Abstract
Objective
The present study was undertaken to analyze the impact of epigenetic alterations with a main focus on nuclear area, aneuploidy, hyperploidy, and proliferation in 70 ovarian cancer specimens.
Methods
Morphometric changes and somatic chromosomal ploidy status were assessed by Feulgen spectrophotometry. DNA-hypomethylation of LINE1 repeats was analyzed by means of MethyLight PCR, and methylation levels of satellite 2 (Sat2) and satellite alpha (Satα) DNA sequences in chromosome 1 were measured by Southern blot analysis. These parameters were analyzed with regard to correlations as well as to recurrence and survival.
Results
We identified a significant association between LINE1 DNA-hypomethylation and patient age (p = 0.029). Furthermore, LINE1 DNA-hypomethylation was positively correlated with the nuclear area (r = 0.47; p<0.001) and the proliferation index (r = 0.36; p<0.001). Univariate survival analysis showed that the nuclear area and LINE1 DNA-hypomethylation were prognostic factors for overall (p=0.015 and = 0.006, respectively) and progression-free survival (p=0.020 and p=0.001 respectively), the percentage of aneuploidy only for overall survival (p=0.031). Subgroup survival analyses revealed that the prognostic value of these factors is strictly confined to mucinous cancers. In serous cancers no prognostic value could be pointed out for any analyzed parameter. Multivariate analysis of the entire cohort showed that the percentage of hyperploidy was an independent prognostic parameter for overall survival (p=0.003) and LINE1 DNA-hypomethylation for progression-free survival (p=0.03). In mucinous cancers nuclear area and LINE1 DNA-hypomethylation were found to be independent predictors of progression-free and overall survival.
Conclusions
In this study we identified the correlations between early cancer-associated genome DNA-hypomethylation, nuclear morphometric changes, somatic chromosomal ploidy status and the proliferation index. Prognostic relevance of nuclear area and LINE1 DNA-hypomethylation was revealed exclusively in mucinous ovarian cancers.
Keywords: DNA ploidy, Nuclear area, Proliferation index, DNA-hypomethylation, Ovarian cancer
1. Introduction
Despite aggressive treatment including highly offensive cytoreductive surgical techniques and multiagent chemotherapy, the prognosis of patients with advanced ovarian cancer remains unacceptably poor. The clinical outcome has not changed considerably over the last decades and appears to be related primarily to individual tumor characteristics. Chromosomal rearrangements represent an early step in the initiation of tumorigenesis and transformation to malignancy is frequently accompanied by quantitative changes in DNA content, which can be evidenced by morphometric alterations of the nucleus in tumor cells. Moreover, also the delicate organization of epigenetic regulatory mechanisms such as characteristic DNA-methylation patterns, that regulate the normal cellular homeostasis of gene expression, often becomes irrecognizable in malignant cells. Aberrant DNA methylation in malignant cells includes both hypermethylation of CpG islands at gene promoters which is mainly responsible for the transcriptional silencing of tumor suppressor genes, as well as global hypomethylation. While a small portion of hypomethylation occurs at gene promoters, resulting in an over-expression of certain oncogenes [1,2], the majority occurs at repetitive elements, such as long interspersed nuclear elements (LINEs) [3]. Since most of the estimated 500,000 copies of LINE1 elements have become nonfunctional during human evolution [4] genome-wide hypomethylation at LINE1 elements during tumorigenesis is presumed to contribute crucially to chromosomal instability [5]. Besides LINE1 elements, the long genomic regions rich in satellite 2 DNA sequences (Sat2) in the juxtacentromeric heterochromatin of chromosomes 1 and 16 which are heavily hypermethylated in normal somatic tissues and are frequently affected by cancer-associated hypomethylation, represent additional surrogate-regions to estimate the global degree of hypomethylation in cancers. Recently, we have revealed Sat2 and Satα DNA-hypomethylation to be an important issue in ovarian carcinogenesis and are associated with poor prognosis at least in patients exhibiting no residual disease after primary surgery [6].
On the other hand the prognostic significance of DNA ploidy remains controversial in ovarian cancer. While in a number of studies DNA-aneuploidy was found to be of independent prognostic value [7–9], other studies were unable to confirm this observation [10–12]. One of the most important reports advocating the introduction of DNA ploidy as a prognostic discriminator for patient recruitment in phase III treatment studies, was that of Kimming et al., who revealed DNA ploidy to be as powerful as or even more powerful than residual disease after primary surgery in multivariate analysis for predicting clinical outcome in advanced ovarian cancer [13]. Prognostic value of proliferative activity and nuclear morphometry in terms of the nuclear area has also been shown in patients with bladder cancer [14,15].
To the best of our knowledge, there are no investigations studying the relationship between DNA-hypomethylation and DNA ploidy status or morphometric characteristics of the nucleus in ovarian cancer cells.
Therefore the aim of the present study was to indirectly analyze genomic instability via morphometric nuclear changes and the chromosomal ploidy status in ovarian cancer cells in the context of the degree of global DNA-hypomethylation in these cells. For this purpose, the morphometric variables nuclear area, aneuploidy, hyperploidy, and the proliferation index were determined with Feulgen spectrophotometry, aberrant global DNA-hypomethylation was measured with MethyLight PCR for LINE1 repeats, and Southern blot analysis was used for Sat2 and Satα juxtacentromeric and centromeric DNA of chromosome 1 determination in 70 ovarian cancer samples. Furthermore, comparisons between serous and mucinous tumors were performed for all the parameters studied. In addition, the prognostic relevance of the various spectrophotometric variables and the global hypomethylation status was assessed with regard to progression-free and overall survival as well for the entire study population as for the patients with serous and mucinous cancers separately.
2. Material and methods
2.1. Patients and samples
All patients were treated at the Department of Obstetrics and Gynecology of the Innsbruck Medical University, Austria between 1989 and 2000.
Frozen ovarian cancer tissue samples and formalin fixed paraffin embedded (FFPE) tissues from 70 patients with ovarian cancer (patients were 24.1 to 83.2 years old; median age at diagnosis, 63.5 years) were analyzed. Staging was done by the International Federation of Gynecology and Obstetrics (FIGO) classification.
Tumor specimens were obtained immediately after surgery and then evaluated by our pathologist. One part of the tissue was pulverized under cooling with liquid nitrogen and stored at −70 °C, and another part was formalin fixed and embedded in paraffin in compliance with and approved by the Institutional Review Board. Clinical, pathological and follow-up data were stored in a database in accordance with hospital privacy rules. A platinum based chemotherapy was part of the treatment for all but 8 cancer patients (7 and 1 who had FIGO stage I and II respectively). After primary treatment, all of the patients were monitored by our department at intervals increasing from 3 months to 1 year until death or the end of the study. Follow-up information was available for all of the patients. Clinico-pathological features are shown in Table 1.
Table 1.
Associations between nuclear area, percentages of ploidy, proliferation, DNA-hypomethylation and clinicopathological features.
| n | Nuclear Area |
Hyperploidy (%) |
Aneuploidy (%) |
Proliferation index |
Chr1 Sat2 hypomethylation |
Chr1 Satα hypomethylation |
LINE1 DNA-Hypomethylation |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | ≤median (n=35) |
>median | P | ≤median (n=35) |
>median (n=35) |
P | ≤median (n=35) |
>median (n=35) |
P | ≤median (n=35) |
>median (n=35) |
P | ≤Score 2 (n=49) |
>Score 2 (n=21) |
P | ≤Score 2 (n=44) |
>Score 2 (n=24) |
P | ≤median (n=35) |
>median (n=35) |
P | |
| Age | ||||||||||||||||||||||
| < 60 yrs. | 29 | 16 | 13 | 0.467 | 14 | 15 | 0.808 | 20 | 9 | 0.008 | 14 | 15 | 0.81 | 23 | 6 | 0.153 | 17 | 11 | 0.564 | 19 | 10 | 0.029 |
| > 60 yrs. | 41 | 19 | 22 | 21 | 20 | 15 | 26 | 21 | 20 | 26 | 15 | 27 | 13 | 16 | 25 | |||||||
| FIGO | ||||||||||||||||||||||
| I/II | 16 | 11 | 5 | 0.088 | 8 | 8 | n.a | 8 | 8 | n.a | 10 | 6 | 0.26 | 14 | 2 | 0.820 | 14 | 1 | 0.020 | 11 | 5 | 0.088 |
| III/IV | 54 | 24 | 30 | 27 | 27 | 27 | 27 | 25 | 29 | 35 | 19 | 30 | 23 | 24 | 30 | |||||||
| Tumor grade | ||||||||||||||||||||||
| I/II | 49 | 27 | 22 | 0.192 | 22 | 27 | 0.192 | 25 | 24 | 0.79 | 25 | 24 | 0.79 | 35 | 14 | 0.690 | 32 | 16 | 0.600 | 27 | 22 | 0.192 |
| III | 21 | 8 | 13 | 13 | 8 | 10 | 11 | 10 | 11 | 14 | 7 | 12 | 8 | 8 | 13 | |||||||
| Histology | ||||||||||||||||||||||
| Serous cancer | 33 | 20 | 13 | 0.075 | 19 | 14 | 0.488 | 19 | 14 | 0.488 | 20 | 13 | 0.243 | 21 | 12 | 0.270 | 18 | 15 | 0.092 | 17 | 16 | 0.917 |
| Mucinous cancer | 30 | 14 | 16 | 13 | 17 | 13 | 17 | 12 | 18 | 24 | 6 | 23 | 6 | 15 | 15 | |||||||
| Endometrioid cancer | 7 | 1 | 6 | 3 | 4 | 3 | 4 | 3 | 4 | 4 | 3 | 3 | 3 | 3 | 4 | |||||||
| Residual disease after surgery | ||||||||||||||||||||||
| No residual disease | 27 | 14 | 13 | 0.880 | 14 | 13 | 0.555 | 12 | 15 | 0.759 | 16 | 11 | 0.054 | 21 | 6 | 0.486 | 19 | 7 | 0.474 | 15 | 12 | 0.759 |
| Residual disease≤2 cm | 19 | 10 | 9 | 11 | 8 | 10 | 9 | 5 | 14 | 13 | 6 | 12 | 7 | 9 | 10 | |||||||
| Residual disease>2 cm | 24 | 11 | 13 | 10 | 14 | 13 | 11 | 14 | 10 | 15 | 9 | 13 | 10 | 11 | 13 | |||||||
Note: Samples with satellite hypomethylation scores of ≤2 had no, slight or only moderate, samples with satellite hypomethylation scores of >2 had much hypomethylation. The significance level (P) was determined by the Chi2-Statistic.
2.2. Analysis of morphometric changes, and somatic chromosomal ploidy status
Three to four paraffin sections of 30 μm thickness were cut from a representative area of each tumor sample and tumor areas were microdissected using syringe needles. Paraffin was removed and section remnants were rehydrated as recently described [16]. Nuclei were separated from the cytoplasm by enzyme digestion and then washed. Preparation of cytospins was performed as described recently [16]. Nuclear area and ploidy determination were carried out by Feulgen spectrophotometry using a SAMBA 200 microscope image processor (Alcatel-TITN, Grenoble, France) as described previously [17–19]. The DNA histogram types were assessed from the nuclear integrated optical density, which relates to nuclear DNA content [19].
2.3. Proliferation analysis
Proliferation indices were calculated as described previously [19].
2.4. DNA isolation and methylation analysis
Genomic DNA from lyophilized, quick-frozen ovarian cancer specimens was isolated using the QIAmp tissue kit (Qiagen, Hilden, Germany). Southern blot analysis for Chromosome 1 satellite 2 and α (Chr1 Sat2, Chr1 Satα) DNA sequences and scoring of Chr1 Sat2 and Chr1 Satα hypomethylation is described in ref. 6.
Sodium bisulfite conversion of genomic DNA and MethyLight assay were performed as previously described [20]. The levels of unmethylated repetitive elements were expressed as percent of unmethylated reference (PUMR) values and were calculated similarly to PMR values except that bisulfite-converted, human sperm DNA was used as an unmethylated reference for PUMR determinations of each repetitive element as described previously [6,21–23], although it is known that this kind of DNA is not 100% hypomethylated. For methylation analysis, ACTB was used as reference gene [21]. The forward and reverse primer and the probe, respectively, specific for an unmethylated LINE1 sequence are as follows: Forward: AACCTCATTACCACCTTACAATTTAATCT, Reverse: GATTGGTTTAAGAAATGGTG TATTATGAGA, 6FAM-CAAAATTCCATAAACATAAAACCCTCTAAACCAA-BHQ-1, (amplicon location: AF149422: 303–431).
2.5. Statistical analysis
For the comparison of the PCR based DNA-hypomethylation analysis (LINE1-PUMR) and the Southern blot based DNA-hypomethylation measurements (DNA-hypomethylation of Chr1 Sat2, Chr1 Satα; Score≤2, Score>2) a logistic regression model, as well as a chi2-test-statistic (LINE1 as dichotomized variable) were used.
Quantitative variables were categorized by their median into two groups. Chi square test and Spearman rank correlation coefficient were used for testing associations between clinicopathological features and nuclear area, percentages of hyper- and aneuploidy, the proliferation index and DNA-hypomethylation of Chr1 Sat2, Chr1 Satα or LINE1 respectively. For univariate survival analysis we calculated the Kaplan–Meier curves and the log-rank test statistic. Multivariate survival analysis was done using a Cox Proportional Hazard Model. For elimination of variables, we applied a backward variable selection procedure. A p-value of 0.1 was used for the exclusion of variables; all other tests were performed using a 0.05% level of significance. All statistical calculations were performed using SPSS, version 16.0.
3. Results
3.1. Association analyses
First we tested the LINE1 MethyLight PCR reaction to measure global DNA-hypomethylation in comparison to dichotomized Southern blot analysis of centromeric DNAs in Chromosome 1 (Chr1 Sat2, Chr1 Satα DNA-hypomethylation) using a logistic regression model. Increasing quantitative LINE1 DNA-hypomethylation showed a high significant association with hypomethylated centromeric DNAs (Chr1 Sat2, Chr1 Satα DNA-hypomethylation; p<0.005 for each). This result was confirmed by a chi2-statistic using LINE1 hypomethlyation as dichotomized variable (Chr1 Sat2, p<0.001; Chr1 Satα DNA-hypomethylation, p=0.001).
Then associations between clinicopathological variables and morpho-metric nuclear characteristics like nuclear area, percentages of hyperploidy and aneuploidy, the proliferation index, and DNA-hypomethylation (Chr1 Sat2, Chr1 Satα DNA-hypomethylation measured by Southern blot analysis and LINE1 DNA-hypomethylation measured by MethyLight PCR) were analyzed in 70 ovarian cancer specimens.
We identified a significant association between LINE1 DNA-hypomethylation and age (p=0.029). Chr1 Satα was found to be significantly associated with FIGO stage (p=0.007). The nuclear area, the percentages of hyperploidy and aneuploidy and the proliferation index showed significant reciprocal positive correlations (Table 1). Interestingly, LINE1 DNA-hypomethylation was highly correlated with nuclear area (r=0.47, p<0.001), proliferation index (r=0.36, p<0.001) and of course with Chr1 Sat2 and Chr1 Satα DNA-hypomethylation (r=0.63 and 0.44 respectively; p<0.001 each; Table 2). In addition, we were interested in possible effects of global DNA-hypomethylation on the ploidy status of the tumors. For this purpose, we categorized the variables of DNA-hypomethylation and those of DNA ploidy along their respective median values into “high” and “low” groups and identified a strong association between a high degree of LINE1 DNA-hypomethylation and higher aneuploidy percentages (p=0.009). However, we did not observe a statistical significant relationship between Chr1 Sat2 or Chr1 Satα DNA-hypomethylation and DNA ploidy. This was also true when Chr1 Sat2 and Chr1 Satα DNA-hypomethylation specimens were grouped into two hypomethylation score groups as described earlier [6]: ≤2 (score 0–2, indicating no, slight or moderate hypomethylation respectively), and score >2 (scores 3–4, indicating high or extreme hypomethylation respectively).
Table 2.
Correlation between nuclear area, percentages of ploidy, proliferation and parameters of DNA-hypomethylation.
| Hyperploidy |
Aneuploidy |
Proliferation index |
Chr1 Sat2 hypomethylation |
Chr1 Sata hypomethylation |
LINE1 DNA- hypomethylation |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ρ | P | ρ | P | ρ | P | ρ | P | ρ | P | ρ | P | |
| Nuclear Area | 0.84 | <0.001 | 0.60 | <0.001 | 0.69 | <0.001 | 0.40 | 0.001 | 0.22 | 0.07 | 0.47 | <0.001 |
| Hyperploidy | 0.70 | <0.001 | 0.55 | <0.001 | 0.15 | 0.21 | 0.05 | 0.66 | 0.22 | 0.06 | ||
| Aneuploidy | 0.40 | 0.001 | 0.19 | 0.11 | 0.09 | 0.47 | 0.21 | 0.09 | ||||
| Proliferation index | 0.41 | 0.001 | 0.20 | 0.1 | 0.36 | 0.002 | ||||||
| Chr1 Sat2 hypomethylation | 0.65 | <0.001 | 0.63 | <0.001 | ||||||||
| Chr1 Satα hypomethylation | 0.44 | <0.001 | ||||||||||
Note: Spearman rank correlation analysis was performed.
Abbreviations: ρ, Spearman rank correlation coefficient.
3.2. Comparison of the studied parameters in serous and mucinous histological subtypes
As serous (n=33) and mucinous (n=30) cancers were almost equally represented in the study population and both cohorts were well balanced with regard to clinicopathologic characteristics (Table 3A), the entire set of morphometric parameters and predictors for global DNA-hypomethylation were compared for both histological subtypes. It is noteworthy that for these variables no statistical differences could be revealed between both histological subtypes with the exception of Chr1 Satα DNA-hypomethylation that was found to be more common and pronounced in serous cancers. (Table 3B).
Table 3.
Comparison of mucinous and serous ovarian tumors analyzed in the present study. (A) Comparison of clinicopathological features. (B) Comparison of analyzed parameters.
| Serous ovarian cancer |
Mucinous ovarian cancer |
P | ||
|---|---|---|---|---|
| Variable | No. Patients | No. Patients | ||
| A | ||||
| Age | ≤60 yrs. | 12 | 12 | 0.49 |
| >60 yrs. | 21 | 18 | ||
| FIGO stage | I/II | 5 | 10 | 0.08 |
| III/IV | 28 | 20 | ||
| Tumor grade | I/II | 22 | 24 | 0.18 |
| III | 11 | 6 | ||
| Residual disease | no | 10 | 13 | 0.21 |
| after surgery | yes | 23 | 17 | |
| Chemotherapy | no | 5 | 3 | 0.41 |
| yes | 28 | 27 | ||
| B | ||||
| Nuclear area | small (≤ median) |
20 | 14 | 0.20 |
| large (> median) |
13 | 16 | ||
| Hyperploidy | low (≤median) |
19 | 13 | 0.19 |
| high (> median) |
14 | 17 | ||
| Aneuploidy | low (≤median) |
19 | 13 | 0.19 |
| high (> median) |
14 | 17 | ||
| Proliferation index | low (≤median) |
20 | 12 | 0.08 |
| high (> median) |
13 | 18 | ||
| Chr1 Sat2 hypomethylation |
≤ score 2 | 21 | 24 | 0.12 |
| > score 2 | 12 | 6 | ||
| Chr1 Sat α hypomethylation |
≤ score 2 | 18 | 23 | 0.04 |
| > score 2 | 15 | 6 | ||
| LINE1 DNA-hypomethylation |
low (≤median) |
17 | 15 | 0.55 |
| high (> median) |
16 | 15 |
Note: The significance level (P) was determined by Chi square test.
3.3. Univariate survival analyses
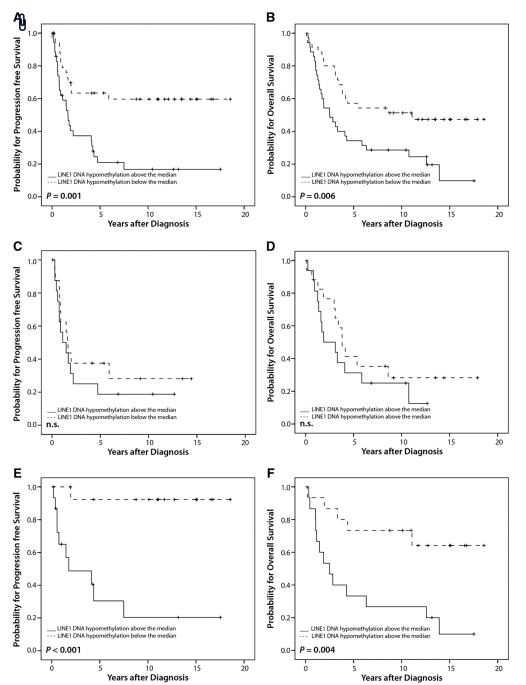
Univariate survival analysis of all 70 ovarian cancer patients showed that the nuclear area was a prognostic factor for overall (p=0.015) and progression-free survival (p=0.02). A large area was associated with poor outcome (Table 4A). Also a high percentage of aneuploidy was related to a poor overall survival (p=0.031) (Table 4A). Since we have demonstrated previously that DNA-hypomethylation of Chr1 Sat2 and Chr1 Satα are prognostically relevant markers, we were interested if the analysis of LINE1 DNA-hypomethylation is more suitable for the prediction of overall and progression-free survival. Indeed, we found that LINE1 DNA-hypomethylation was a better prognostic factor for overall (p=0.006) and progression-free survival (p=0.001) than the two Chr1 Satellite DNA markers (Table 4A). Kaplan–Meier curves for overall and progression-free survival for LINE1 DNA-hypomethylation are shown in Fig. 1A and B.
Table 4.
Univariate survival analysis of patients with primary ovarian cancer. (A) All 70 ovarian cancer patients. (B) Patients stratified by histology.
| A | |||||
|---|---|---|---|---|---|
| Progression-free survival |
Overall survival |
||||
| Variable | No. Patients (relapsed/total) | P | No. Patients (died/total) | P | |
| Age | ≤60 yrs. | 17/29 | 0.33 | 14/29 | 0.002 |
| >60 yrs. | 23/41 | 32/41 | |||
| FIGO stage | I/II | 2/16 | 0.001 | 6/16 | 0.012 |
| III/IV | 38/54 | 40/54 | |||
| Tumor grade | I/II | 22/49 | 0.01 | 28/49 | 0.032 |
| III | 18/21 | 18/21 | |||
| Residual disease | no | 8/27 | <0.001 | 10/27 | <0.001 |
| after surgery | yes | 32/43 | 36/43 | ||
| Histology | serous | 24/33 | 0.02 | 25/33 | 0.215 |
| mucinous | 11/30 | 18/30 | |||
| endometroid | 5/7 | 3/7 | |||
| Chemotherapy | no | 1/8 | 0.09 | 4/8 | 0.544 |
| yes | 39/62 | 42/62 | |||
| Nuclear area | small (≤median) | 16/35 | 0.02 | 19/35 | 0.015 |
| large (>median) | 24/35 | 27/35 | |||
| Hyperploidy | low (≤median) | 19/35 | 0.43 | 20/35 | 0.069 |
| high (>median) | 21/35 | 26/35 | |||
| Aneuploidy | low (≤median) | 17/35 | 0.07 | 19/35 | 0.031 |
| high (>median) | 23/35 | 27/35 | |||
| Proliferation index | low (≤median) | 18/35 | 0.32 | 21/35 | 0.220 |
| high (>median) | 22/35 | 25/35 | |||
| Chr1 Sat2 hypomethylation | ≤score 2 | 18/35 | 0.02 | 29/49 | 0.017 |
| >score 2 | 22/35 | 17/21 | |||
| Chr1 Sat α hypomethylation | ≤score 2 | 20/44 | 0.048 | 27/44 | 0.460 |
| >score 2 | 19/24 | 18/24 | |||
| LINE1 DNA-hypomethylation | low (≤median) | 13/35 | 0.001 | 18/35 | 0.006 |
| high (>median) | 27/35 | 28/35 | |||
| B | |||||
|---|---|---|---|---|---|
| Serous ovarian cancer |
Mucinous ovarian cancer |
||||
| Variable | Progression-free survival |
Overall survival |
Progression-free suvival |
Overall survival |
|
| P | P | P | P | ||
| Nuclear area | small (≤median) large (>median) |
0.465 | 0.304 | 0.006 | 0.003 |
| Hyperploidy | low (≤median) high (>median) |
0.899 | 0.647 | 0.010 | < 0.001 |
| Aneuploidy | low (≤median) high (>median) |
0.101 | 0.303 | 0.020 | 0.019 |
| Proliferation | low (≤median) high (>median) |
0.594 | 0.641 | 0.010 | 0.056 |
| Chr1 Sat2 hypomethylation | ≤score 2 >score 2 |
0.344 | 0.087 | 0.045 | 0.018 |
| Chr1 Sat α hypomethylation | ≤score 2 >score 2 |
0.717 | 0.941 | 0.18 | 0.42 |
| LINE1 DNA-hypomethylation | low (≤median) high (>median) |
0.278 | 0.313 | < 0.001 | 0.004 |
Note: The significance level (P) was determined by log-rank test.
Fig. 1.
Kaplan–Meier survival analysis and LINE1 DNA-hypomethylation. (A) Progression-free survival, (B) overall survival in 70 ovarian cancer patients. (C) Progression-free survival (D) Overall survival in 33 ovarian cancer patients with serous tumors. (E) Progression-free survival, (F) overall survival and nuclear size in 30 ovarian cancer patients with mucinous tumors.
Interestingly, the prognostic value of LINE1 DNA-hypomethylation and of the nuclear area was totally abrogated when progression-free and overall survival was analyzed in patients with serous histological subtype separately. In contrary, in mucinous cancers prognostic relevance of LINE1 hypomethylation and the nuclear area was found to be more prominent as compared with that obtained in the whole cohort of 70 patients (Table 4A and B). The prognostic role of LINE1 hypomethylation in serous and mucinous cancers is depicted in the Kaplan–Meier curves 1C to 1F. Other significant predictors for progression-free and/or overall survival in mucinous cancers only are listed in Table 4B.
3.4. Multivariate survival analyses
Multivariate analysis of all 70 ovarian cancer patients revealed that the percentage of hyperploidy was an independent prognostic parameter for overall survival [Relative risk (RR) for death 2.7 (95% CI 1.4–5.3; p=0.003)] and LINE1 DNA-hypomethylation was identified to be an independent prognostic markers for progression-free survival [RR for relapse 2.2 (95% CI 1.1–4.3; p=0.03); (Table 5)]. When multivariate adjustment for age, FIGO stage, grading and residual disease was performed for the subgroup of mucinous cancers, LINE1 DNA-hypomethylation retained significant prognostic relevance for progression-free survival [RR of relapse 15.66 (95% CI 1.6–152.4; p=0.02)] and retained borderline significance in predicting overall survival [RR for death 3.35 (95% CI 1.0–11.5; p=0.05)]. In addition, the nuclear area turned out to be another independent predictor for progression-free survival [RR for relapse 5.25 (95% CI 1.1–25.9; p=0.04)] and overall survival [RR for death 4.16 (95% CI 1.2–14.4; p=0.02)] in mucinous ovarian cancers.
Table 5.
Multivariate survival analysis of 70 patients with primary ovarian cancer.
| Progression-free survival |
Overall survival |
||||
|---|---|---|---|---|---|
| Variable | RR of progression (95% CI) | P | RR of death (95% CI) | P | |
| Age | ≤ 60 vs. ≥ 60 yrs. | – | n.s. | 3.3 (1.6–6.5) | < 0.001 |
| Tumor grade | I/II vs. III | 2.0 (1.03–4.0) | 0.04 | 2.0 (1.03–3.8) | 0.04 |
| Residual disease after surgery | no vs. yes | 3.4 (1.4–7.8) | 0.01 | 4.1 (2.0–8.5) | < 0.001 |
| Hyperploidy | low vs. high percentages | – | n.s. | 2.7 (1.4–5.3) | 0.003 |
| LINE1 DNA-hypomethylation | low vs. high PUMR values | 2.2 (1.1–4.3) | 0.03 | – | n.s. |
Note : The significance level was determined by Cox PH Model, backward elimination of variables. Variables at regression start: Age, FIGO stage, Grade, Residual disease after surgery, Nuclear Area, Percentage of Hyperploidies, Percentage of Aneuploidies, Proliferation index, Chr1 Sat2 hypomethylation, Chr1 Sat α hypomethylation, LINE1 DNA-hypomethylation. RR, relative risk.
4. Discussion
Our study demonstrates associations between the degree of genome-wide DNA-hypomethylation and nuclear morphometric changes on the one hand and the somatic chromosomal ploidy status on the other hand. Hence the question arises whether these associations are based on a causal or non-causal relationship. It has been shown recently that DNA-hypomethylation promotes cancer through effects on chromosomal stability [5]. The dramatic changes of the methylation pattern in transformed cells may lead to global chromosomal instability associated with reactivation of transposable elements, loss of imprinting, illegitimate expression, aneuploidy, and mutations [24]. It is worth mentioning that nonetheless we could not identify a significant association between tumor grading and global DNA-hypomethylation assessed either by LINE1 MethyLight PCR or Chr1 Sat2 and Satα Southern blot analysis. However, the analysis of LINE1 DNA-hypomethylation and the percentage of aneuploidy, revealed a strong statistical association, when both variables were categorized along their respective median values into “low” and “high” subgroups.
The identified relationship between nuclear size and DNA-hypomethylation (LINE1 and Chr1 Sat2) may reflect the chromatin decondensation induced by an overall decrease in the level of 5-methylcytosines. It has been shown that methylated cytosines of silenced promoters bind specific repressive proteins, the so called methyl-CpG-binding-domain proteins, in a complex that also includes histone deacetylases which leads to chromatin condensation [25,26].
In contrast, we were unable to identify an association between Chr1 Satα DNA-hypomethylation and nuclear area.
Interestingly, we found LINE1 hypomethylation to be positively correlated with the age of ovarian cancer patients. This finding is tempting to speculate that tumors in older patients are more prone to be affected by an extreme genome-wide hypomethylation. Although global hypomethylation has been implicated in mechanisms of senescence [27] age-dependency for the extent of hypomethylation in cancers might not be true for all tumor entities. In contrast to what we have found in ovarian cancer, a relationship between LINE1 extreme hypomethylation and younger age at diagnosis was revealed in colorectal cancers [28]. Furthermore, it is worth mentioning that a recent study aimed to validate LINE1 methylation status as a predictor for bladder cancer risk revealed that healthy women were more likely to have lower LINE1 methylation than men when peripheral blood-derived DNA was analyzed. Indeed this trial pointed out that the degree of LINE1 hypomethylation is associated with increased risk for bladder cancer, but interestingly this risk association was by far stronger in women than in men [29]. All this may point to a certain gender specificity of LINE1 methylation status and may therefore represent an issue of particular interest in the tumorigenesis of ovarian cancer but also of other gynecologic cancers.
Moreover, in the entire training set of 70 patients, we were able to show that LINE1 DNA-hypomethylation is an independent prognostic factor for poor progression-free survival in ovarian cancer, but not for overall survival. Although in the univariate survival analysis, Chr1 Sat2 DNA-hypomethylation was of prognostic value in predicting as well progression-free as overall survival, this prognostic effect could not be retained in the multivariate Cox regression model. Recently, we found Chr1 Sat2 DNA-hypomethylation to be an independent prognostic factor for poor overall and progression-free survival, but this was only true in patients who were debulked to no macroscopic residual disease during primary surgery [6]. Unfortunately in this study, we were not able to perform the multivariate survival analysis restricted to patients without macroscopic residual disease after surgery because of the limited size of the study collective.
The prognostic significance of DNA ploidy remains disputable in ovarian cancer [7–13]. However, the majority of studies found at least DNA-aneuploidy to be an independent prognosticator and even one group of investigators revealed DNA-ploidy measurements optimized by cytokeratin gating to be superior to residual disease in predicting clinical outcome in advanced FIGO III cancers [13]. Other trials nonetheless were unable to confirm these promising results. Even though in the present investigation we found DNA-aneuploidy to be of prognostic relevance in univariate analysis for overall survival, this prognostic significance was completely lost in the multivariate model. From all the spectrophotometric variables obtained in the training set of 70 patients, only the percentage of hyperploidy was identified as an independent predictor for poor overall survival.
The heterogeneity of histological types and especially the over-representation of mucinous cancers in our relatively small study population account for some limitations in the interpretation of the herein obtained results. To avoid histology-related biases, it has been proposed to study the various histological subtypes in completely separated studies. Nonetheless, when in the present study both serous and mucinous cancers were compared; no differences in the investigated variables could be pointed out with the exception of Chr1 Satα DNA-hypomethylation, which was more common in serous cancers. However, the most meaningful and unexpected finding of this pilot investigation was, that subgroup survival analyses based on histology revealed that prognostic relevance either of surrogate parameters for global DNA-hypomethylation or of spectrometric variables was exclusively observed in mucinous and not in serous or endometrioid (data not shown) cancers. It is obvious that the significant prognostic relevance observed for the various variables in the entire cohort of 70 patients is essentially due to their tremendous prognostic impact in mucinous tumors. Therefore the herein observed findings on prognosis related to the entire cohort should be interpreted with caution because it is more than questionable that these results could be verified in a larger collective of ovarian cancer patients, in which histological subtypes, especially mucinous cancers are differently represented.
Although both LINE1 DNA-hypomethylation and the nuclear areas in tumor cells are not different in serous compared with mucinous cancers, it appears that the clinical and even biological role of these parameters may be totally different in mucinous compared to serous cancers. Nonetheless, a proper interpretation of these preliminary data requests further validations, which are ongoing in a larger study collective in order to increase statistical power and to include herein unconsidered histological types (e.g. undifferentiated and clear-cell cancer). Furthermore, elucidation of the molecular mechanisms responsible for the different function of LINE1 DNA-hypomethylation in serous compared to mucinous ovarian cancers is certainly warranted.
In conclusion, this pilot approach outlined associations between aberrations in nuclear morphometry, DNA ploidy and global DNA-hypomethylation in ovarian cancer and we furthermore, identified LINE1 DNA-hypomethylation and the nuclear area of cancer cells as two independent prognosticators, whose relevance however, is strictly confined to the histological subtype of mucinous ovarian cancer.
Acknowledgments
We thank I. Gaugg and M. Fleischer for their excellent technical assistance.
Footnotes
Conflict of interest statement The authors declare that there are no conflicts of interest.
References
- [1].Lipsanen V, Leinonen P, Alhonen L, Janne J. Hypomethylation of ornithine decarboxylase gene and erb-A1 oncogene in human chronic lymphatic leukemia. Blood. 1988;72:2042–4. [PubMed] [Google Scholar]
- [2].Hanada M, Delia D, Aiello A, Stadtmauer E, Reed JC. bcl-2 gene hypomethylation and high-level expression in B-cell chronic lymphocytic leukemia. Blood. 1993;82:1820–8. [PubMed] [Google Scholar]
- [3].Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–96. [PubMed] [Google Scholar]
- [4].Ovchinnikov I, Rubin A, Swergold GD. Tracing the LINEs of human evolution. Proc Natl Acad Sci USA. 2002;99:10522–7. doi: 10.1073/pnas.152346799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- [6].Widschwendter M, Jiang G, Woods C, Müller HM, Fiegl H, Goebel G, et al. DNA hypomethylation and ovarian cancer biology. Cancer Res. 2004;64:4472–80. doi: 10.1158/0008-5472.CAN-04-0238. [DOI] [PubMed] [Google Scholar]
- [7].Blumenfeld D, Braly PS, Ben-Ezra J, Klevecz RR. Tumor DNA content as a prognostic feature in advanced epithelial ovarian carcinoma. Gynecol Oncol. 1987;27:389–402. doi: 10.1016/0090-8258(87)90264-2. [DOI] [PubMed] [Google Scholar]
- [8].Friedlander ML, Hedley DW, Swanson C, Russell P. Prediction of long-term survival by flow cytometric analysis of cellular DNA content in patients with advanced ovarian cancer. J Clin Oncol. 1988;6:282–90. doi: 10.1200/JCO.1988.6.2.282. [DOI] [PubMed] [Google Scholar]
- [9].Trope C, Kaern J, Hogberg T, Abeler V, Hagen B, Kristensen G, et al. Randomized study on adjuvant chemotherapy in stage I high-risk ovarian cancer with evaluation of DNA-ploidy as prognostic instrument. Ann Oncol. 2000;11:281–8. doi: 10.1023/a:1008399414923. [DOI] [PubMed] [Google Scholar]
- [10].Bernabei VM, Miller DS, Bauer KD, Murad TM, Rademaker AW, Lurain JR. Flow cytometric evaluation of epithelial ovarian cancer. Am J Obstet Gynecol. 1990;162:1584–90. doi: 10.1016/0002-9378(90)90924-v. [DOI] [PubMed] [Google Scholar]
- [11].Pfisterer J, Kommoss F, Sauerbrei W, Renz H, du Bois A, Kiechle-Schwarz M, et al. Cellular DNA content and survival in advanced ovarian carcinoma. Cancer. 1994;74:2509–15. doi: 10.1002/1097-0142(19941101)74:9<2509::aid-cncr2820740919>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- [12].Coley HM, Sargent JM, Titley J, Taylor CG. Lack of prognostic significance of ploidy and S-phase measurements in advanced ovarian cancer. Anticancer Res. 1999;19:2111–6. [PubMed] [Google Scholar]
- [13].Kimmig R, Wimberger P, Hillemanns P, Kapsner T, Caspari C, Hepp H. Multivariate analysis of the prognostic significance of DNA-ploidy and S-phase fraction in ovarian cancer determined by flow cytometry following detection of cytokeratin-labeled tumor cells. Gynecol Oncol. 2002;84:21–31. doi: 10.1006/gyno.2001.6440. [DOI] [PubMed] [Google Scholar]
- [14].Yusup A, Kanamaru H, Aoki Y, Saikawa S, Moriyama N, Oyama N, et al. Prognostic value of nuclear area index in patients with bladder cancer. Int J Urol. 2004;11:374–8. doi: 10.1111/j.1442-2042.2004.00813.x. [DOI] [PubMed] [Google Scholar]
- [15].Bol MG, Baak JP, Rep S, Marx WL, Kruse AJ, Bos SD, et al. Prognostic value of proliferative activity and nuclear morphometry for progression in TaT1 urothelial cell carcinomas of the urinary bladder. Urology. 2002;60:1124–30. doi: 10.1016/s0090-4295(02)01906-4. [DOI] [PubMed] [Google Scholar]
- [16].Klein M, Graf AH, Rosen A, Lahousen M, Hacker GW. Tumor progression, histologic grading and DNA-ploidy as predictive factors of lymphogenous metastasis in primary carcinoma of the Fallopian tube. Cancer Lett. 2002;177:209–14. doi: 10.1016/s0304-3835(01)00801-1. [DOI] [PubMed] [Google Scholar]
- [17].Colomb E, Kopp F, Spyratos F, Martin PM. Cell-cycle studies by multiparametric automatic scanning of topographically preserved cells in culture. Cytometry. 1989;10:263–72. doi: 10.1002/cyto.990100305. [DOI] [PubMed] [Google Scholar]
- [18].Charpin C, Martin PM, Lissitzky JC, Jacquemier J, Kopp F, Pourreau-Schneider N, et al. Estrogen receptor immunocytochemical assay (ERICA) and laminin detection in 130 breast carcinomas and computerized (Samba 200) multi-parametric quantitative analysis on tissue sections. Bull Cancer. 1986;73:651–64. [PubMed] [Google Scholar]
- [19].van Velthoven R, Petein M, Oosterlinck WJ, Zandona C, Zlotta A, Van der Meijden AP, et al. Image cytometry determination of ploidy level, proliferative activity, and nuclear size in a series of 314 transitional bladder cell carcinomas. Hum Pathol. 1995;26:3–11. doi: 10.1016/0046-8177(95)90108-6. [DOI] [PubMed] [Google Scholar]
- [20].Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–36. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ehrlich M, Hopkins NE, Jiang G, Dome JS, Yu MC, Woods CB, et al. Satellite DNA hypomethylation in karyotyped Wilms’ tumors. Cancer Genet Cytogenet. 2003;141:97–105. doi: 10.1016/s0165-4608(02)00668-4. [DOI] [PubMed] [Google Scholar]
- [23].Narayan A, Ji W, Zhang XY, Marrogi A, Graff JR, Baylin SB, et al. Hypomethylation of pericentromeric DNA in breast adenocarcinomas. Int J Cancer. 1998;77:833–8. doi: 10.1002/(sici)1097-0215(19980911)77:6<833::aid-ijc6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- [24].Esteller M, Herman JG. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol. 2002;196:1–7. doi: 10.1002/path.1024. [DOI] [PubMed] [Google Scholar]
- [25].Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–81. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- [26].Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–91. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- [27].Jintaridth P, Mutirangura A. Distinctive patterns of age-dependent hypomethylation in interspersed repetitive sequences. Physiol Genomics. 2010 doi: 10.1152/physiolgenomics.00146.2009. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- [28].Baba Y, Huttenhower C, Nosho K, Tanaka N, Shima K, Hazra A, et al. Epigenomic diversity of colorectal cancer indicated by LINE1 methylation in a database of 869 tumors. Mol Cancer. 2010;9:125. doi: 10.1186/1476-4598-9-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wilhelm CS, Kelsey KT, Butler R, Plaza S, Gagne L, Zens MS, et al. Implications of LINE1 methylation for bladder cancer risk in women. Clin Cancer Res. 2010;16:1682–9. doi: 10.1158/1078-0432.CCR-09-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]