Abstract
DNA repair genes are important for maintaining genomic stability and limiting carcinogenesis. We analyzed all single nucleotide polymorphisms (SNPs) of 125 DNA repair genes covered by the Illumina HumanHap300 (v1.1) BeadChips in a previously conducted genome-wide association study (GWAS) of 1,154 lung cancer cases and 1,137 controls and replicated the top-hits of XRCC4 SNPs in an independent set of 597 cases and 611 controls in Texas populations. We found that six of 20 XRCC4 SNPs were associated with a decreased risk of lung cancer with a P value of 0.01 or lower in the discovery dataset, of which the most significant SNP was rs10040363 (P for allelic test = 4.89 ×10−4). Moreover, the data in this region allowed us to impute a potentially functional SNP rs2075685 (imputed P for allelic test = 1.3 ×10−3). A luciferase reporter assay demonstrated that the rs2075685G>T change in the XRCC4 promoter increased expression of the gene. In the replication study of rs10040363, rs1478486, rs9293329, and rs2075685, however, only rs10040363 achieved a borderline association with a decreased risk of lung cancer in a dominant model (adjusted OR = 0.80, 95% CI = 0.62–1.03, P = 0.079). In the final combined analysis of both the Texas GWAS discovery and replication datasets, the strength of the association was increased for rs10040363 (adjusted OR = 0.77, 95% CI = 0.66–0.89, Pdominant = 5×10−4 and P for trend = 5×10−4) and rs1478486 (adjusted OR = 0.82, 95% CI = 0.71 −0.94, Pdominant = 6×10−3 and P for trend = 3.5×10−3). Finally, we conducted a meta-analysis of these XRCC4 SNPs with available data from published GWA studies of lung cancer with a total of 12,312 cases and 47,921 controls, in which none of these XRCC4 SNPs was associated with lung cancer risk. It appeared that rs2075685, although associated with increased expression of a reporter gene and lung cancer risk in the Texas populations, did not have an effect on lung cancer risk in other populations. This study underscores the importance of replication using published data in larger populations.
Keywords: XRCC4, variant, Genetic susceptibility, genome-wide association study, replication study
1. Introduction
Lung cancer remains the leading cause of cancer-related deaths in both men and women, with an estimated 157,300 deaths and 222,520 new cases, accounting for about 28% of all cancer deaths and 15% of all new cancer cases, in the United States in 2010 [1]. Although major risk factor for lung cancer is cigarette smoking, exposure to ionizing radiation, such as radon and medical imaging, is also the recognized risk factor for lung cancer [2–5]. Environmental carcinogenic agents, however, cause lung cancer only in a minority of exposed individuals, suggesting that inherited susceptibility might contribute to the variation in lung cancer risk [6, 7]. Numerous studies [8–12] have shown that individuals with a familial history of lung cancer have an increased risk, which further supports an etiological role of genetic factors in lung cancer risk.
Cellular DNA integrity is constantly threatened by various assaults. DNA damage is caused by both endogenous metabolites, such as reactive oxygen, nitrogen species and lipid peroxidation products, and environmental carcinogens, such as those found in tobacco smoke. DNA damage, if left unrepaired or repaired incorrectly, may result in genetic instability and mutation fixation, subsequently leading to cancer development [13, 14]. Therefore, DNA damage has emerged as a major culprit in cancer [15]. In humans, at least four major repair pathways have been evolved to repair most of the DNA lesions according to their chemical and physical properties [16]. The nucleotide-excision repair (NER) pathway mainly repairs bulk lesions, whereas the base-excision repair (BER) recognizes and removes incorrect and damaged bases. The mismatch repair (MMR) is responsible for correcting replication errors, whereas the DNA double-strand break (DSB) repair involves two major pathways, the non-homologous end joining (NHEJ) and the homologous recombination (HR). The DSBs are the most toxic and mutagenic DNA lesions in human cells, because a single DSB can potentially lead to loss of more than 100 million base pairs of genetic information [17].
Genetic variants in DNA repair genes have been investigated in many association studies of cancer based on either a candidate or pathway approach, with inconsistent results and failure to replicate in later studies, particularly in lung cancer [18–20]. Recently, genome-wide association study (GWAS) has emerged as a powerful agnostic approach for identifying novel susceptibility loci involved in human diseases [21]. Several recent GWA studies have identified some loci associated with lung cancer, including CHRNA3/5 at chromosome 15q25.1, TERT and CLPTM1L at 5p15.33, BAT3-MSH5 at 6p21.33, a common variant rs1051730 in the nicotinic acetylcholine receptor gene cluster on chromosome 15q24, and HLADQA1 at 6p21.31 [22–28]. However, most of the previously studied candidate genes, including DNA repair genes, were not among the top-hit loci in these GWAS datasets. It is possible that contribution of each of a large number of genetic variants to lung cancer susceptibility is weak to be detected in GWA studies.
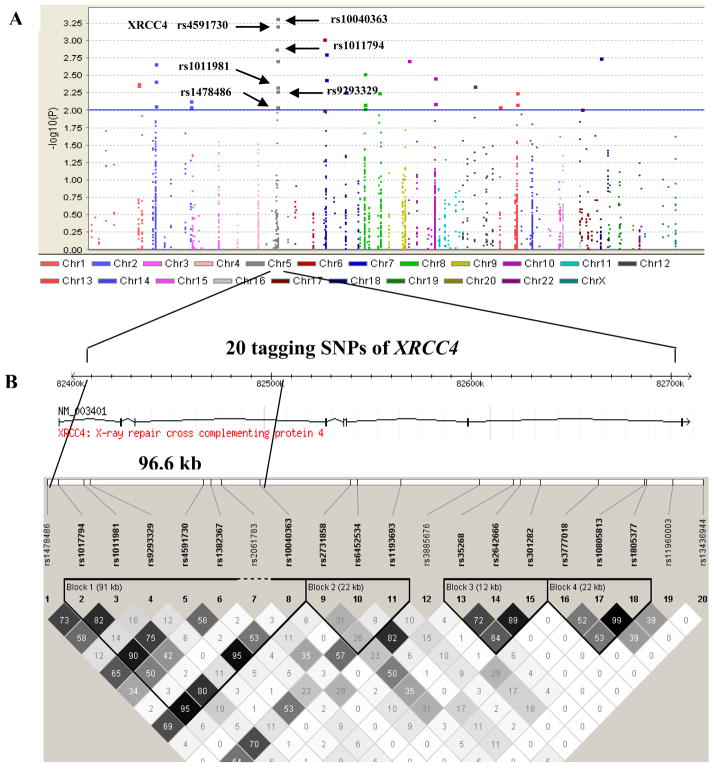
In the present study, we analyzed all 1,806 SNPs in 125 DNA repair genes covered by the Illumina HumamHap300 (v1.1) BeadChip in 1,154 lung cancer cases and 1,137 controls in a Texas population [23]. Although none of the SNPs achieved genome-wide significance (i.e., a P value < 10−7) for an association with lung cancer risk, 32 SNPs had P value of < 10−2, of which 6 SNPs (rs10040363, rs4591730, rs1017794, rs1011981, rs9293329 and rs1478486) were located in the XRCC4 gene region (Fig. 1 and Table 1), suggesting that these XRCC4 variants may be associated lung cancer susceptibility.
Fig. 1.
Tagging SNPs in the XRCC4 gene region in the Texas lung cancer genome-wide association study (GWAS). Associations were expressed as–log (P); P values were obtained from χ2 tests for allele differences. All statistical tests were two sided. (A) Tagging SNPs in the DNA repair pathway associated with lung cancer risk. (B) Tagging SNPs in XRCC4 associated with lung cancer risk. There were 20 tagging SNPs in the XRCC4 region and their LD structure at 5q13-q14, from around 82,412 kb to 82,710 kb in controls from this GWAS. The color of each square represents the pairwise r2; the darker, the stronger r2, with dark black representing r2 = 1 and pure white representing r2 = 0.
Table 1.
Associations between 32 tagging SNPs with P value < 0.01 from DNA repair pathways and the risk of lung cancer based on the Texas lung cancer genome-wide association study
| Gene | SNP | Minor Allele (A1) | Major Allele (A2) | No. of cases A1/A2 | No. of controls A1/A2 | HWEa | MAFb | P valuec |
|---|---|---|---|---|---|---|---|---|
| Base excision repair (BER) | ||||||||
| PARP1 | rs2099380 | A | G | 304/1970 | 245/2063 | 0.7003 | 0.1337 | 0.004113 |
| PARP1 | rs12033699 | G | A | 470/1836 | 543/1731 | 0.9351 | 0.2388 | 0.004358 |
| SMUG1 | rs10161263 | A | G | 711/1597 | 790/1484 | 0.1019 | 0.3474 | 0.004549 |
| Nucleotide excision repair (NER) | ||||||||
| GTF2H5 | rs9348153 | A | G | 148/2160 | 205/2069 | 0.5855 | 0.0902 | 0.000956 |
| GTF2H5 | rs9347131 | G | A | 164/2144 | 209/2065 | 1.0 | 0.0919 | 0.009861 |
| RPA3 | rs7456324 | A | C | 185/2101 | 244/2006 | 0.3574 | 0.1084 | 0.001543 |
| RPA3 | rs1012997 | A | C | 247/2055 | 307/1961 | 0.7049 | 0.1354 | 0.003659 |
| Homologous recombination (HR) | ||||||||
| RBBP8 | rs299247 | A | G | 313/1995 | 240/2034 | 0.1888 | 0.1055 | 0.001781 |
| SHFM1 | rs10808101 | A | G | 338/1966 | 402/1870 | 1.0 | 0.1769 | 0.005479 |
| RAD54B | rs2445729 | A | G | 341/1965 | 405/1869 | 0.1057 | 0.1781 | 0.005611 |
| BRCA2 | rs144848 | C | A | 684/1624 | 595/1679 | 0.8181 | 0.2617 | 0.008829 |
| Non-homologous end joining (NHEJ) | ||||||||
| XRCC4 | rs10040363 | G | A | 1002/1306 | 1104/1170 | 0.9665 | 0.4596 | 0.000489 |
| XRCC4 | rs4591730 | A | G | 976/1330 | 1076/1196 | 0.8658 | 0.4482 | 0.000615 |
| XRCC4 | rs1017794 | A | C | 1020/1288 | 1108/1164 | 0.8666 | 0.4646 | 0.001917 |
| XRCC4 | rs1011981 | G | A | 928/1378 | 1009/1265 | 0.9659 | 0.4229 | 0.00469 |
| XRCC4 | rs9293329 | A | G | 214/2008 | 269/2005 | 0.7394 | 0.1056 | 0.005297 |
| XRCC4 | rs1478486 | A | G | 949/1359 | 1022/1252 | 0.5227 | 0.4302 | 0.00893 |
| DCLRE1C | rs1151737 | A | C | 285/2007 | 354/1912 | 0.5739 | 0.1562 | 0.00194 |
| LIG4 | rs9301233 | G | A | 1066/1242 | 958/1316 | 0.8553 | 0.4213 | 0.005672 |
| LIG4 | rs9555369 | A | G | 728/1580 | 636/1638 | 0.4619 | 0.2797 | 0.008157 |
| XRCC5 | rs828699 | A | C | 961/1347 | 1036/1238 | 0.0559 | 0.4556 | 0.007451 |
| XRCC5 | rs828701 | G | A | 972/1336 | 1044/1228 | 0.1204 | 0.4595 | 0.008924 |
| Mismatch excision repair (MMR) | ||||||||
| MSH3 | rs4703820 | G | A | 270/2038 | 339/1933 | 0.2539 | 0.1492 | 0.001322 |
| Fanconi anemia (FA) | ||||||||
| FANCL | rs6743916 | A | G | 649/1659 | 734/1540 | 0.3423 | 0.3228 | 0.002171 |
| FANCL | rs17269126 | G | A | 322/1952 | 261/2047 | 0.7685 | 0.1416 | 0.003778 |
| FANCL | rs13387871 | A | G | 419/1887 | 483/1789 | 0.0760 | 0.2126 | 0.008612 |
| Direct reversal of DNA damage | ||||||||
| MGMT | rs2308321 | G | A | 245/1983 | 315/1955 | 0.541 | 0.11 | 0.00344 |
| MGMT | rs2246538 | C | A | 704/1604 | 613/1661 | 0.2595 | 0.2696 | 0.008011 |
| Genes defective in diseases associated with sensitivity to DNA damaging agents | ||||||||
| WRN | rs1369887 | A | C | 615/1693 | 696/1578 | 1.0 | 0.3061 | 0.003019 |
| WRN | rs4452759 | A | G | 1057/1249 | 1131/1143 | 0.9527 | 0.4974 | 0.008257 |
| WRN | rs4634594 | A | G | 1056/1252 | 953/1319 | 0.4285 | 0.4195 | 0.009405 |
| Other conserved DNA damage response genes | ||||||||
| PER1 | rs2585408 | A | G | 1023/1285 | 922/1352 | 0.2187 | 0.4055 | 0.009668 |
Hardy-weinberg equilibrium.
Minor allele frequency in controls.
χ2 test for the difference in the distributions of alleles.
To further test for the significance of loci in XRCC4 associated with lung cancer susceptibility, we did following as the validation and replication of the finding: 1) we used genotype imputation to infer untyped XRCC4 SNPs, thereby increasing the chance to capture putative, untyped causal and functional SNPs; 2) we performed additional experiments to identify functional relevance of the observed significant SNPs, 3) we performed a replication study of the observed significant SNPs in an independent set of 597 cases and 611 controls from a similar Texas population to that used in the discovery phase, and 4) we conducted a meta-analysis of the Texas GWA and replication studies with four additional GWA studies of a total of 12,312 primary lung cancer cases and 47,921 controls.
2. Materials and Methods
2.1 Study participants
The study protocols were approved by the Institutional Review Board of the University of Texas M. D. Anderson Cancer Center. Informed consent was obtained from all participants.
Texas GWA study
The discovery phase included a subset of Texas populations in our ongoing lung cancer case-control studies at The University of Texas M. D. Anderson Cancer Center [23]. The ascertainment and matching criteria of cases and controls and the methods of recruitment, genotyping, and quality control have been described in details elsewhere [23, 29]. Briefly, the GWAS dataset included 1154 lung cancer cases and 1137 controls. Cases were patients with newly diagnosed and histopathologically-confirmed lung cancer, and controls were healthy individuals seen for routine care at a multispecialty physician practice in Houston, with a frequency matching to the cases by age (in 5 year categories), sex and smoking status. The demographic characteristics of this study population are shown as Table 2.
Table 2.
Characteristics of study population
| Texas discovery set |
Texas replication set |
|||||
|---|---|---|---|---|---|---|
| Characteristics | Cases | Controls | Pa | Cases | Controls | Pa |
| (n =1154, % ) | (n =1137, %) | (n=597, % ) | (n=611, %) | |||
| Age (years) | < 0.0001 | 0.275 | ||||
| Range | 31–92 | 31–86 | 21–94 | 28–92 | ||
| Mean | 62.1± 10.75 | 61.1± 8.91 | 61.8± 11.99 | 61.6± 11.12 | ||
| ≤50 | 195(16.90) | 137 (12.05) | 109 (18.26) | 91 (14.89) | ||
| 50 – 60 | 286 (24.78) | 359 (31.57) | 133 (22.28) | 137 (22.42) | ||
| >60 | 673 (53.82) | 641 (56.38) | 355 (59.46) | 383 (62.68) | ||
| Sex | 0.855 | 0.547 | ||||
| Male | 658 (57.02) | 644 (56.64) | 318 (53.27) | 336 (54.99) | ||
| Female | 496 (42.98) | 493 (43.36) | 279 (46.73) | 375 (45.01) | ||
| Smoking Status | 0.0054 | 0.536 | ||||
| Current smoker | 600 (51.99) | 657 (57.78) | 304 (50.92) | 322 (52.70) | ||
| Former smoker | 554 (48.01) | 480 (42.22) | 293 (49.08) | 289 (47.30) | ||
| Pack-years smoked b | <0.0001 | <0.0001 | ||||
| 0– 25 | 209 (18.11) | 288 (25.33) | 130 (21.78) | 207 (33.88) | ||
| 26–50 | 458 (39.69) | 492 (43.27) | 226 (37.86) | 229 (37.48) | ||
| > 50 | 487 (42.20) | 357 (31.40) | 241 (40.37) | 175 (28.64) | ||
Two-sided χ2 test.
Cigarettes per day × years smoked.
Replication study
The replication study was an independent sample drawn from the same case-control populations as we did in the GWAS discovery phase, including 602 cases with histopathologically confirmed lung cancer, and 618 healthy controls with the same matching criteria.
Meta-analysis
The meta-analysis of our GWA study and replication study with genotyping data from four additional GWA studies was conducted. These four additional GWA studies were from the National Cancer Institute (NCI), UK, the International Agency for Research on Cancer (IARC) and deCODE Genetics. We communicated with the corresponding or principal investigators of these studies to request data on genotype frequencies in cancer cases and non-cancer controls for the XRCC4 variants. Details of case and control ascertainment and matching criteria as well as the genotyping in each of these studies have been published previously [24, 25, 27, 28]. Briefly, the NCI study, which included 5,739 cases and 5,848 controls, was based on one population-based case-control study in Italy, a cohort study in Finland and two cohort studies in all U.S. states; The UK study included 1,978 cases from Genetic Lung Cancer Predisposition Study (GELCAPS), and 1,438 controls from the 1958 Birth cohort; The IARC study, which included 1968 case and 2598 control, was based on a lung cancer case-control study conducted in 6 central European countries (Czech Republic, Hungary, Poland, Romania, Russia and Slovakia); The 876 cases and 36,272 controls in the deCODE Genetics study were drawn from one population-based case-control study from deCODE Genetics in Iceland. Together, the meta-analysis included a total of 12,312 primary lung cancer cases and 47,921 controls, which were all European descent. Genotyping of all subjects from GWA studies of IARC and deCODE Genetics was conducted using the HumanHap300 K, while that from UK conducted using the HumanHap550K. Three illumina platforms, i.e., the HumanHap550K, the HumanHap610 and the HumanHap 1 Million Chips, were used in the NCI GWAS.
2.2 SNPs selection and genotyping
The discovery dataset had data on 317,498 tagging SNPs [23], of which 20 SNPs in XRCC4 were covered and screened for replication using Applied Biosystems TaqMan genotyping platform according to the manufacture’s recommendations. Briefly, the reactions were prepared by using TaqMan Universal Master Mix, 80×SNP Genotyping Assay Mix, Dnase-free water, and 10-ng genomic DNA in a final volume of 5 μL per reaction. Both negative and positive controls were included in each plate to ensure the accuracy of the genotyping. The PCR amplification was run, and the plate was read using a TaqMan 7900 HT sequence detection system (Applied Biosystems). The analyzed fluorescence results were then auto-called in to the genotypes using the built-in SDS2.3 software of the system.
2.3 Genotype imputation
To infer potentially functional SNPs tagged by the 20 genotyped XRCC4 SNPs covered by the HumanHap300 (v1.1) BeadChips, we imputed genotypes using these 20 SNPs within a 307 kb region on chromosome 5q 14.2 from 82,407,760 bp to 82,715,135 bp, covering the whole region of the XRCC4 gene. The imputation was conducted using a Hidden Markov Model programmed in MACHv1.0 (http://www.sph.umich.edu/csg/abecasis/MACH/) [30]. The imputation method combined the observed GWAS genotype data with the HapMap CEU genotype data as a reference panel (Release 24/Phase II Apr07, on NCBI B36 assembly, dbSNP b126, http://www.hapmap.org/cgi-perl/gbrowse/hapmap24_B36/) and then inferred the untyped genotypes probabilistically. A minor allele frequency [MAF] > 0.01 and an estimated r 2 > 0.3 were chosen as thresholds to flag and discard low quality imputed SNPs without removing many successfully imputed markers [31, 32]. We found that only one imputed SNP, rs2075685 was located in the promoter region (−651G>T) of XRCC4 and therefore is likely to affect its function. We therefore studied the effect of variation in this SNP for additional functional studies.
2.4 Cell lines
To explore a potential function of the imputed SNP rs2075685 located in the promoter region of XRCC4, we further performed a luciferase reporter assay. The human colon cancer cell line HCT116 was obtained from John Hopkins University School of Medicine (a gift from Dr. Bert Vogelstein). The human non-small cell lung carcinoma cell line H1299 and human cervical cancer Hela cell line were obtained from the American Type Culture Collection (ATCC, Manassas, VA). HCT116 and Hela cells were cultured in 1X Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (Sigma-Aldrich, MO), and H1299 cells were cultured in RPMI1640 medium with 10% fetal bovine serum at 37ºC in 5% CO2.
2.5 Construction of luciferase reporter plasmid
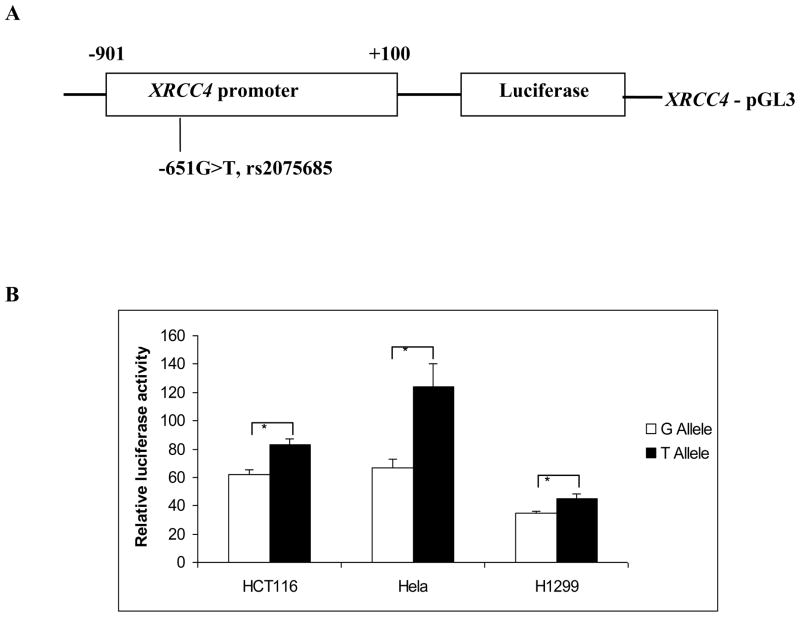
The 1001-bp XRCC4 promoter (from −901 to +100 bp relative to the translation start site) was cloned by PCR with the primers of 5'-AAGGTACCCCAGGTGGTAAATTCGTCCA-3' (forward) and 5'-AAGCTAGCATCTAAATCCCGCCTTTTCC-3' (reverse), including the KpnI and NheI restriction sites. The difference between G and T alleles in the promoters was obtained by using the DNA template from subjects homozygous (GG and TT) for the -651G>T (rs2075685) variant. The PCR products were cloned into the basic-pGL3 firefly luciferase vector (Promega, Madison, WI) at the KpnI and NheI restriction sites. The -651G and -651T constructs were sequenced to confirm the orientation and integrity of each insert (Fig. 2A).
Fig. 2.
The luciferase reporter gene assay using constructs with different alleles of the XRCC4 promoter. (A) Schematic presentation of the position of rs2075533 SNP relative to the transcription start site. (B) Luciferase expression of the constructs in HCT116, H1299 and Hela cell lines. Values represented fold change of the luciferase activity relative to the wild-type construct as 1. Each bar represented the mean of triplicate transfection plates plus standard deviation. * P < 0.01 compared with the wild-type construct.
2.6 Transient transfection and luciferase reporter assays
The cultured cells were transiently transfected with 1 μg of each of the XRCC4-reporter plasmids by FuGENE HD (Roche Applied Science, IN) in 12-well culture plates. The p-TK renilla luciferase (pRL-TK) (Promega) was co-transfected as an internal control. The pGL3 basic vector without the insert was used as a negative assay control. The cells were collected 48 hours after transfection and analyzed for luciferase activity with a Dual-Luciferase Reporter Assay System (Promega). The relative luciferase activity was calculated according to the manufacturer’s instruction using a luminometer (TD-20/20 DLReasy, Promega). Promoter activity was calculated for each of the constructs as a ratio of the luciferase activity to that of the pGL3 basic vector. All transfections were performed in triplicate.
2.7 Statistical methods
The Hardy-Weinberg equilibrium (HWE) was tested using a χ2 test with one degree of freedom for each SNP. The differences in the distributions of demographic characteristics, selected variables, and genotypes between cases and controls were examined using the χ2 test. Haploview program (v 4.1) (http://www.broad.mit.edu/mpg) was used to infer the linkage disequilibrium (LD) structure among the 20 SNPs. Logistic regression was used to calculate the odds ratios (ORs) and 95% confidence intervals (CIs) for the association between a single locus and lung cancer risk with and without adjustment for age, sex, smoking status. The case-control data sets were tested for heterogeneity using the Breslow-Day test. As no significant heterogeneity was identified between the two study population subsets, all raw genotype data were combined for the final analysis. All the P values reported here were two sided. Statistical tests were performed using the SAS statistical software (version 9.13; SAS Institute, Cray, NC) and PLINK software (version 1.06; http://pngu.mgh.harvard.edu/purcell/plink/) [33]. In the meta-analysis, we combined data from our own studies and from an additional four published GWA studies using Review Manager (RevMan) version 4.3 (Cochrane Collaboration, Copenhagen, Denmark). A random-effects model was used to pool the results depending on the heterogeneity between studies [34]. Between-study heterogeneity was tested by the χ2-based Q-statistic and the I2 statistic, where I2 greater than 50% is considered significantly heterogeneous [35].
3. Results
3.1 Single marker association analysis of the GWAS data
From the initial screening analysis for significant SNPs of the 125 DNA repair genes (Fig. 1 and Table 1), we identified 32 SNPs of 17 genes with a P value for an association with risk of lung cancer. These genes included two in BER, two in NER, four in HR and one in MMR. We chose the XRCC4 gene for further investigation, because this gene had four SNPs associated with lung cancer in the study population, and these associations have not been reported before. Therefore, we further assessed the risk associated with all 20 SNPs in XRCC4 covered by the Illumina HumanHap300 (v1.1) BeadChips, which represents a region mapping to a 234-kb region of chromosome 5 spanning between 82,412,760 bp and 82,710,135 bp. The 20 SNPs were distributed across intronic (n = 18; 90%), or flanking intergenic regions (n = 2, 10%).
Genotype distributions of all 20 SNPs were in Hardy-Weinberg equilibrium among the controls (P > 0.05). Further examination of the LD pattern among the six top-hit SNPs revealed that except for rs1478486 and rs9293329, the other four SNPs (i.e., rs10040363, rs4591730, rs1017794, and rs1011981) were in strong LD (r2 = 0.95 and D’ = 1.0 for rs10040363 and rs4591730, r2 = 0.95 and D’ = 0.98 for rs10040363 and rs1017794, r2 = 0.80 and D’ = 0.97 for rs10040363 and rs1011981, and r2 = 0.83 and D’ = 1.0 for rs1017794 and rs1011981) in the controls (Fig. 1B). We then evaluated associations between these three SNPs (i.e., rs10040363, rs1478486 and rs9293329) and lung cancer risk in both additive and dominant models and found that except for rs9293329, which was associated with a decreased lung cancer risk in a dominant model, other two SNPs (rs10040363 and rs1478486) were nominally significantly associated with a decreased risk of lung cancer in either an additive or a dominant model with adjustment for age, sex, smoking status, and pack-years of smoking (Table 3).
Table 3.
Results from association testing of the top SNPs in XRCC4
| Texas discovery a | Texas Replication a | Combined data set | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Genotype | Cases/Controls | OR (95%CI)b | P value | Cases/Controls | OR (95%CI)b | P value | Cases/Controls | OR (95%CI)b | P value |
| All subjects | 1154/1137 | 597/611 | 1751/1748 | |||||||
| rs10040363 | AA | 370/298 | Reference | 188/165 | Reference | 558/463 | Reference | |||
| AG | 563/574 | 0.80 (0.66–0.97) | 0.024 | 282/318 | 0.77 (0.59–1.00) | 0.053 | 845/892 | 0.79 (0.68–0.92) | 0.003 | |
| GG | 218/265 | 0.66 (0.52–0.84) | 0.0006 | 127/128 | 0.87(0.63–1.21) | 0.417 | 345/393 | 0.72 (0.60–0.87) | 0.0009 | |
| AG+GG | 781/839 | 0.76 (0.63–0.91) | 0.003 | 409/446 | 0.80 (0.62–1.03) | 0.079 | 1190/1285 | 0.77 (0.66–0.89) | 0.0005 | |
| P-trend b | 0.0005 | 0.314 | 0.0005 | |||||||
| rs1478486 | GG | 398/338 | Reference | 0.007 | 199/180 | Reference | 597/518 | Reference | ||
| AG | 561/576 | 0.84 (0.70–1.02) | 0.069 | 292/316 | 0.83 (0.64–1.08) | 0.168 | 853/892 | 0.84 (0.72–0.98) | 0.025 | |
| AA | 192/223 | 0.72 (0.57–0.92) | 0.009 | 106/115 | 0.82 (0.59–1.15) | 0.257 | 298/338 | 0.76 (0.62–0.92) | 0.006 | |
| AG+AA | 753/799 | 0.81 (0.68–0.97) | 0.018 | 398/431 | 0.83 (0.65–1.06) | 0.139 | 1151/1230 | 0.82 (0.71–0.94) | 0.006 | |
| P-trend b | 0.007 | 0.197 | 0.0035 | |||||||
| rs9293329 | GG | 948/884 | Reference | 489/502 | Reference | 1437/1386 | Reference | |||
| AG | 192/237 | 0.75 (0.60–0.93) | 0.008 | 99/100 | 1.02 (0.75–1.38) | 0.925 | 291/337 | 0.83 (0.69–0.98) | 0.035 | |
| AA | 11/16 | 0.72 (0.57–0.92) | 0.344 | 9/9 | 0.99 (0.38–2.53) | 0.975 | 20/25 | 0.79 (0.44–1.44) | 0.444 | |
| AG+AA | 203/253 | 0.69 (0.32–1.49) | 0.006 | 108/109 | 1.01 (0.75–1.36) | 0.937 | 311/362 | 0.82 (0.70–0.98) | 0.028 | |
| P-trend b | 0.006 | 0.951 | 0.030 | |||||||
Three cases in the discovery set, five cases and seven controls in the replication set were removed from the current analysis because of calls missing or unclear identity for their genotypes.
Adjusted for age in years, sex, smoking status and pack-years.
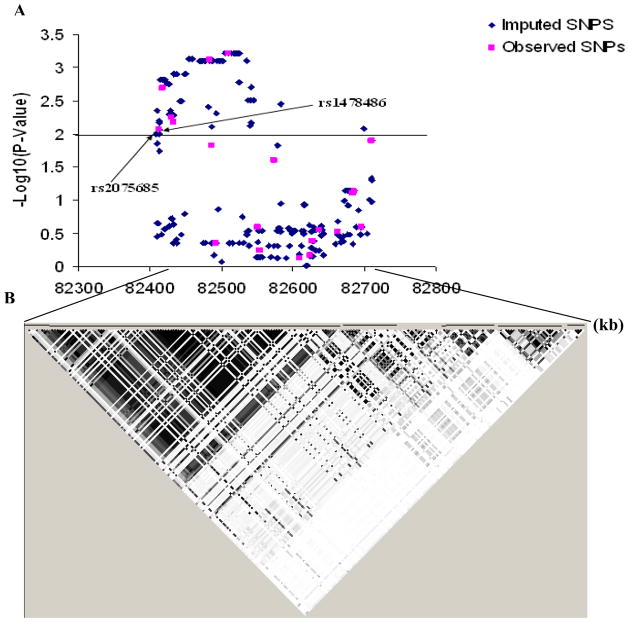
These six top-hit SNPs map to about a 96.6-kb region of chromosome 5 extending from 82,412,760 bp to 82,509,401 bp (Fig. 1B), located in the intronic regions of XRCC4, with two SNPs (rs10040363 and rs4591730) in intron 3 and the other four SNPs in intron 1. To identify potential functional SNPs that may be linked with these tagging SNPs, we used imputation to further map the associated SNPs across a 307-kb region on chromosome 5q14.2 from 82,407,760 bp to 82,715,135 bp, spanning the whole region of the XRCC4 gene. Imputation expanded the number of SNPs from 20 observed SNPs to a total of 236 SNPs in the 307-kb region. After exclusion of imputed SNPs based on quality control measures (r 2 = 0.3 and MAF > 0.01), 218 SNPs remained in the final imputed dataset (Fig. 2), of which only two imputed SNPs (rs2075685 and rs1056503) were located in the functional regions of XRCC4. The imputed SNP rs1056503 is located in exon 8 of XRCC4 and in high LD (r2 = 1.0 and D’ = 1.0 in the CEU component of HapMap) with the observed SNP rs1805377, but it was not associated with lung cancer risk (imputed P for allelic test = 0.1151) (data not shown). Notably, the other imputed SNP, rs2075685, located in the promoter region of XRCC4 was found to be in high LD (r2 = 0.90 and D’ = 1.0) with the observed top SNP rs1478486 (imputed P for allelic test = 0.0013) (data not shown) and was predicted to be functional via the analysis of the TRANSFAC (Searching Transcription Factor Binding Sites) program [36].
To further determine the allele-specific effects of −651G>T (rs2075685, which is in strong LD with the top-hit SNP rs1478486 (r2 = 0.90 and D’ = 1.0) on the XRCC4 promoter activity, we generated two luciferase reporter gene constructs with 1,001 bp of the XRCC4 promoter region (from −901 to +100), containing either G or T at the −651 position (Fig. 3A). As shown in Fig. 3B, the XRCC4 promoter containing the protective T allele displayed a significantly higher reporter gene expression, compared with the G allele in the human cervical cancer cell line Hela (P < 0.01), the human non-small cell lung carcinoma cell line H1299 (P < 0.01), and the human colon adenocarcinoma cell line HCT116 (P < 0.01), suggesting that G to T allele change at XRCC4-651 may increase the XRCC4 promoter activity in a non-tissue specific manner.
Fig. 3.
The associations between lung cancer risk and the SNPs within the 307-kb region on chromosome 5q13-q14 between 82,407-kb and 82,715-kp, covering the whole region of the XRCC4 gene. (A) Associations between all 218 SNPs and lung cancer risk. The pink points were for the observed SNPs; the blue ones were for the imputed SNPs. P values were estimated using additive models in logistic regression with adjustment for age, sex, smoking status and pack-years. (B) Inferred haplotype blocks of these SNPs were estimated in controls using the Haploview program (v 4.1) (http://www.broad.mit.edu/mpg). The color of each square represents the pairwise r2; the darker, the stronger r2, with dark black representing r2 = 1 and pure white representing r2 = 0.
3.2 Replication study and combined analysis of all genotype data
Among the six top-hit SNPs, four SNPs (i.e., rs10040363, rs4591730, rs1017794 and rs1011981) are in strong LD, and rs10040363 showed the strongest evidence of an association with lung cancer risk in the Texas GWAS discovery dataset. Hence, we selected rs10040363, other two observed top-hit SNPs (rs1478486 and rs9293329), and the imputed functional SNP rs2075685 for further replication in an additional 602 cases and 618 controls from the same Texas population. Among these subjects, five case and seven controls failed in the genotyping assays. Thus, the final analysis included 597 lung cancer cases and 611 controls. The frequency distribution of selected characteristics of the cases and controls is presented in Table 2. Because of frequency matching, there was no statistically significant difference in the distributions of age, sex and smoking status between cases and controls in the replication population that was similar to the population used in the discovery GWAS.
The genotype frequencies of these four SNPs among the controls were all in agreement with the Hardy-Weinberg equilibrium (chi-square test: P = 0.269 for rs10040363, P = 0.253 for rs1478486, P = 0.126 for rs9293329, P = 0.382 for rs2075685). The SNP rs10040363 G allele was associated with a decreased risk of lung cancer with a borderline statistical significance (adjusted OR = 0.80, 95%CI = 0. 62–1.03, P = 0.079) (Table 3) in the replication set. There was a similar, but non-significant association with the rs1478486 A allele (adjusted OR = 0.83 and 95% CI = 0.65–1.06), but rs9293329 was not associated with significant altered risk (Table 3). Except for rs9293329, the trends for association between rs10040363 and rs1478486 and lung cancer risk were in the identical direction in both the discovery and replication sets, and therefore, the lack of significance of the replication was likely due to limited power because of smaller sample size of the replication set. For the imputed functional rs2075685, the T allele also tended to be associated with decreased lung cancer risk (adjusted OR = 0.83 and 95% CI = 0.64–1.07, P = 0.152) (data not shown).
Because the observed risks associated with the replicated SNPs were in the same direction in both the discovery and replication datasets, we then combined the two datasets to increase study power. Because rs2075685, which is in strong LD with the top-hit SNP rs1478486 (r2 = 0.90 and D’ = 1.0) was imputed and not directly genotyped, we did not include this SNP in the final combined analysis. Using the Breslow-Day test, we found no statistically significant evidence of heterogeneity between the GWAS population and the replication population for rs10040363, rs1478486, and rs9293329. As shown in Table 3, the strength of association for either rs10040363 G allele or rs1478486 A allele was substantially enhanced (adjusted OR = 0.77, 95% CI = 0.66–0.89, P = 5×10−4 and P for trend = 5×10−4 for rs10040363 and adjusted OR = 0.82, 95% CI = 0.71 −0.94, P = 6×10−3, and P for trend = 3.5×10−3 for rs1478486).
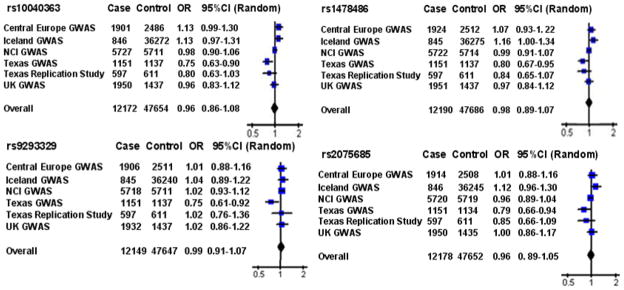
We subsequently conducted a meta-analysis of these four replicated XRCC4 SNPs with a total of 12,312 cases and 47,921 controls using the combined data from the two Texas populations and four published GWA studies. However, the results of the meta-analysis did not show an association with risk of lung cancer for any of these SNPs (Fig. 4). The overall ORs for rs10040363, rs1478486, rs9293329, and rs2075685 were 0.96 (95% CI: 0.86–1.08; Pheterogeneity = 0.002), 0.98 (95% CI: 0.89–1.07; Pheterogeneity = 0.02), 0.99 (95% CI: 0.91 –1.07; Pheterogeneity = 0.15), and 0.96 (95% CI: 0.89 –1.05; Pheterogeneity = 0.07) by random effects, respectively.
Fig. 4.
Forest plot showing association between the four variants in XRCC4 and lung cancer risk. The ORs and 95% CIs are obtained from the dominant model. The x axis corresponds to the OR. The diamonds and the horizontal bars represent the over all ORs with 95%CIs given by their width.
4. Discussion
Using the published Texas lung cancer GWAS discovery dataset, we first analyzed 1,806 SNPs of 125 DNA repair genes, among which 32 SNPs of 17 genes were found to have an allele effect on cancer risk with a P value of <0.01, although no genome-wide significant association was identified. We then assessed the associations between 20 SNPs of XRCC4 (the top-hit gene in the list of 17 genes) and lung cancer risk. We found that, of 20 SNPs, six (i.e. rs10040363, rs4591730, rs1017794, rs1011981, rs1478486, and rs9293329) were associated with risk of lung cancer with a P value of < 1×10−2, and the most significant SNP was rs10040363 (P value for allelic test = 4.89 ×10−4) with an imputed functional rs2075685 SNP (the imputed P value for allelic test = 1.3 ×10−3). The minor alleles of the six top-hit observed SNPs appeared to be protective against lung cancer risk, which is consistent with the data from the luciferase reported assay that further demonstrated that the rs2075685G>T change in the XRCC4 promoter increased XRCC4 expression.
In our replication study of three independent top-hit SNPs and one imputed functional SNP, we found that the rs10040363 G allele was associated with decreased risk of lung cancer with a borderline statistical significance, whereas all the three SNP, i.e.rs1478486, rs9293329 and rs2075685, were not. It is noted, however, that both rs1478486 A and rs2075685 T alleles exhibited reduced lung cancer risk. In the combined analysis of both GWAS discovery and replication datasets, the strength for an association was increased for rs10040363 (Pdominant = 5.0×10−4 and P for trend = 5×10−4) and rs1478486 (Pdominant = 6.0×10−3 and P for trend = 3.5×10−3), and the trends of the risk were consistently in the same direction in both discovery and the replication datasets. In the meta-analysis, however, we did not find evidence of an association between overall lung cancer risk and any of the four XRCC4 SNPs. This underscores the importance of replication of any findings of an effect of a low-penetrance locus on cancer risk, particularly from a GWA study, in different study populations.
XRCC4 is a limiting factor in the NHEJ [37], which is required for both normal development and suppression of tumors. It has been recognized that mouse embryonic cells with disruption of XRCC4 show reduced proliferation, radiation hypersensitivity, chromosomal instability, and severely impaired V(D)J recombination [38]. A deficiency in XRCC4 results not only in an increased sensitivity of cells to X-ray but also may give rise to immunodeficiency in animals [39]. Although our experimental data showed that the rs2075685G>T change in the XRCC4 promoter region increased XRCC4 expression, the association with risk for this functional SNP did not achieve statistical significance in our replication dataset (despite its association with a similarly decreased risk). Since rs2075685 is located in the promoter region in XRCC4 and the G>T change in this SNP increases XRCC4 expression, if rs1478486, one of the observed top-hit SNPs in high LD with rs2075685, contributes to the risk of lung cancer, rs2075685 could be the causal SNP linked to rs1478486. It is also likely that rs10040363 in XRCC4, though intronic, could also be linked to other untyped causal SNPs. It is plausible, however, that disease-associated variants with modest effects may be distributed proportionately between coding and noncoding sequences of the genome [40]. Indeed, several studies have found that ‘functional’ intronic variants are associated with disease occurrence [41, 42]. For example, an intronic SNP in a RUNX1 binding site of SLC22A4, which affects the transcriptional efficiency of SLC22A4, is strongly associated with rheumatoid arthritis [42]. The results from our replication dataset might represent an association of mild to modest effect, but such a weak association was not supported by the meta-analysis.
So far, at least two small studies have reported that rs10040363 and rs2075685 are involved in the susceptibility to lung cancer [43, 44]. A French study in 151 cases and 172 controls found that variant alleles of rs10040363 and rs2075685 were associated with decreased risk of lung cancer. This direction of association is the same as seen in our data. However, a candidate gene study from Taiwan of 164 lung cancer patients and 649 healthy controls found that rs2075685 was not associated with lung cancer risk, and this discrepancy could be due to ethnic differences, genotyping platform and study sizes. Again, these discrepancies further underscore the importance of replication to rule out a chance finding, particularly from under-powered studies.
The limitations of the present study include: 1) our analysis was limited to individuals of non-Hispanic whites, the controls were frequency-matched to the cases by age (within 5 year categories), sex, smoking status, and all the subjects in the discovery and replication datasets were ever smokers; 2) the sample size in the replication phase was smaller than the original discovery dataset; and 3) the subgroup meta-analysis was not conducted because only genotyping data was available from other four GWA studies.
5. Conclusions
In summary, using the data on 1,806 SNPs of 125 DNA repair genes from a published GWAS with a replication study in Texas populations, we first identified rs10040363 in XRCC4 that was associated with lung cancer risk in the study populations. We then identified another variant rs2075685, tagged by rs1478486, in the XRCC4 promoter, that might increase XRCC4 expression. However, the evidence supporting such findings is lacking from both our replication in an independent Texas population and from the meta-analysis of four previously published GWA studies. It appeared that rs2075685, although associated with increased expression of a reporter gene and lung cancer risk in the Texas populations, did not have an effect on lung cancer risk in other populations.
Acknowledgments
We thank Min Zhao, Jianzhong He and Kejing Xu for their laboratory assistance, and Dakai Zhu for his technical support. This study was supported in part by National Institutes of Health grants ES11740 and CA131274 (to Q. W.), CA86390 and CA55769 (to M. R. S.), CA121197 (to C.A.), and CA 16672 (to The University of Texas M. D. Anderson Cancer Center). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Einstein AJ. Medical imaging: the radiation issue. Nat Rev Cardiol. 2009;6:436–438. doi: 10.1038/nrcardio.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert ES. Ionising radiation and cancer risks: what have we learned from epidemiology? Int J Radiat Biol. 2009;85:467–482. doi: 10.1080/09553000902883836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gray A, Read S, McGale P, Darby S. Lung cancer deaths from indoor radon and the cost effectiveness and potential of policies to reduce them. BMJ. 2009;338:a3110. doi: 10.1136/bmj.a3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wakeford R. The cancer epidemiology of radiation. Oncogene. 2004;23:6404–6428. doi: 10.1038/sj.onc.1207896. [DOI] [PubMed] [Google Scholar]
- 6.Alberg AJ, Ford JG, Samet JM. Chest. 2. Vol. 132. 2007. Epidemiology of lung cancer: ACCP evidence-based clinical practice guidelines; pp. 29S–55S. [DOI] [PubMed] [Google Scholar]
- 7.Wright GS, Gruidl ME. Early detection and prevention of lung cancer. Curr Opin Oncol. 2000;12:143–148. doi: 10.1097/00001622-200003000-00008. [DOI] [PubMed] [Google Scholar]
- 8.Bromen K, Pohlabeln H, Jahn I, Ahrens W, Jockel KH. Aggregation of lung cancer in families: results from a population-based case-control study in Germany. Am J Epidemiol. 2000;152:497–505. doi: 10.1093/aje/152.6.497. [DOI] [PubMed] [Google Scholar]
- 9.Liu P, Vikis HG, Wang D, Lu Y, Wang Y, Schwartz AG, Pinney SM, Yang P, de Andrade M, Petersen GM, Wiest JS, Fain PR, Gazdar A, Gaba C, Rothschild H, Mandal D, Coons T, Lee J, Kupert E, Seminara D, Minna J, Bailey-Wilson JE, Wu X, Spitz MR, Eisen T, Houlston RS, Amos CI, Anderson MW, You M. Familial aggregation of common sequence variants on 15q24-25.1 in lung cancer. J Natl Cancer Inst. 2008;100:1326–1330. doi: 10.1093/jnci/djn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matakidou A, Eisen T, Houlston RS. Systematic review of the relationship between family history and lung cancer risk. Br J Cancer. 2005;93:825–833. doi: 10.1038/sj.bjc.6602769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaw GL, Falk RT, Pickle LW, Mason TJ, Buffler PA. Lung cancer risk associated with cancer in relatives. J Clin Epidemiol. 1991;44:429–437. doi: 10.1016/0895-4356(91)90082-k. [DOI] [PubMed] [Google Scholar]
- 12.Tokuhata GK, Lilienfeld AM. Familial aggregation of lung cancer in humans. J Natl Cancer Inst. 1963;30:289–312. [PubMed] [Google Scholar]
- 13.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 14.Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 15.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 16.Nagy Z, Soutoglou E. DNA repair: easy to visualize, difficult to elucidate. Trends Cell Biol. 2009;19:617–629. doi: 10.1016/j.tcb.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Dong LM, Potter JD, White E, Ulrich CM, Cardon LR, Peters U. Genetic susceptibility to cancer: the role of polymorphisms in candidate genes. JAMA. 2008;299:2423–2436. doi: 10.1001/jama.299.20.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hung RJ, Christiani DC, Risch A, Popanda O, Haugen A, Zienolddiny S, Benhamou S, Bouchardy C, Lan Q, Spitz MR, Wichmann HE, LeMarchand L, Vineis P, Matullo G, Kiyohara C, Zhang ZF, Pezeshki B, Harris C, Mechanic L, Seow A, Ng DP, Szeszenia-Dabrowska N, Zaridze D, Lissowska J, Rudnai P, Fabianova E, Mates D, Foretova L, Janout V, Bencko V, Caporaso N, Chen C, Duell EJ, Goodman G, Field JK, Houlston RS, Hong YC, Landi MT, Lazarus P, Muscat J, McLaughlin J, Schwartz AG, Shen H, Stucker I, Tajima K, Matsuo K, Thun M, Yang P, Wiencke J, Andrew AS, Monnier S, Boffetta P, Brennan P. International Lung Cancer Consortium: pooled analysis of sequence variants in DNA repair and cell cycle pathways. Cancer Epidemiol Biomarkers Prev. 2008;17:3081–3089. doi: 10.1158/1055-9965.EPI-08-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vineis P, Manuguerra M, Kavvoura FK, Guarrera S, Allione A, Rosa F, Di Gregorio A, Polidoro S, Saletta F, Ioannidis JP, Matullo G. A field synopsis on low-penetrance variants in DNA repair genes and cancer susceptibility. J Natl Cancer Inst. 2009;101:24–36. doi: 10.1093/jnci/djn437. [DOI] [PubMed] [Google Scholar]
- 21.Easton DF, Eeles RA. Genome-wide association studies in cancer. Hum Mol Genet. 2008;17:R109–115. doi: 10.1093/hmg/ddn287. [DOI] [PubMed] [Google Scholar]
- 22.Spitz MR, Amos CI, Dong Q, Lin J, Wu X. The CHRNA5-A3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. J Natl Cancer Inst. 2008;100:1552–1556. doi: 10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, Sullivan K, Matakidou A, Wang Y, Mills G, Doheny K, Tsai YY, Chen WV, Shete S, Spitz MR, Houlston RS. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, Mirabello L, Jacobs K, Wheeler W, Yeager M, Bergen AW, Li Q, Consonni D, Pesatori AC, Wacholder S, Thun M, Diver R, Oken M, Virtamo J, Albanes D, Wang Z, Burdette L, Doheny KF, Pugh EW, Laurie C, Brennan P, Hung R, Gaborieau V, McKay JD, Lathrop M, McLaughlin J, Wang Y, Tsao MS, Spitz MR, Krokan H, Vatten L, Skorpen F, Arnesen E, Benhamou S, Bouchard C, Metsapalu A, Vooder T, Nelis M, Valk K, Field JK, Chen C, Goodman G, Sulem P, Thorleifsson G, Rafnar T, Eisen T, Sauter W, Rosenberger A, Bickeboller H, Risch A, Chang-Claude J, Wichmann HE, Stefansson K, Houlston R, Amos CI, Fraumeni JF, Jr, Savage SA, Bertazzi PA, Tucker MA, Chanock S, Caporaso NE. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, Qureshi M, Dong Q, Gu X, Chen WV, Spitz MR, Eisen T, Amos CI, Houlston RS. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40:1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohno T, Kunitoh H, Shimada Y, Shiraishi K, Ishii Y, Goto K, Ohe Y, Nishiwaki Y, Kuchiba A, Yamamoto S, Hirose H, Oka A, Yanagitani N, Saito R, Inoko H, Yokota J. Individuals susceptible to lung adenocarcinoma defined by combined HLA-DQA1 and TERT genotypes. Carcinogenesis. 2010 doi: 10.1093/carcin/bgq003. [DOI] [PubMed] [Google Scholar]
- 27.McKay JD, Hung RJ, Gaborieau V, Boffetta P, Chabrier A, Byrnes G, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, McLaughlin J, Shepherd F, Montpetit A, Narod S, Krokan HE, Skorpen F, Elvestad MB, Vatten L, Njolstad I, Axelsson T, Chen C, Goodman G, Barnett M, Loomis MM, Lubinski J, Matyjasik J, Lener M, Oszutowska D, Field J, Liloglou T, Xinarianos G, Cassidy A, Vineis P, Clavel-Chapelon F, Palli D, Tumino R, Krogh V, Panico S, Gonzalez CA, Ramon Quiros J, Martinez C, Navarro C, Ardanaz E, Larranaga N, Kham KT, Key T, Bueno-de-Mesquita HB, Peeters PH, Trichopoulou A, Linseisen J, Boeing H, Hallmans G, Overvad K, Tjonneland A, Kumle M, Riboli E, Zelenika D, Boland A, Delepine M, Foglio M, Lechner D, Matsuda F, Blanche H, Gut I, Heath S, Lathrop M, Brennan P. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40:1404–1406. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, Stacey SN, Bergthorsson JT, Thorlacius S, Gudmundsson J, Jonsson T, Jakobsdottir M, Saemundsdottir J, Olafsdottir O, Gudmundsson LJ, Bjornsdottir G, Kristjansson K, Skuladottir H, Isaksson HJ, Gudbjartsson T, Jones GT, Mueller T, Gottsater A, Flex A, Aben KK, de Vegt F, Mulders PF, Isla D, Vidal MJ, Asin L, Saez B, Murillo L, Blondal T, Kolbeinsson H, Stefansson JG, Hansdottir I, Runarsdottir V, Pola R, Lindblad B, van Rij AM, Dieplinger B, Haltmayer M, Mayordomo JI, Kiemeney LA, Matthiasson SE, Oskarsson H, Tyrfingsson T, Gudbjartsson DF, Gulcher JR, Jonsson S, Thorsteinsdottir U, Kong A, Stefansson K. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li D, Firozi PF, Wang LE, Bosken CH, Spitz MR, Hong WK, Wei Q. Sensitivity to DNA damage induced by benzo(a)pyrene diol epoxide and risk of lung cancer: a case-control analysis. Cancer Res. 2001;61:1445–1450. [PubMed] [Google Scholar]
- 30.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanna S, Jackson AU, Nagaraja R, Willer CJ, Chen WM, Bonnycastle LL, Shen H, Timpson N, Lettre G, Usala G, Chines PS, Stringham HM, Scott LJ, Dei M, Lai S, Albai G, Crisponi L, Naitza S, Doheny KF, Pugh EW, Ben-Shlomo Y, Ebrahim S, Lawlor DA, Bergman RN, Watanabe RM, Uda M, Tuomilehto J, Coresh J, Hirschhorn JN, Shuldiner AR, Schlessinger D, Collins FS, Davey Smith G, Boerwinkle E, Cao A, Boehnke M, Abecasis GR, Mohlke KL. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kavvoura FK, Ioannidis JP. Methods for meta-analysis in genetic association studies: a review of their potential and pitfalls. Hum Genet. 2008;123:1–14. doi: 10.1007/s00439-007-0445-9. [DOI] [PubMed] [Google Scholar]
- 35.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 36.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khanna KK, Jackson SP. DNA double–strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 38.Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, Cheng HL, Davidson L, Kangaloo L, Alt FW. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 39.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 40.Carlson CS, Eberle MA, Kruglyak L, Nickerson DA. Mapping complex disease loci in whole–genome association studies. Nature. 2004;429:446–452. doi: 10.1038/nature02623. [DOI] [PubMed] [Google Scholar]
- 41.Naukkarinen J, Gentile M, Soro-Paavonen A, Saarela J, Koistinen HA, Pajukanta P, Taskinen MR, Peltonen L. USF1 and dyslipidemias: converging evidence for a functional intronic variant. Hum Mol Genet. 2005;14:2595–2605. doi: 10.1093/hmg/ddi294. [DOI] [PubMed] [Google Scholar]
- 42.Tokuhiro S, Yamada R, Chang X, Suzuki A, Kochi Y, Sawada T, Suzuki M, Nagasaki M, Ohtsuki M, Ono M, Furukawa H, Nagashima M, Yoshino S, Mabuchi A, Sekine A, Saito S, Takahashi A, Tsunoda T, Nakamura Y, Yamamoto K. An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat Genet. 2003;35:341–348. doi: 10.1038/ng1267. [DOI] [PubMed] [Google Scholar]
- 43.Danoy P, Michiels S, Dessen P, Pignat C, Boulet T, Monet M, Bouchardy C, Lathrop M, Sarasin A, Benhamou S. Variants in DNA double-strand break repair and DNA damage-response genes and susceptibility to lung and head and neck cancers. Int J Cancer. 2008;123:457–463. doi: 10.1002/ijc.23524. [DOI] [PubMed] [Google Scholar]
- 44.Hsu NY, Wang HC, Wang CH, Chang CL, Chiu CF, Lee HZ, Tsai CW, Bau DT. Lung cancer susceptibility and genetic polymorphism of DNA repair gene XRCC4 in Taiwan. Cancer Biomark. 2009;5:159–165. doi: 10.3233/CBM-2009-0617. [DOI] [PubMed] [Google Scholar]