histone H3 lysine 9 (H3K9) methyltransferase Suppressor of variegation 3-9 (suv39) homologs. While structurally and functionally much more is understood of the HP1/CBX proteins in their involvement in methylated histone H3K9 or K27 recognition, a methylated histone binding role is expected for other uncharacterized chromodomains within this subset given their ability in vitro to bind histone methylated lysines, and their colocalization with these histone marks (5-8). The subfamily defined by the ATP-dependent chromatin remodeling CHD proteins (9) are unique to the superfamily by possessing two tandem chromodomains. Earlier work (10, 11) has determined that these chromodomains could work cooperatively to recognize methylated histones. Within the CHD subfamily are three distinct groups, i.e. CHD1/2, CHD3/4/5, and CHD6/7/8/9. CHD1 is structurally better characterized than the others, and together with CHD2 are also unique in their extended linker sequences found between the first two β-strands (β1, β2) of the first chromodomain (Figure 1B). Furthermore, the first chromodomains of CHD1 and CHD2 seem closely related to the second chromodomains of the other CHD proteins, although the significance of this has yet to be established.
Figure 1.
Chromodomains and chromo barrel domains in the human genome. A, Unrooted phylogenetic tree of chromodomains and chromo barrel domains (in green). B, Alignment of sequences used to generate the tree in A. Residue numbers of first and last amino acids in the alignment are noted. Secondary structure elements (orange, denoted α for helices; cyan, denoted β for strands) defined for HP1β (from PDB code 3F2U), Pc (1PDQ) and CHD1 (2B2W) are shown at top, and those defined for MSL3 chromo barrel domain (3M9P) are shown at bottom. Subgroups within the chromodomain superfamily are colored separately. Residues highlighted in yellow form the aromatic cage; those in magenta are additional aromatic residues known from structural studies to supplement the coordination of the methylated lysine; those in bold are well-conserved residues structurally on the periphery of the aromatic cage, with non-conserved residues highlighted in black; those in green are conserved phosphorylatable residues at the position of a residue known to be phosphorylated by casein kinase II in HP1. Those marked with an asterisk are notable residues conserved in chromodomains but not among chromo barrel domains.
The third subfamily is characterized by several proteins that contain domains inconsistently classified as chromodomains by numerous automated domain predictor programs (12), but have been described structurally as chromo barrel domains (13). These domains of Mortality factor 4-like protein 1 (MORF4L1; MRG15 or Eaf3 in yeast) and Male-specific lethal 3 (MSL3) strongly resemble the chromodomain and interact with methylated histone peptides, although their coordination is distinct from that of the classical chromodomain (discussed below). AT rich interactive domain 4A and 4B (ARID4A and ARID4B), of which little is known, might be expected to have similar methylated lysine binding properties based on their sequence alignment, which may provide clues to their function. Key residues that are not conserved in these domains but are found in HP1 and related proteins provided the first indication that the fold was unlikely to be a canonical chromodomain (14) (see below and Figure 1B).
We note that the related chromoshadow domain, found C-terminal to the chromodomain of HP1 isoforms, would reside on a separate branch of the phylogenetic tree if included with the chromodomains, despite differing only slightly from the canonical topology with the addition of an α-helix N-terminal to β1 (15, 16). Only the second of the three aromatic cage residues is conserved, and the domain does not recognize methylated lysines. Instead, it forms a dimer to function as a protein-protein interaction domain.
Chromodomain-containing proteins are generally involved in chromatin remodeling
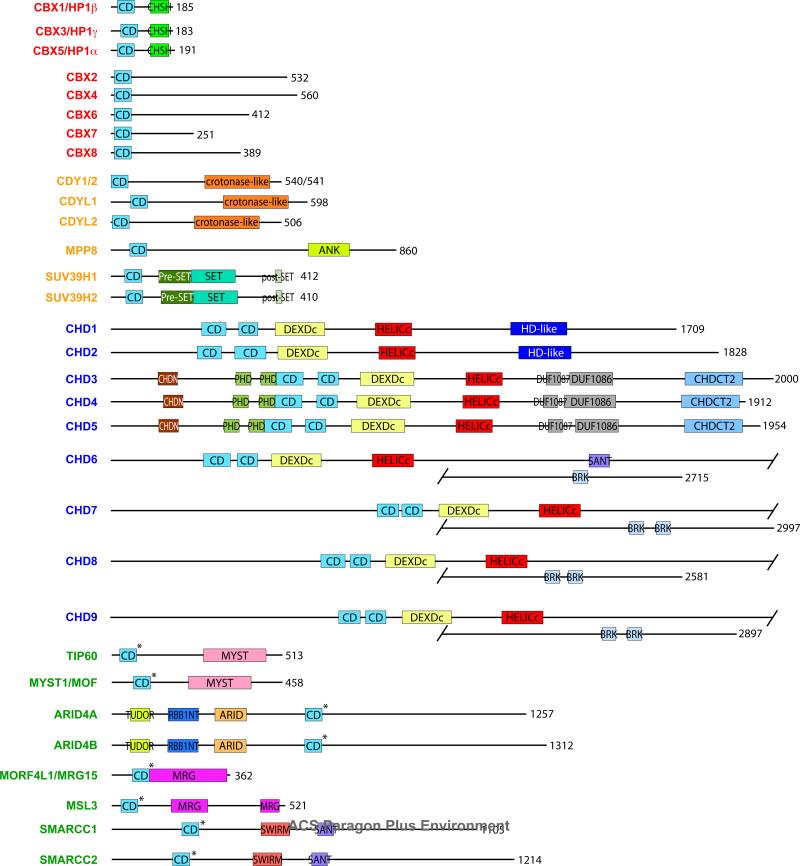
Chromodomain-containing proteins generally are modular and many contain domain repeats in tandem (Figure 2). We observe that chromodomains belonging to the same subfamily (Figure 1) exhibit similar domain layouts, including the CHD protein subgroups (11). It is interesting to note that all of the proteins contain chromodomains at their N-terminus, with an exception of ARID4A and ARID4B, which have them in the middle of their sequences, and CHD3 and CHD4, whose tandem chromodomains are preceded by an N-terminal NUC034/HMG-box helicase (CHDN) domain and a tandem pair of Plant homeodomain (PHD) fingers.
Figure 2.
Domain layout of human chromodomain-containing proteins. Chromo barrel domains are denoted by asterisks. CD: chromodomain. RBB1NT: N-terminal to ARID/BRIGHT domain in Rb-binding protein 1 family. CHSH: chromoshadow domain. ANK: four ankyrin repeats. DEXDc: DEAD-like helicase domain. HELICc: helicase domain. HD-like: homeodomain-like. CHDN: N-terminal NUC034/HMG-box helicase domain. DUF1087, DUF1086: domains of unknown function. CHDCT2: C-terminal NUC038 helicase domain.
Some proteins contain related folds within their sequences. ARID4A and ARID4B contain an N-terminal Tudor domain, which is structurally related to the chromodomain and can recognize methylated lysines on both histone and non-histone proteins; and the HP1 proteins contain the chromoshadow domain at its C-terminus. Other proteins contain domains that have been associated with chromatin remodeling, such as PHD fingers (found in CHD3 and CHD4), which can recognize methylated and unmethylated histone H3K4; Swi3p, Rsc8p and Moira (SWIRM) and Swi3, Ada2, N-CorR, TFIIIB (SANT) domains (found in SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1 and 2, or SMARCC1 and SMARCC2), which recognize nucleosomal DNA, and the pre-Suv39, Enhancerof-zeste, Trithorax (SET), SET and post-SET domains (found in Suv39 homologs 1 and 2) which are associated with the catalytic activity of histone methyltransferases.
The functions of human chromodomain-containing proteins are predominantly related to chromatin remodeling and gene transcription, although several proteins are known for their involvement in DNA damage repair, differentiation and senescence (Table 1). Most function as part of larger multiprotein complexes that effect transcriptional repression. Given these essential roles, up- or down-regulation of gene expression is frequently observed in cancer and numerous other diseases. Beyond the human genome, interestingly, very few chromodomains have been found in proteins outside the nucleus, with one notable exception being the chloroplast signal recognition particle protein cpSRP43 in Arabidopsis (17). Its three chromodomains are most closely related to the second chromodomains of CHD6-9, although no methylated or unmethylated substrate has been identified.
Table 1.
Functions of Chromodomain-containing Proteins
| Proteina | Functions in mammalsb | Functional Contributions of CDs | Human Disease Relevance |
|---|---|---|---|
| CBX1 (HP1β) Refs (19, 58, 71, 78-83) | (All HP1 isoforms) Heterochromatin formation and transcriptional repression; interacts with SUV39H1; interacts with KAP1, DNMT1 for euchromatic gene silencing; recruited to UV-induced DNA damage and double strand breaks; dynamic exchange with other HP1 isoforms for maintaining stable heterochromatic state; cell cycle-dependent localization. | (All HP1 isoforms) H3K9me binding, ejected by H3S10ph during M phase; possible localization by binding to H1.4K26me, ejected by H1.4S27ph; binds H3K23me1; multimerization mediated by CD for heterochromatin spreading (yeast). (CBX1-specific) T51 phosphorylation with mobilization releases HP1β from chromatin. | Low expression associated with melanoma progression. |
| CBX3 (HP1γ) Ref (19, 58, 79, 81-84) | See CBX1. (CBX3-specific) Regulation of cell differentiation. | See CBX1. | |
| CBX5 (HP1α) Refs (19, 58, 79, 81-83, 85, 86) | See CBX1. (CBX5-specific) Interacts with linker H1; neuronal terminal differentiation; interacts with BRG1 to negatively regulate SWI/SNF chromatin remodeling. | See CBX1. | Alters invasive potential of breast cancer cells requiring HP1 dimerization but not PXVXL interaction. |
| CBX2 (Pc1/M33) Refs (5, 35, 72, 87-89) | (All Pc isoforms) Chromatin recruitment module of Polycomb Repressive Complex 1, interacts with Ring1B; different isoforms have distinct localization and mobility patterns within chromatin; enriched on Xi (except CBX4). (CBX2-specific) Repression of ovarian development in XY gonads possibly by regulation of SRY or WT1. | (All Pc isoforms) H3K27me reader of PRC1. Differential affinities for H3K9me, H3K27me and RNA (except CBX2 cannot bind RNA). (CBX2-specific) S42 phosphorylation leads to minor changes in affinity for H3K9me and H3K27me. | Overexpression in diploid breast carcinoma. |
| CBX4 (Pc2) Refs (5, 35, 87, 90, 91) | See CBX2. (CBX4-specific) Target of SENP2 desumoylation enzyme in cardiac development; SUMO E3 ligase; tumor suppressor. | See CBX2. | |
| CBX6 Refs (5, 35, 87, 92) | See CBX2. (CBX6-specific) Distinct distribution and mobility properties, weaker interaction with endogenous Ring1B and Polycomb group target genes suggest a different role from other Pc isoforms, although chromobox can still bind Ring1B. | Binds H3K27me3 weakly. | |
| CBX7 Refs (5, 93) | See CBX2. (CBX7-specific) Cellular lifespan extension and senescence; regulation of Ink4a/ARF locus; binds Xist RNA in X inactivation. | See CBX2. (CBX7-specific) Association with RNA. | Up- or downregulation in several cancers. Marker of poor prognosis. Initiates repression of genes silenced with cancer-specific DNA hypermethylation. |
| CBX8 (Pc3) Refs (5, 35, 94) | See CBX2. (CBX8-specific) Regulation of Ink4a/ARF locus, dependent on Bmi1. | See CBX2. (CBX8-specific) Necessary for nuclear localization. | |
| CDY1/2 Refs (6, 7, 95) | HAT with preference for H4, H4 hyperacetylation during spermatogenesis; colocalizes with CBX1/HP1b. | Binds H3K9me1/2/3, H1.4K26me3, H3K27me2/3, G9aK185me1/3. | Marker for various male sex chromosomal abnormalities. |
| CDYL1 (CDYL) Refs (95-97) | Spermatogenesis; transcriptional co-repressor, binds HDACs and CoA; HAT activity in vitro; REST corepressor that interacts with REST and G9a. | Methylation by G9a outside CD abolishes H3K9me3 interaction. | |
| CDYL2 Refs (7, 96) | Spermatogenesis. | Binds H3K9me1/2/3, H1.4K26me3, H3K27me2/3, G9aK185me1/3. | |
| MPP8 Refs (98) | Probable involvement in M-phase functions, phosphorylation-dependent; localized to nucleus during interphase, throughout the cell during M phase. | Binds H3K4me3; H3K9me2/3 binding leads to recruitment of E-cadherin, then DNMT3A. | Up-regulated in carcinomas, function in tumor progression, repress tumor suppressor gene expression. |
| SUV39H1 (KMT1A) Refs (99-101) | HMT for H3K9me3 to establish constitutive pericentric heterochromatin; S-phase gene silencing during differentiation. | Binding to H3K9me essential for spreading of heterochromatin (yeast Clr4). | ERalpha transcription in breast cancer; RB1 mutants found in human cancers can't bind SUV39H1; higher expression in colorectal tumors; lymphomagenesis. |
| SUV39H2 (KMT1B) Refs (100, 102) | Formation of pericentric heterochromatin via H3K9 trimethylation. | SNP in 3’-UTR associated with increase in lung cancer risk. | |
| CHD1 Refs (10, 103-105) | Regulation of RNA polymerase II transcription; ATP-dependent chromatin assembly; pluripotency of ESCs. | Tandem CDs recognize H3K4me3. Acidic linker helix gates DNA access to ATPase motor. | |
| CHD2 Refs (11, 106, 107) | Development, hematopoiesis, tumor suppression; kidney function. | Binds H3K4me3 more weakly than CHD1, possibly gated by phosphorylation. | Differential expression in urinary bladder cancer; translocation disruption results in scoliosis. |
| CHD3 (Mi-2a) Refs (10, 46, 108-110) | NuRD HDAC and repressive complex; transcriptional repression or co-activator of c-Myb. | DNA binding by Drosophila Mi-2 CDs in nucleosome binding and mobilization. | Dermatomyositis autoimmune disease resulting in autoantibodies against CHD3/4. |
| CHD4 (Mi-2b) Refs (46, 108-111) | NuRD complex; checkpoint signaling and DNA damage repair; promotes CD4 gene expression during T cell development. | See CHD3. | See CHD3. |
| CHD5 Refs (46, 112) | Tumor suppressor that controls proliferation, apoptosis and senescence via the p19ARF/p53 pathway. | See CHD3. | Downregulated through promoter hypermethylation, mutation in several types of cancer. |
| CHD6 Refs (11, 113, 114) | Interacts with Nrf2 transcription factor in cellular redox homeostasis; cell proliferation and radiosensitivity; transcription preinitiation and elongation via RNA pol II. | Possible shared coordination of methylated lysine by both CDs. | TCF4 translocation leads to mild retardation related to Pitt-Hopkins syndrome. |
| CHD7 Refs (11, 115-118) | Neural crest formation and cell motility; inner ear development; ribosomal RNA biogenesis; enhancer mediated transcription. | See CHD6. | Mutations in CHARGE and Kallmann syndromes; idiopathic scoliosis. |
| CHD8 Refs (119-122) | AR-mediated or beta catenin-mediated transcription; HOXA2 and cyclin E2 expression; interacts with CHD7; suppresses p53-mediated apoptosis. | H3K4me2 binding to both CDs similar to CHD1, possible chromatin recruitment. | Idiopathic developmental delay and cognitive impairment. |
| CHD9 Ref (11, 123) | Osteogenic cell differentiation. | See CHD6. | |
| TIP60 (KAT5) Refs (124, 125) | NuA4 HAT complex; tumor suppressor, apoptosis, DNA repair, cell cycle progression; H2A and H4 HAT, acetylation and activation of ATM. | H3K9me3 binding activates HAT, dependent on DNA damage-induced displacement of HP1b from H3K9me3. | Low levels in several cancers; prostate cancer cell proliferation; implicated in Alzheimer's disease |
| MYST1 (MOF/MOZ) Refs (126-128) | MSL H4K16 HAT complex; cell cycle, DNA repair; associates with H3K4 HMT MLL1 for transcriptional activation. | Interaction with ATM. | |
| ARID4A (RBBP1) Refs (129-131) | mSIN3 HDAC complex; binds Rb, promotes repression and growth arrest. | Leukemia suppressor gene | |
| ARID4B (RBBP1L1) Refs (130, 131) | mSIN3 HDAC complex | Molecular marker for several cancers; leukemia suppressor gene | |
| MORF4L1 (Eaf3/MRG15) Refs (132, 133) | NuA4 HAT and mSIN3 complexes; transcriptional repression, cell proliferation and aging, DNA damage repair, gene splicing. | Assembly of MAF2 complex (with MOF) and HAT activity | |
| MSL3 (MSL3L1) Refs (128, 134-136) | MSL H4K16 HAT complex; X inactivation. | Nucleic acid (RNA, ssDNA) binding. H3K36me3 binding, X chromosome gene association, MSL complex spreading to active genes in cis. | |
| SMARCC1 (BAF155) Refs (137, 138) | SWI/SNF, WINAC, BRG complexes; transcriptional activation and repression, heterochromatin formation, chromatin compaction during differentiation. | Up-regulated in prostate cancer, tumor recurrence. | |
| SMARCC2 (BAF170) Refs (138-140) | SWI/SNF, WINAC, BRG complexes; transcriptional activation and repression. | Reduced cell viability in primary chronic lymphocytic leukemia cells. Dysregulation in testicular germ cell, squamous non-small cell lung cancers. |
References cited are representative only. We apologize to all authors whose work are not cited due to space limitations.
Abbreviations: CD, chromodomain. HAT, histone acetyltransferase. HMT, histone methyltransferase. HDAC, histone deacetylase.
Contributions of the chromodomain to protein function
The prototypical chromodomain, described within the HP1 proteins, recognizes di- and tri-methylated histone H3K9 (18, 19), which is essential for the recruitment of HP1 to sites of repressed chromatin (e.g. heterochromatin, for which HP1 is named). Similarly, the CBX proteins, which contain a sequence- and structurally-similar chromodomain at their N-termini, are known to be recruited to di- or tri-methylated H3K27 and function in a Polycomb repressive multi-protein complex involved in the regulation of genes associated with development and differentiation (20). CDY1 was also shown to bind to H3K9me (7), which points to a role in gene repression. Human CHD1 has been shown to recognize H3K4me, although, surprisingly, a single mark is bound by the tandem chromodomains (10) (see below). Unlike H3K9me and H3K27me, this mark is traditionally associated with active transcription. There is conflicting evidence regarding the ability of Chd1 to recognize H3K4me in yeast (10, 11, 21-24). However, it appears that its localization and function is somewhat independent of H3K4 methylation (25-30). Nevertheless, the chromodomains appear to be important for association with actively transcribed chromatin, and this is independent of either methylation at H3K4 or K36, although these modifications may yet be important for Chd1/Rpd3S-mediated maintenance of chromatin structure during transcription (28). It is interesting to note that Rpd3S subunit Eaf3 contains a chromo barrel domain capable of binding H3K36me3 (see below). Additionally, the chromodomains of Chd1, independent of direct histone modification interactions, counteract the activity of Facilitates chromatin transcription (FACT) complex (25).
Thus far, the chromodomain has been predominantly characterized as a methylated histone lysine binding module. The chromodomain of HP1 has also been shown in vitro to bind to nonhistone protein G9a, a histone methyltransferase, bearing a similar, conserved sequence flanking a self-methylated lysine (31), although the function of this interaction is unclear. Nevertheless, this study shows that binding partners for the chromodomain are not necessarily restricted to histones, as has been shown for other structural folds. All Royal domains (except the plant Agenet domain) have been characterized as general methylated substrate binding modules, suggesting that their common ancestor also recognized methylated ligands (3). While tempting to speculate that ligands for as-yet uncharacterized chromodomains will be methylated lysines, we note that the Mi-2/nucleosome remodeling and deacetylase (NuRD) chromatin remodeling complex, of which CHD3 and CHD4 are a part, preferentially binds the unmodified H3 tail (32, 33), while neither pair of chromodomains in these proteins appear to have the residues expected to be important for methylated lysine binding (24).
Chromodomains for which a binding substrate has not yet been identified also highlight the possibility of novel functions for the proteins that contain them. For example, the retrotransposon Tf1 integrase from Schizosaccharomyces pombe, which integrates into Pol II promoter regions, contains a chromodomain that has been directly implicated in the frequency of integration by mediating Tf1 binding to DNA, as well as the selectivity of integration site targeting (34). However, since the chromodomain is unable to directly associate with either DNA or histones, it is possible that Tf1 is regulated by the recognition of an as-yet unidentified substrate and/or modification on chromatin.
The fact that the principal function defined for the chromodomain is the recognition of methylated lysine points to the possibility of cross-talk or competition between chromodomain-containing proteins and a requirement for additional interactions to confer specificity. For example, several CBX isoforms are able to bind methylated H3K27, yet they control the transcription of non-redundant genes and have unique chromatin localization patterns that depend upon multiple factors such as differentiation state and cell cycle stage (35, 36). It is believed that their unique C-terminal domains help establish this specificity. In S. pombe, the Suv39 homolog Chp1 (which contains an H3K9me-recognizing chromodomain) assists in methylation of H3K9 on newly deposited nucleosomes and recruits the RNA-induced transcriptional silencing (RITS) complex to chromatin. Acetylation at H3K4 ejects Chp1 from transcriptionally active heterochromatin, thus enabling the recruitment of Chp2/Swi6 (homologs of HP1), which is insensitive to the acetylation mark, to enable re-assembly of heterochromatin at S/G2 (37).
Structural insights into the chromodomain interaction with methylated lysine
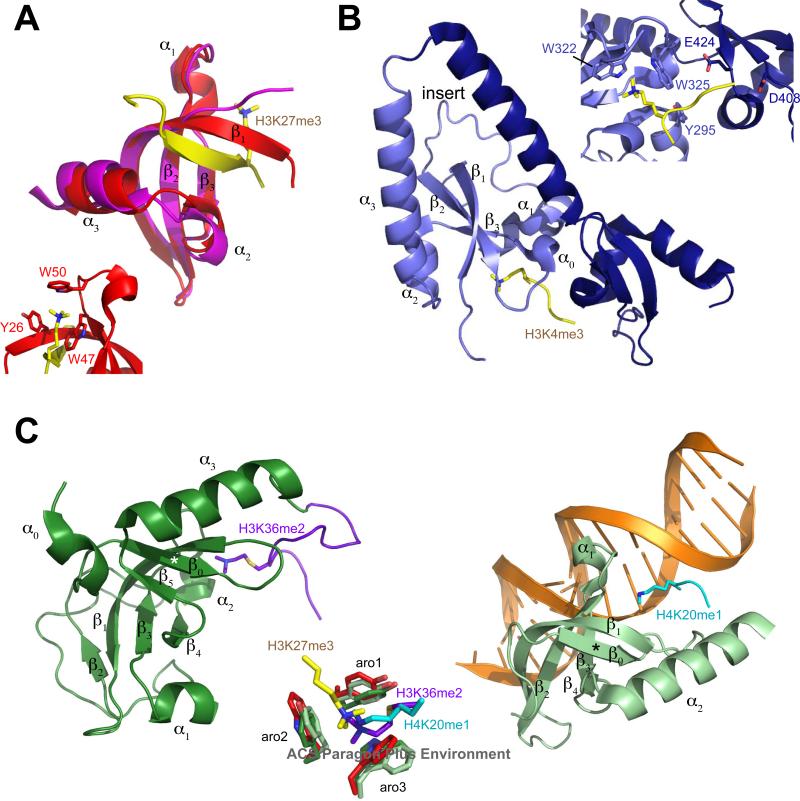
The prototypical chromodomain of HP1 was the first to be characterized structurally at an atomic level and provided insight into its function as a methylated histone lysine binding domain (14, 38, 39) (Figure 3A). The fold is a curved anti-parallel β-sheet comprising three strands (β1, β2, β3) with helix α1 occurring between β2 and β3 and helices α2 and α3 following β3 (Figure 1B). The methylated histone peptide in the complex structure is accommodated by a hydrophobic groove and forms a β-strand that either contributes to the curved β-sheet (40), mimicking the four-stranded barrel found in Tudor domains, or forms a second smaller β-sheet with β3 and the N-terminus of β1 (39) (Figure 3A). Histone recognition is accomplished by a conserved “cage” of three aromatic residue side chains (Y21, W42, F45 in HP1β; Figure 1B) that interacts with the methylated lysine moiety via cation-π interactions (41). In vitro measured affinities for di- or tri-methylated lysine ligands by a chromodomain is typically in the micromolar range for most proteins and their presumed in vivo substrates (10, 42). The HP1/CBX chromodomain interaction is virtually the same for di- or tri-methylated lysines, although the additional methyl group improves the affinity by enabling more polar and van der Waals interactions with the chromodomain binding pocket (39, 41, 43). The second and third aromatic residues are within a highly conserved loop sequence of hydrophobic and basic residues between β2 and β3, and these two are far more conserved among all chromodomains than the first aromatic residue (Figure 1B). Interestingly, the first aromatic residue, which resides at the beginning of β1, appears to be within a highly flexible region in the absence of a ligand, suggesting that the cage may not be preformed in the absence of the ligand (K. Yap and M.-M. Zhou, unpublished observations).
Figure 3.
Structures of chromodomains and chromo barrel domains. Secondary structure elements are labeled as defined in Figure 1B and the text. Residues interacting with the methylated lysine are labeled in inset images. A, Chromodomain of uncomplexed HP1β (PDB code 3F2U, magenta) and Drosophila Pc (1PDQ, red) in complex with H3K27me3 peptide (yellow). B, CHD1 (2B2W) chromodomains 1 (light purple) and 2 (dark blue) in complex with H3K4me3 (yellow). C, Left, chromo barrel domain of Eaf3 (2K3Y, green) in complex with fused H3K36me2 peptide (purple). Right, MSL3 chromo barrel domain (3M9P, light green) in complex with H4K20me1 peptide (cyan) and DNA (orange). β-strand that mimics the H3 peptide of the Pc complex is indicated with an asterisk. Center, aromatic cage residues that coordinate the methylated lysine of Pc+H3K27me3 (red/yellow), Eaf3+H3K36me2 (green/purple) and MSL3+H4K20me1 (light green/cyan) complex structures.
The interaction between chromodomain and methylated histone lysines extends beyond the methylated moiety. In all complex structures, but especially for the CBX chromodomains, several contacts are observed between the binding pocket of the chromodomain and residues up to three positions N-terminal of the methylated residue on the histone peptide. The interaction is more extensive for the CBX chromodomains, with a larger surface area buried by peptide binding (40). Additional hydrogen bonds stabilize the β-strand formed by the histone peptide to extend the β-sheet (44). In contrast, methylated ligands do not penetrate deeply into the pockets of the related MBT domains, which bind lower-state methylated lysines via a “cavity insertion recognition” mode (45).
The CHD proteins contain two tandem chromodomains at their N-terminus and have been implicated in chromatin remodeling. CHD1 is structurally the best characterized of the nine isoforms found in human, and its two chromodomains bind to a single histone H3 peptide methylated at K4 (10) (Figure 3B). The methylated lysine is coordinated in part by the latter two of the three aromatic residues (W322, W325) that are conserved in most chromodomains (Figure 1B), although E272, which aligns with the first cage residue, faces toward the methyl-lysine moiety. Y295 within helix α0, inserted between β1 and β2 of the first chromodomain, contributes to the binding of H3T3, K4me and Q5. The equivalent “cage” residues of the second chromodomain (Q389, W413, L416) do not participate in peptide coordination; instead, acidic residues within the insert (D408) and β3 (E424) are in contact with H3A1. CHD2, which is closely related to CHD1 yet contains a longer insert within its second chromodomain (Figure 1), binds H3K4me with much lower affinity and has been hypothesized to require an additional factor or modification to fully enable its interaction with H3K4me (11). CHD7 and CHD9 tandem chromodomains, which are in a subclass distinct from CHD1 and CHD2 and are related to CHD6 and CHD8 (Figure 1A) have also been demonstrated to interact in vitro with methylated histone peptides, while the chromodomains of CHD3 and CHD4, which are in a subclass with CHD5, appear to have greater affinity for DNA and do not require binding to histone tails to associate with chromatin (46).
Chromo barrel domains
The chromo barrel domain is often misidentified as a chromodomain in literature, despite their clear distinction within the phylogenetic tree (Figure 1A). SMART identifies some occurrences of these domains as Tudor-knot folds. The chromo barrel domain contains a strand (β0 in Figure 3C) that precedes the chromodomain fold, mimicking the strand formed by the methylated histone peptide in the chromodomain complex; another strand, β4, in the place of α2 of the chromodomain; the N-terminal tail displaces the C-terminal helix such that it is packed against the β-barrel (Figure 3C). The latter two of three aromatic cage residues are conserved, and the first aromatic is found only in ARID4A/B and MORF4L1/MSL3 chromo barrel domains. Previously the chromo barrel domain was characterized as an RNA-binding module, and mutation of the second aromatic cage residue of histone H4K16 acetyltransferase Males absent on first (MOF/MYST1) was shown to abolish binding to rox2 non-coding RNA (47); however the interaction of the chromo barrel domain is believed necessary but not sufficient for MOF interaction with RNA, while other regions outside the chromo barrel domain may be important (48). Furthermore, no binding to modified histones was observed, leading to the suggestion that the lack of conservation of the first and third aromatic residues may distinguish this and other chromo barrel domains in terms of functionality (13). Indeed, it was shown that other chromo barrel domains with these conserved residues could interact with methylated histone peptides in a manner somewhat similar to that of the chromodomains (49-51) but with ~10-100 fold weaker affinities. To obtain a complex structure and circumvent the problem of poor affinity, an H3K36me2 peptide was fused to the C-terminus of the chromo barrel domain of yeast Eaf3 (50) (Figure 3C, left). The structure illustrates how the methylated lysine still occupies the binding cleft coordinated by an aromatic cage, but the rest of the peptide is oriented on the periphery of the surface, and the methylated lysine side chain is rotated 100° away from its position in the chromodomain complex structure (Figure 3C, center). A fourth aromatic residue that lies just outside the third cage residue and is otherwise only conserved in the CBX chromodomains provides additional contacts with the methylated lysine. Another study showed that the Rpd3S complex requires both the PHD finger from the Rco1 subunit and the chromodomain of Eaf3 to recognize H3K36-methylated nucleosomes (52) suggesting how the interaction would occur in vivo. Recently, it was reported that the chromo barrel domain of MSL3 (a mammalian homolog of Eaf3) interacts with monomethylated H4K20, but only in the presence of DNA (53), although a longer construct or induced dimerization of the domain may alleviate this requirement (54). Unlike H3K36me2, this modification has been associated with active transcription (55, 56), genomic stability and entry into prometaphase (57). Moreover, the MSL3 chromo barrel domain in the presence of DNA does not interact with either H3K36me3 or H4K20me3 (54). The ternary structure of MSL3 chromo barrel domain, H4K20me1 peptide and DNA (Figure 3C, right) shows that the monomethyl group inserts into a four-residue cage and is additionally hydrogen-bonded by a water molecule. Despite the requirement for DNA, which packs against the histone peptide, β3 and loop between β3 and β4 and the third and fourth cage residues, the position of the methylated peptide is very similar to that of the Eaf3-fused H3K36me2 complex structure. Eaf3 contains an extensive linker that contributes an additional strand and helix folding against the β-barrel (49, 50) and would prevent a DNA interaction as observed in the MSL3 ternary complex. Another mammalian homolog of Eaf3, Mortality factor related gene 15 (MRG15), which does not contain this linker, has also been shown to bind di- or trimethylated H3K36 (51), but its DNA-binding capability has not been assessed, so it is unclear if this is a general mode of recognition for these chromo barrel domains.
Altering specificity and functionality of the chromodomain
The binary switch
Histone lysine residues that are methylated often occur adjacent to a serine or threonine residue, phosphorylation of which can block binding of the effector protein. This “binary methylation-phosphorylation switch” mechanism allows for the cell cycle-dependent regulation of chromatin binding proteins while maintaining the methylation throughout multiple generations. The binding of HP1 isoforms to chromatin is regulated in this manner; phosphorylation of H3S10 by the kinase Aurora B prevents interaction of the chromodomain with H3K9me3 during mitosis. Removal of HP1 from chromosomes during mitosis is thought to be a necessary step to allow for proper chromosome condensation and segregation. This enables the H3K9me3 mark to remain in place and recruit back HP1 when S10 is dephosphorylated at the end of mitosis (58). This binary mechanism is not restricted to interactions involving H3K9me3, as H3T3 phosophorylation has been found to regulate H3K4 methyl interactions (10, 59), while H3S28 phosphorylation has been speculated to regulate H3K27 methyl interactions with Polycomb Group proteins (60).
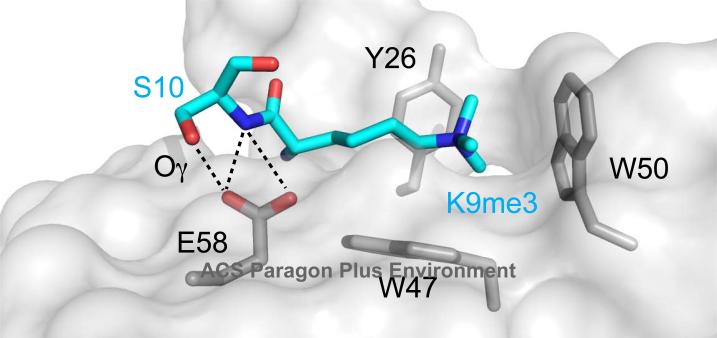
The molecular mechanism of the “binary switch” is explained by structures of HP1 chromodomain-H3K9me complexes that reveal how a bulky and negatively charged phosphate on S10 would disrupt the interaction and lead to expulsion from chromatin (Figure 4). In addition, it was proposed that the two modifications together on the tail might also assume a different conformation (61) that might further prevent association with the chromodomain. Furthermore, phosphorylation of H3T3 reduces affinity of the CHD1 tandem chromodomains for an H3K4me3 peptide about 30 fold, and the structure shows that H3T3ph inserts into a cleft formed between the two chromodomains (10). Phosphorylation of T3 during mitosis disrupts CHD1 binding to H3K4me3 in vivo, with dephosphorylation enabling CHD1 to bind again during telophase, suggesting the presence of another binary switch.
Figure 4.
Depiction of H3K9me3 and S10 side chains in the Pc-H3K27me3 structure, with Pc depicted in surface representation. Polar contacts made between the side chain of Pc E58 and H3S10 (backbone nitrogen in blue, side chain oxygen in red and denoted Oγ) illustrate how phosphorylation at S10 could disrupt the interaction. Aromatic cage residue side chains are also shown in stick representation.
Dimerization via the chromodomain
Recently, it was reported that HP1 homolog Swi6 in S. pombe binds to mononucleosomes as a tetramer, with dimerization occurring via both chromo shadow and chromodomains to establish specificity for the H3K9me mark on chromatin (62). Mutations made to residues V82, immediately adjacent to the first aromatic cage residue Y81 in β1, and Y131 within α3, result in strengthening the chromodomain-chromodomain association and increasing specificity significantly. Dimerization via the chromodomain may only occur in HP1, while Y131 is well conserved across all HP1/CBX and some CHD chromodomains, a hydrophobic residue at the V82 position is unique to HP1 isoforms. Notably, it has also been shown that both H3K9 methylation binding and oligomerization of CDYL1b is necessary for heterochromatin association, although how this would be mediated is unknown (63).
Post-translational modifications to the chromodomain
Little has been reported about post-translational modifications to the chromodomain affecting its activity, although phosphorylation, sumoylation and acetylation have been reported to occur on many chromodomain-containing proteins. Phosphorylation and N6-acetylation sites found within chromodomains are for the most part the result of genomic-scale mass spectrometry studies, functionally uncharacterized, and often on non-conserved residues. Previously a number of post-translational modifications within the HP1 isoforms were characterized, including lysine acetylation and methylation throughout both chromodomain and chromoshadow domains, and formylation at highly conserved lysines adjacent to the second aromatic cage residue (64).
Casein kinase II (CKII) is a kinase found associated with several chromodomain-containing proteins, including HP1 (65), CBX8 (66, 67) and CHD1 (68). Phosphorylation at S15 at the N-terminus of Drosophila HP1 by CKII leads to a large decrease of HP1 binding to heterochromatin (65, 69); however, this residue is not conserved in mammalian HP1 or CBX proteins. Conversely, phosphorylation at two serine residues (S11, S14) by CKII at the N-terminus of mammalian HP1α was shown to increase affinity for methylated H3K9, although the effect may be predominantly due to HP1α interaction directly with K14 and K18 on the H3 tail, C-terminal to methylated K9 (70). Phosphorylation by CKII of a well-conserved threonine, T51, on HP1β (Figure 1B) that lies just outside the aromatic cage is important for HP1β mobilization, leads to phosphorylation of histone H2AX and initiation of the DNA damage response (71). Phosphorylation or mutation of this residue has been shown to abolish binding to H3K9me3, potentially due to the disruption of hydrogen bonds that normally stabilize interaction with the methylated lysine. In contrast, another study showed that CKII treatment of mouse CBX2 results in modestly increased affinity for H3K27me3, and slightly decreased affinity for H3K9me3 (72). However, because CKII may phosphorylate CBX2 at multiple sites, this change in binding affinities may not be due to phosphorylation at the site equivalent to HP1β T51.
Binding of nucleic acids to the chromodomain
In addition to the characterization of some chromo barrel domains as RNA binding modules (see above), a handful of chromodomains have been characterized as nucleic acid binding domains, which had been suggested given the chromodomain's structural similarity to the oligonucleotide/oligosaccaride binding (OB)-fold (73). The tandem domains of Drosophila Mi-2 (a homolog of CHD3) are required for its ATPase activity and associate with nucleosomes independent of histone tail binding, instead preferentially binding DNA (46). HP1 recruitment has been suggested to be dependent on RNA transcripts, although it has not been demonstrated that any binding is direct (74) and if so, whether the interaction would occur through the chromodomain; a hinge region between the chromodomain and chromoshadow domain has been shown to bind RNA that, coordinated with methylated lysine binding by the chromodomain, enables HP1α to be recruited to heterochromatin (75). Furthermore, this hinge region may be important for non-specific DNA and linker histone binding in native chromatin in which the H3K9me mark is inaccessible (76).
The chromodomains of several mammalian CBX isoforms were shown to interact with RNA, suggesting a mechanism for how these proteins can associate with inactive X chromosome in silencing, and mutations within the aromatic cage of CBX7 result in disruption of Xi localization, suggesting that binding the non-coding RNA is important for CBX7 recruitment to target loci (5). Recently we reported that both H3K27me and RNA interactions with the CBX7 chromodomain, which employ slightly overlapping yet distinct surfaces for binding, are necessary for its function in controlling senescence and modulating p16INK4a repression (77). Binding of the two ligands is negatively cooperative, suggesting that local concentrations of either the H3K27me mark or RNA at the target locus may dictate what is bound to CBX7. Although non-coding RNA has been increasingly recognized as an important factor in transcriptional control, to our knowledge this is the only chromodomain that has been functionally characterized as both a methylated lysine binding module and an RNA binding domain, for which both interactions appear to be important for its targeting and retention at its target locus. Although structurally distinct from a chromodomain, the recent study of the ternary complex of MSL3 chromo barrel domain, H4K20me1 and DNA suggests a novel multi-nucleosomal interaction with the MSL3 protein (53) that offers yet another tantalizing model for chromodomain-mediated gene recruitment and transcriptional regulation.
Conclusions
Most chromodomains found in the human genome function as methylated histone lysine binding domains via conserved residues, facilitating recruitment to chromatin. Other functions, such as nucleic acid recognition, may be mediated by residues unique to each chromodomain. More diverse functions are associated with chromodomain-containing proteins due to other modular domains that work in concert with the methylated lysine binding capacity. It remains to be seen if other histone modifications are capable of being recognized by the chromodomain, as is the case for the PHD finger (77), but evolutionary evidence suggests this is less likely. Instead, we may expect to see more examples of nucleic acid and histone methylation co-recognition, or post-translational modifications to the chromodomain, as more accurate portrayals of chromodomain function in vivo.
ABBREVIATIONS
- ARID4A
AT rich interactive domain 4A
- CBX
Chromobox
- CDY
Chromodomain-Y-linked
- CHD
Chromodomain helicase DNA-binding
- CHDN
N-terminal NUC034/HMG-box helicase domain
- CKII
Casein kinase II
- FACT
Facilitates chromatin transcription
- HP1
Heterochromatin protein 1
- MBT
Malignant brain tumor
- MOF
Males absent on first
- MORF4L1
Mortality factor 4-like protein 1
- MRG15
Mortality factor related gene 15
- MSL3
Male-specific lethal 3
- NuRD
Mi-2/nucleosome remodeling and deacetylase
- OB-fold
Oligonucleotide/oligosaccaride binding-fold
- Pc
Polycomb
- PHD
Plant homeodomain
- RITS
RNA-induced transcriptional silencing
- SANT
Swi3, Ada2, N-CorR, TFIIIB
- SET
Su(var)3-9, Enhancer-of-zeste, Trithorax
- SMARCC1
SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1
- Su(var)3-9
Suppressor of variegation 3-9
- SWIRM
Swi3p, Rsc8p and Moira
Footnotes
The work in the authors’ laboratory is currently supported by the U.S. National Institutes of Health (HG004508-03, CA87658-10, CA80058-11, and DA028776-01 to M.-M.Z.), and the New York State Department of Health NYSTEM (N08G-498 to M.-M.Z.).
References
- 1.Yap KL, Zhou MM. Keeping it in the family: diverse histone recognition by conserved structural folds. Crit Rev Biochem Mol Biol. 2010;45:488–505. doi: 10.3109/10409238.2010.512001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paro R, Hogness D. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci U S A. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maurer-Stroh S, Dickens N, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting C. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 4.Sanchez R, Zhou MM. The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Devel. 2009;12:659–665. [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse Polycomb Proteins Bind Differentially to Methylated Histone H3 and RNA and Are Enriched in Facultative Heterochromatin. Mol. Cell. Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischle W, Franz H, Jacobs SA, Allis CD, Khorasanizadeh S. Specificity of the chromodomain Y chromosome family of chromodomains for lysine-methylated ARK(S/T) motifs. J Biol Chem. 2008;283:19626–19635. doi: 10.1074/jbc.M802655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schalch T, Job G, Noffsinger VJ, Shanker S, Kuscu C, Joshua-Tor L, Partridge JF. High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin. Mol Cell. 2009;34:36–46. doi: 10.1016/j.molcel.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delmas V, Stokes DG, Perry RP. A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SWI2-like helicase domain. Proc Natl Acad Sci USA. 1993;90:2414–2418. doi: 10.1073/pnas.90.6.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 11.Flanagan JF, Blus BJ, Kim D, Clines KL, Rastinejad F, Khorasanizadeh S. Molecular implications of evolutionary differences in CHD double chromodomains. J Mol Biol. 2007;369:334–342. doi: 10.1016/j.jmb.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brehm A, Tufteland KR, Aasland R, Becker PB. The many colours of chromodomains. Bioessays. 2004;26:133–140. doi: 10.1002/bies.10392. [DOI] [PubMed] [Google Scholar]
- 13.Nielsen P, Nietlispach D, Buscaino A, Warner R, Akhtar A, Murzin A, Murzina N, Laue E. Structure of the chromo barrel domain from the MOF acetyltransferase. J Biol Chem. 2005;280:32326–32331. doi: 10.1074/jbc.M501347200. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen P, Nietlispach D, Mott H, Callaghan J, Bannister A, Kouzarides T, Murzin A, Murzina N, Laue E. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 15.Brasher S, Smith B, Fogh R, Nietlispach D, Thiru A, Nielsen P, Broadhurst R, Ball L, Murzina N, Laue E. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cowieson NP, Partridge JF, Allshire RC, McLaughlin PJ. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr Biol. 2000;10:517–525. doi: 10.1016/s0960-9822(00)00467-x. [DOI] [PubMed] [Google Scholar]
- 17.Stengel KF, Holdermann I, Cain P, Robinson C, Wild K, Sinning I. Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43. Science. 2008;321:253–256. doi: 10.1126/science.1158640. [DOI] [PubMed] [Google Scholar]
- 18.Bannister A, Zegerman P, Partridge J, Miska E, Thomas J, Allshire R, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 19.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 20.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDaniel IE, Lee JM, Berger MS, Hanagami CK, Armstrong JA. Investigations of CHD1 function in transcription and development of Drosophila melanogaster. Genetics. 2008;178:583–587. doi: 10.1534/genetics.107.079038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pray-Grant M, Daniel J, Schieltz D, Yates J, Grant P. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 23.Santos-Rosa H, Schneider R, Bernstein BE, Karabetsou N, Morillon A, Weise C, Schreiber SL, Mellor J, Kouzarides T. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Molecular Cell. 2003;12:1325–1332. doi: 10.1016/s1097-2765(03)00438-6. [DOI] [PubMed] [Google Scholar]
- 24.Sims RJ, 3rd, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biswas D, Dutta-Biswas R, Stillman DJ. Chd1 and yFACT act in opposition in regulating transcription. Mol Cell Biol. 2007;27:6279–6287. doi: 10.1128/MCB.00978-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biswas D, Takahata S, Xin H, Dutta-Biswas R, Yu Y, Formosa T, Stillman DJ. A role for Chd1 and Set2 in negatively regulating DNA replication in Saccharomyces cerevisiae. Genetics. 2008;178:649–659. doi: 10.1534/genetics.107.084202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eissenberg JC, Lee MG, Schneider J, Ilvarsonn A, Shiekhattar R, Shilatifard A. The trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase. Nature structural & molecular biology. 2007;14:344–346. doi: 10.1038/nsmb1217. [DOI] [PubMed] [Google Scholar]
- 28.Quan TK, Hartzog GA. Histone H3K4 and K36 methylation, Chd1 and Rpd3S oppose the functions of Saccharomyces cerevisiae Spt4-Spt5 in transcription. Genetics. 2010;184:321–334. doi: 10.1534/genetics.109.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srinivasan S, Armstrong JA, Deuring R, Dahlsveen IK, McNeill H, Tamkun JW. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development. 2005;132:1623–1635. doi: 10.1242/dev.01713. [DOI] [PubMed] [Google Scholar]
- 30.Srinivasan S, Dorighi KM, Tamkun JW. Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet. 2008;4:e1000217. doi: 10.1371/journal.pgen.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sampath SC, Marazzi I, Yap KL, Sampath SC, Krutchinsky AN, Mecklenbräuker I, Viale A, Rudensky E, Zhou M-M, Chait BT, Tarakhovsky A. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol Cell. 2007;27:596–608. doi: 10.1016/j.molcel.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 32.Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis C, Tempst P, Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zegerman P, Canas B, Pappin D, Kouzarides T. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J Biol Chem. 2002;277:11621–11624. doi: 10.1074/jbc.C200045200. [DOI] [PubMed] [Google Scholar]
- 34.Chatterjee AG, Leem YE, Kelly FD, Levin HL. The chromodomain of Tf1 integrase promotes binding to cDNA and mediates target site selection. J Virol. 2009;83:2675–2685. doi: 10.1128/JVI.01588-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren X, Vincenz C, Kerppola TK. Changes in the distributions and dynamics of polycomb repressive complexes during embryonic stem cell differentiation. Mol Cell Biol. 2008;28:2884–2895. doi: 10.1128/MCB.00949-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincenz C, Kerppola TK. Different polycomb group CBX family proteins associate with distinct regions of chromatin using nonhomologous protein sequences. Proc Natl Acad Sci USA. 2008;105:16572–16577. doi: 10.1073/pnas.0805317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xhemalce B, Kouzarides T. A chromodomain switch mediated by histone H3 Lys 4 acetylation regulates heterochromatin assembly. Genes Dev. 2010;24:647–652. doi: 10.1101/gad.1881710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ball LJ, Murzina NV, Broadhurst RW, Raine AR, Archer SJ, Stott FJ, Murzin AG, Singh PB, Domaille PJ, Laue ED. Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. EMBO J. 1997;16:2473–2481. doi: 10.1093/emboj/16.9.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobs S, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 40.Fischle W, Wang Y, Jacobs S, Kim Y, Allis C, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hughes R, Benshoff M, Waters M. Effects of chain length and N-methylation on a cation-pi interaction in a beta-hairpin peptide. Chemistry (Weinheim an der Bergstrasse, Germany) 2007;13:5753–5764. doi: 10.1002/chem.200601753. [DOI] [PubMed] [Google Scholar]
- 42.Bernstein B, Mikkelsen T, Xie X, Kamal M, Huebert D, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber S, Lander E. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 43.Hughes RM, Wiggins KR, Khorasanizadeh S, Waters ML. Recognition of trimethyllysine by a chromodomain is not driven by the hydrophobic effect. Proc Natl Acad Sci USA. 2007;104:11184–11188. doi: 10.1073/pnas.0610850104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouazoune K, Mitterweger A, Längst G, Imhof A, Akhtar A, Becker PB, Brehm A. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J. 2002;21:2430–2440. doi: 10.1093/emboj/21.10.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akhtar A, Zink D, Becker P. Chromodomains are protein-RNA interaction modules. Nature. 2000;407:405–409. doi: 10.1038/35030169. [DOI] [PubMed] [Google Scholar]
- 48.Buscaino A, Kocher T, Kind JH, Holz H, Taipale M, Wagner K, Wilm M, Akhtar A. MOF-regulated acetylation of MSL-3 in the Drosophila dosage compensation complex. Mol Cell. 2003;11:1265–1277. doi: 10.1016/s1097-2765(03)00140-0. [DOI] [PubMed] [Google Scholar]
- 49.Sun B, Hong J, Zhang P, Dong X, Shen X, Lin D, Ding J. Molecular basis of the interaction of Saccharomyces cerevisiae Eaf3 chromo domain with methylated H3K36. J Biol Chem. 2008;283:36504–36512. doi: 10.1074/jbc.M806564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu C, Cui G, Botuyan MV, Mer G. Structural basis for the recognition of methylated histone H3K36 by the Eaf3 subunit of histone deacetylase complex Rpd3S. Structure. 2008;16:1740–1750. doi: 10.1016/j.str.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang P, Du J, Sun B, Dong X, Xu G, Zhou J, Huang Q, Liu Q, Hao Q, Ding J. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res. 2006;34:6621–6628. doi: 10.1093/nar/gkl989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li B, Gogol M, Carey M, Lee D, Seidel C, Workman J. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 53.Kim D, Blus BJ, Chandra V, Huang P, Rastinejad F, Khorasanizadeh S. Corecognition of DNA and a methylated histone tail by the MSL3 chromodomain. Nat Struct Mol Biol. 17:1027–1029. doi: 10.1038/nsmb.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moore SA, Ferhatoglu Y, Jia Y, Al-Jiab RA, Scott MJ. Structural and biochemical studies on the chromo-barrel domain of male specific lethal 3 (MSL3) reveal a binding preference for mono or dimethyl lysine 20 on histone H4. J Biol Chem. doi: 10.1074/jbc.M110.134312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barski A, Cuddapah S, Cui K, Roh T, Schones D, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 56.Vakoc C, Mandat S, Olenchock B, Blobel G. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 57.Houston SI, McManus KJ, Adams MM, Sims JK, Carpenter PB, Hendzel MJ, Rice JC. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem. 2008;283:19478–19488. doi: 10.1074/jbc.M710579200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 59.Varier RA, Outchkourov NS, de Graaf P, van Schaik FM, Ensing HJ, Wang F, Higgins JM, Kops GJ, Timmers HT. A phospho/methyl switch at histone H3 regulates TFIID association with mitotic chromosomes. EMBO J. 2010;29:3967–3978. doi: 10.1038/emboj.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Niessen HE, Demmers JA, Voncken JW. Talking to chromatin: post-translational modulation of polycomb group function. Epigenetics Chromatin. 2009;2:10. doi: 10.1186/1756-8935-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eberlin A, Grauffel C, Oulad-Abdelghani M, Robert F, Torres-Padilla ME, Lambrot R, Spehner D, Ponce-Perez L, Wurtz JM, Stote RH, Kimmins S, Schultz P, Dejaegere A, Tora L. Histone H3 tails containing dimethylated lysine and adjacent phosphorylated serine modifications adopt a specific conformation during mitosis and meiosis. Mol Cell Biol. 2008;28:1739–1754. doi: 10.1128/MCB.01180-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Canzio D, Chang EY, Shankar S, Kuchenbecker KM, Simon MD, Madhani HD, Narlikar GJ, Al-Sady B. Chromodomain-Mediated Oligomerization of HP1 Suggests a Nucleosome-Bridging Mechanism for Heterochromatin Assembly. Mol Cell. 2011;41:67–81. doi: 10.1016/j.molcel.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Franz H, Mosch K, Soeroes S, Urlaub H, Fischle W. Multimerization and H3K9me3 binding are required for CDYL1b heterochromatin association. J Biol Chem. 2009;284:35049–35059. doi: 10.1074/jbc.M109.052332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.LeRoy G, Weston JT, Zee BM, Young NL, Plazas-Mayorca MD, Garcia BA. Heterochromatin protein 1 is extensively decorated with histone code-like post-translational modifications. Mol Cell Proteomics. 2009;8:2432–2442. doi: 10.1074/mcp.M900160-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao T, Eissenberg JC. Phosphorylation of heterochromatin protein 1 by casein kinase II is required for efficient heterochromatin binding in Drosophila. J Biol Chem. 1999;274:15095–15100. doi: 10.1074/jbc.274.21.15095. [DOI] [PubMed] [Google Scholar]
- 66.Dietrich N, Bracken AP, Trinh E, Schjerling CK, Koseki H, Rappsilber J, Helin K, Hansen KH. Bypass of senescence by the polycomb group protein CBX8 through direct binding to the INK4A-ARF locus. Embo J. 2007;26:1637–1648. doi: 10.1038/sj.emboj.7601632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sánchez C, Sánchez I, Demmers JAA, Rodriguez P, Strouboulis J, Vidal M. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- 68.Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao T, Heyduk T, Eissenberg JC. Phosphorylation site mutations in heterochromatin protein 1 (HP1) reduce or eliminate silencing activity. J Biol Chem. 2001;276:9512–9518. doi: 10.1074/jbc.M010098200. [DOI] [PubMed] [Google Scholar]
- 70.Hiragami-Hamada K, Shinmyozu K, Hamada D, Tatsu Y, Uegaki K, Fujiwara S, Nakayama JI. N-terminal phosphorylation of HP1{alpha} promotes its chromatin binding. Mol Cell Biol. 2011 doi: 10.1128/MCB.01012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 72.Hatano A, Matsumoto M, Higashinakagawa T, Nakayama KI. Phosphorylation of the chromodomain changes the binding specificity of Cbx2 for methylated histone H3. Biochem Biophys Res Commun. 2010;397:93–99. doi: 10.1016/j.bbrc.2010.05.074. [DOI] [PubMed] [Google Scholar]
- 73.Eissenberg JC. Molecular biology of the chromo domain: an ancient chromatin module comes of age. Gene. 2001;275:19–29. doi: 10.1016/s0378-1119(01)00628-x. [DOI] [PubMed] [Google Scholar]
- 74.Piacentini L, Fanti L, Berloco M, Perrini B, Pimpinelli S. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J Cell Biol. 2003;161:707–714. doi: 10.1083/jcb.200303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Muchardt C, Guilleme M, Seeler J-S, Trouche D, Dejean A, Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 2002;3:975–981. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meehan R, Kao C, Pennings S. HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. EMBO J. 2003;22:3164–3174. doi: 10.1093/emboj/cdg306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nishimura K, Hirokawa YS, Mizutani H, Shiraishi T. Reduced heterochromatin protein 1-beta (HP1beta) expression is correlated with increased invasive activity in human melanoma cells. Anticancer Res. 2006;26:4349–4356. [PubMed] [Google Scholar]
- 79.Smallwood A, Esteve PO, Pradhan S, Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 2007;21:1169–1178. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, Warmerdam DO, Lindh M, Brink MC, Dobrucki JW, Aten JA, Fousteri MI, Jansen G, Dantuma NP, Vermeulen W, Mullenders LH, Houtsmuller AB, Verschure PJ, van Driel R. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol. 2009;185:577–586. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, Reuter G, Jenuwein T. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 83.Ayyanathan K, Lechner MS, Bell P, Maul GG, Schultz DC, Yamada Y, Tanaka K, Torigoe K, Rauscher FJ., 3rd Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takanashi M, Oikawa K, Fujita K, Kudo M, Kinoshita M, Kuroda M. Heterochromatin protein 1gamma epigenetically regulates cell differentiation and exhibits potential as a therapeutic target for various types of cancers. Am J Pathol. 2009;174:309–316. doi: 10.2353/ajpath.2009.080148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Norwood LE, Moss TJ, Margaryan NV, Cook SL, Wright L, Seftor EA, Hendrix MJ, Kirschmann DA, Wallrath LL. A requirement for dimerization of HP1Hsalpha in suppression of breast cancer invasion. J Biol Chem. 2006;281:18668–18676. doi: 10.1074/jbc.M512454200. [DOI] [PubMed] [Google Scholar]
- 86.Lavigne M, Eskeland R, Azebi S, Saint-Andre V, Jang SM, Batsche E, Fan HY, Kingston RE, Imhof A, Muchardt C. Interaction of HP1 and Brg1/Brm with the globular domain of histone H3 is required for HP1-mediated repression. PLoS Genet. 2009;5:e1000769. doi: 10.1371/journal.pgen.1000769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Satijn DP, Otte AP. RING1 interacts with multiple Polycomb-group proteins and displays tumorigenic activity. Mol Cell Biol. 1999;19:57–68. doi: 10.1128/mcb.19.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Biason-Lauber A, Konrad D, Meyer M, DeBeaufort C, Schoenle EJ. Ovaries and female phenotype in a girl with 46,XY karyotype and mutations in the CBX2 gene. Am J Hum Genet. 2009;84:658–663. doi: 10.1016/j.ajhg.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Parris TZ, Danielsson A, Nemes S, Kovacs A, Delle U, Fallenius G, Mollerstrom E, Karlsson P, Helou K. Clinical implications of gene dosage and gene expression patterns in diploid breast carcinoma. Clin Cancer Res. 2010;16:3860–3874. doi: 10.1158/1078-0432.CCR-10-0889. [DOI] [PubMed] [Google Scholar]
- 90.Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 91.Kang X, Qi Y, Zuo Y, Wang Q, Zou Y, Schwartz RJ, Cheng J, Yeh ET. SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol Cell. 2010;38:191–201. doi: 10.1016/j.molcel.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bezsonova I, Walker JR, Bacik JP, Duan S, Dhe-Paganon S, Arrowsmith CH. Ring1B contains a ubiquitin-like docking module for interaction with Cbx proteins. Biochemistry. 2009;48:10542–10548. doi: 10.1021/bi901131u. [DOI] [PubMed] [Google Scholar]
- 93.Gil J, Bernard D, Martinez D, Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- 94.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Mönch K, Minucci S, Porse BT, Marine J-C, Hansen KH, Helin K. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lahn BT, Tang ZL, Zhou J, Barndt RJ, Parvinen M, Allis CD, Page DC. Previously uncharacterized histone acetyltransferases implicated in mammalian spermatogenesis. Proc Natl Acad Sci U S A. 2002;99:8707–8712. doi: 10.1073/pnas.082248899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Caron C, Pivot-Pajot C, van Grunsven LA, Col E, Lestrat C, Rousseaux S, Khochbin S. Cdyl: a new transcriptional co-repressor. EMBO Rep. 2003;4:877–882. doi: 10.1038/sj.embor.embor917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, Komatsu Y, Shinkai Y, Cheng X, Jeltsch A. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol. 2008;4:344–346. doi: 10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kokura K, Sun L, Bedford MT, Fang J. Methyl-H3K9-binding protein MPP8 mediates E-cadherin gene silencing and promotes tumour cell motility and invasion. EMBO J. 2010;29:3673–3687. doi: 10.1038/emboj.2010.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, Kouzarides T. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 100.Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 101.Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 102.Yoon KA, Hwangbo B, Kim IJ, Park S, Kim HS, Kee HJ, Lee JE, Jang YK, Park JG, Lee JS. Novel polymorphisms in the SUV39H2 histone methyltransferase and the risk of lung cancer. Carcinogenesis. 2006;27:2217–2222. doi: 10.1093/carcin/bgl084. [DOI] [PubMed] [Google Scholar]
- 103.Lusser A, Urwin DL, Kadonaga JT. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol. 2005;12:160–166. doi: 10.1038/nsmb884. [DOI] [PubMed] [Google Scholar]
- 104.Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, Ramalho-Santos M. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hauk G, McKnight JN, Nodelman IM, Bowman GD. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell. 2010;39:711–723. doi: 10.1016/j.molcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Osman I, Bajorin DF, Sun TT, Zhong H, Douglas D, Scattergood J, Zheng R, Han M, Marshall KW, Liew CC. Novel blood biomarkers of human urinary bladder cancer. Clin Cancer Res. 2006;12:3374–3380. doi: 10.1158/1078-0432.CCR-05-2081. [DOI] [PubMed] [Google Scholar]
- 107.Nagarajan P, Onami TM, Rajagopalan S, Kania S, Donnell R, Venkatachalam S. Role of chromodomain helicase DNA-binding protein 2 in DNA damage response signaling and tumorigenesis. Oncogene. 2009;28:1053–1062. doi: 10.1038/onc.2008.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 109.Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 110.Burd CJ, Kinyamu HK, Miller FW, Archer TK. UV radiation regulates Mi-2 through protein translation and stability. J Biol Chem. 2008;283:34976–34982. doi: 10.1074/jbc.M805383200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29:3130–3139. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, Francis D, Bredel M, Vogel H, Mills AA. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–475. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 113.Lutz T, Stoger R, Nieto A. CHD6 is a DNA-dependent ATPase and localizes at nuclear sites of mRNA synthesis. FEBS Lett. 2006;580:5851–5857. doi: 10.1016/j.febslet.2006.09.049. [DOI] [PubMed] [Google Scholar]
- 114.Kalscheuer VM, Feenstra I, Van Ravenswaaij-Arts CM, Smeets DF, Menzel C, Ullmann R, Musante L, Ropers HH. Disruption of the TCF4 gene in a girl with mental retardation but without the classical Pitt-Hopkins syndrome. Am J Med Genet A. 2008;146A:2053–2059. doi: 10.1002/ajmg.a.32419. [DOI] [PubMed] [Google Scholar]
- 115.Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB, Janssen IM, van der Vliet WA, Huys EH, de Jong PJ, Hamel BC, Schoenmakers EF, Brunner HG, Veltman JA, van Kessel AG. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 116.Gao X, Gordon D, Zhang D, Browne R, Helms C, Gillum J, Weber S, Devroy S, Swaney S, Dobbs M, Morcuende J, Sheffield V, Lovett M, Bowcock A, Herring J, Wise C. CHD7 gene polymorphisms are associated with susceptibility to idiopathic scoliosis. Am J Hum Genet. 2007;80:957–965. doi: 10.1086/513571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kim HG, Kurth I, Lan F, Meliciani I, Wenzel W, Eom SH, Kang GB, Rosenberger G, Tekin M, Ozata M, Bick DP, Sherins RJ, Walker SL, Shi Y, Gusella JF, Layman LC. Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am J Hum Genet. 2008;83:511–519. doi: 10.1016/j.ajhg.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T, Wysocka J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sakamoto I, Kishida S, Fukui A, Kishida M, Yamamoto H, Hino S, Michiue T, Takada S, Asashima M, Kikuchi A. A novel beta-catenin-binding protein inhibits beta-catenin-dependent Tcf activation and axis formation. J Biol Chem. 2000;275:32871–32878. doi: 10.1074/jbc.M004089200. [DOI] [PubMed] [Google Scholar]
- 120.Zahir F, Firth HV, Baross A, Delaney AD, Eydoux P, Gibson WT, Langlois S, Martin H, Willatt L, Marra MA, Friedman JM. Novel deletions of 14q11.2 associated with developmental delay, cognitive impairment and similar minor anomalies in three children. J Med Genet. 2007;44:556–561. doi: 10.1136/jmg.2007.050823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nishiyama M, Oshikawa K, Tsukada Y, Nakagawa T, Iemura S, Natsume T, Fan Y, Kikuchi A, Skoultchi AI, Nakayama KI. CHD8 suppresses p53-mediated apoptosis through histone H1 recruitment during early embryogenesis. Nat Cell Biol. 2009;11:172–182. doi: 10.1038/ncb1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rodriguez-Paredes M, Ceballos-Chavez M, Esteller M, Garcia-Dominguez M, Reyes JC. The chromatin remodeling factor CHD8 interacts with elongating RNA polymerase II and controls expression of the cyclin E2 gene. Nucleic Acids Res. 2009;37:2449–2460. doi: 10.1093/nar/gkp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shur I, Solomon R, Benayahu D. Dynamic interactions of chromatin-related mesenchymal modulator, a chromodomain helicase-DNA-binding protein, with promoters in osteoprogenitors. Stem Cells. 2006;24:1288–1293. doi: 10.1634/stemcells.2005-0300. [DOI] [PubMed] [Google Scholar]
- 124.Sun Y, Jiang X, Xu Y, Ayrapetov M, Moreau L, Whetstine J, Price B. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009;11:1376–1382. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sun Y, Jiang X, Price BD. Tip60: connecting chromatin to DNA damage signaling. Cell Cycle. 2010;9:930–936. doi: 10.4161/cc.9.5.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 127.Gupta A, Sharma GG, Young CS, Agarwal M, Smith ER, Paull TT, Lucchesi JC, Khanna KK, Ludwig T, Pandita TK. Involvement of human MOF in ATM function. Mol Cell Biol. 2005;25:5292–5305. doi: 10.1128/MCB.25.12.5292-5305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Smith ER, Cayrou C, Huang R, Lane WS, Cote J, Lucchesi JC. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol. 2005;25:9175–9188. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lai A, Marcellus RC, Corbeil HB, Branton PE. RBP1 induces growth arrest by repression of E2F-dependent transcription. Oncogene. 1999;18:2091–2100. doi: 10.1038/sj.onc.1202520. [DOI] [PubMed] [Google Scholar]
- 130.Fleischer TC, Yun UJ, Ayer DE. Identification and characterization of three new components of the mSin3A corepressor complex. Mol Cell Biol. 2003;23:3456–3467. doi: 10.1128/MCB.23.10.3456-3467.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wu MY, Eldin KW, Beaudet AL. Identification of chromatin remodeling genes Arid4a and Arid4b as leukemia suppressor genes. J Natl Cancer Inst. 2008;100:1247–1259. doi: 10.1093/jnci/djn253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pardo PS, Leung JK, Lucchesi JC, Pereira-Smith OM. MRG15, a novel chromodomain protein, is present in two distinct multiprotein complexes involved in transcriptional activation. J Biol Chem. 2002;277:50860–50866. doi: 10.1074/jbc.M203839200. [DOI] [PubMed] [Google Scholar]
- 133.Chen M, Tominaga K, Pereira-Smith OM. Emerging role of the MORF/MRG gene family in various biological processes, including aging. Ann N Y Acad Sci. 2010;1197:134–141. doi: 10.1111/j.1749-6632.2010.05197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Morales V, Regnard C, Izzo A, Vetter I, Becker PB. The MRG domain mediates the functional integration of MSL3 into the dosage compensation complex. Mol Cell Biol. 2005;25:5947–5954. doi: 10.1128/MCB.25.14.5947-5954.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 136.Sural TH, Peng S, Li B, Workman JL, Park PJ, Kuroda MI. The MSL3 chromodomain directs a key targeting step for dosage compensation of the Drosophila melanogaster X chromosome. Nat Struct Mol Biol. 2008;15:1318–1325. doi: 10.1038/nsmb.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Andersen CL, Christensen LL, Thorsen K, Schepeler T, Sorensen FB, Verspaget HW, Simon R, Kruhoffer M, Aaltonen LA, Laurberg S, Orntoft TF. Dysregulation of the transcription factors SOX4, CBFB and SMARCC1 correlates with outcome of colorectal cancer. Br J Cancer. 2009;100:511–523. doi: 10.1038/sj.bjc.6604884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schaniel C, Ang YS, Ratnakumar K, Cormier C, James T, Bernstein E, Lemischka IR, Paddison PJ. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27:2979–2991. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Remmelink M, Mijatovic T, Gustin A, Mathieu A, Rombaut K, Kiss R, Salmon I, Decaestecker C. Identification by means of cDNA microarray analyses of gene expression modifications in squamous non-small cell lung cancers as compared to normal bronchial epithelial tissue. Int J Oncol. 2005;26:247–258. [PubMed] [Google Scholar]
- 140.Lind GE, Skotheim RI, Fraga MF, Abeler VM, Esteller M, Lothe RA. Novel epigenetically deregulated genes in testicular cancer include homeobox genes and SCGB3A1 (HIN-1) J Pathol. 2006;210:441–449. doi: 10.1002/path.2064. [DOI] [PubMed] [Google Scholar]