Abstract
Recombination between inverted repeats is RAD52 dependent, but reduced only modestly in the rad51Δ mutant. RAD59 is required for RAD51-independent inverted-repeat recombination, but no clear mechanism for how recombination occurs in the absence of RAD51 has emerged. Because Rad59 is thought to function as an accessory factor for the single-strand annealing activity of Rad52 one possible mechanism for spontaneous recombination could be by strand annealing between repeats at a stalled replication fork. Here we demonstrate the importance of the Rad52 single-strand annealing activity for generating recombinants by showing suppression of the rad52Δ, rad51Δ rad52Δ and rad52Δ rad59Δ inverted-repeat recombination defects by the rfa1-D228Y mutation. In addition, formation of recombinants in the rad51Δ mutant was sensitive to the distance between the inverted repeats, consistent with a replication-based mechanism. Deletion of RAD5 or RAD18, which are required for error-free post-replication repair, reduced the recombination rate in the rad59Δ mutant, but not in wild type. These data are consistent with RAD51-independent recombinants arising by a faulty template switch mechanism that is distinct from nascent strand template switching.
1. Introduction
Homologous recombination (HR) is an important repair mechanism to eliminate DNA double-strand breaks (DSBs) and to bypass lesions that block the replicative polymerases during DNA synthesis. The RAD52 epistasis group genes (MRE11, RAD50, RAD51, RAD52, RAD54, RDH54, RAD55, RAD57, RAD59 and XRS2) encode the proteins responsible for HR, although some participate in other DNA repair processes as well [1]. Of this group, the rad52Δ mutant has the most severe recombination defect, while the rad51Δ mutant is still proficient for some types of HR. Rad52 functions as a mediator for the Rad51 strand exchange protein by facilitating nucleation of Rad51 on RPA-coated single-stranded DNA (ssDNA) [2–6]. The Rad51 nucleoprotein filament catalyzes homologous pairing and invasion of a donor duplex to initiate DNA repair synthesis. In addition, Rad52 has a potent ssDNA annealing activity that is thought to be important for Rad51-independent recombination events that occur by the single-strand annealing (SSA) mechanism [7, 8].
Spontaneous recombination (recombination during normal growth with no inducing agent) most likely occurs during S phase as a mechanism to restart replication when progression of the DNA polymerase is blocked by a DNA lesion or bound protein. HR between sister chromatids is normally genetically silent, but can be detected between misaligned repeats using mutant alleles (heteroalleles) of a selectable gene to allow selection for recombinants that arise during growth of a culture [1]. Gene conversion between direct repeats is RAD51 dependent, but deletion of one repeat and the intervening DNA can occur in the absence of RAD51 [9–12]. DSB-induced deletion between direct repeats can occur by the SSA mechanism, involving resection of the DNA ends, annealing of the exposed complementary sequences, flap trimming and ligation [1]. Because the SSA mechanism lacks a strand invasion step it is Rad51-independent, but requires Rad52-catalyzed strand annealing. This requirement for Rad52 can be alleviated by a mutation in the RFA1 gene (rfa1-D228Y) that results in lower levels of the mutant replication protein A (RPA) complex [13, 14]. RPA-coated ssDNA is refractory to strand annealing and the Rad52 protein overcomes this barrier in vitro [8]. Smith and Rothstein suggested that the reduced level of the RPAD228Y complex bound to ssDNA allows spontaneous annealing, thus bypassing the requirement for Rad52-promoted annealing [13].
Inverted-repeat recombination assays were developed with the expectation that SSA is unlikely to be an interfering mechanism. Although spontaneous recombination between inverted repeats is reduced by 50 to 3,000-fold in the rad52Δ mutant, only a 5 to 10-fold reduction is observed in the rad51Δ mutant [15–18]. We identified the RAD59 gene by its requirement for RAD51-independent recombination between inverted ade2 heteroalleles [19]. The rad51Δ and rad59Δ single mutants both exhibit modest decreases in the rate of recombination between inverted repeats, while the rad51Δ rad59Δ double mutant is very defective, similar to the rad52Δ mutant [18, 19]. Rad59 is homologous to the N-terminal DNA binding and multimerization domain of Rad52, and interacts directly with Rad52 [20, 21]. Although Rad59 exhibits ssDNA-annealing activity in vitro, unlike Rad52 it cannot overcome the inhibitory effect of RPA on ssDNA annealing and is thought to augment the activity of Rad52 [20, 22, 23]. The rad59Δ mutant exhibits decreased efficiency of SSA, particularly when the direct repeats are short [20, 24, 25]. Additional studies suggest Rad59 might be important to counteract the negative effect of Rad51 on Rad52-mediated strand annealing [26, 27].
The minimal requirement for RAD51, and dependence on RAD52 and RAD59, suggests spontaneous inverted-repeat recombination can occur without Rad51-catalyzed strand invasion. DSB-induced recombination between chromosomal inverted repeats requires RAD51, consistent with a Rad51-catalyzed strand invasion step for DSB initiated events, and suggests that RAD51-independent spontaneous events do not initiate from DSBs [17]. Thus, we considered the possibility that spontaneous RAD51-independent events occur by a strand annealing mechanism during DNA synthesis to account for the requirement for RAD52 and RAD59. In support of this hypothesis, we show the inverted-repeat recombination defect of rad52Δ mutants is suppressed by the rfa1-D228Y mutation. Furthermore, increasing the distance between the repeats reduced the rate of RAD51-independent recombination and altered the spectrum of products, consistent with a replication-based mechanism. In addition, we show the error-free mode of post-replication repair plays a minor role in the formation of inverted-repeat recombinants.
2. Materials and methods
2.1. Yeast strains
All yeast strains are derived from W303, corrected for the rad5-535 mutation by crossing to generate either RAD5 or rad5::URA3 derivatives as appropriate [28] (Table 1). The ade2 inverted-repeat recombination reporter (ade2-IR) (Figure 1A) was described previously [18, 19]. The ade2::hisG-URA3-hisG cassette replaces ade2-1 in strain B404-3C as described in Rattray and Symington [18]. A Ura- derivative of B404-3C was first selected on 5-fluoorotic acid (5-FOA) for loss of URA3 before construction of any of the strains described in this study. The ade2-IR strains used are ade2::hisG, with one exception (rad18, LSY2002-2B). The strains containing the rfa1-D228Y allele were derived from crosses with a 5-FOA resistant isolate (LSY2236) of strain U859 (a gift from the Rothstein Lab). Determination of the presence of the rfa1-D228Y allele by PCR and restriction digestion has been described [13].
Table 1.
Yeast strains
| Strain | Relevant genotype* | Source |
|---|---|---|
| U859/LSY2235 | MATa rfa1-D228Y sup4+::HIS3+ rad5-535 ade2-1 | [13] |
| B404-3C |
MATa his3::ade2-5'delta-TRP1-ade2-n::his3 ade2::hisG rad5-535 rad51::HIS3 rad59::LEU2 |
[19] |
| HKY578-6B | MATα rad5::URA3 ade2-1 | H. Klein |
| HKY1331-5D | MATα rad18::LEU2 ade2-1 | H. Klein |
| LSY2001-41B |
MATα his3::ade2-5'Δ-TRP1-ade2-n ade2::hisG rad5::URA3 |
This study |
| LSY2001-46A |
MATα his3::ade2-5'Δ-TRP1-ade2-n ade2::hisG rad5::URA3 rad51::HIS3 |
This study |
| LSY2001-49A |
MATa his3::ade2-5'Δ-TRP1-ade2-n ade2::hisG rad5::URA3 rad59::LEU2 |
This study |
| LSY2002-9D | MATa his3::ade2-5'Δ-TRP1-ade2-n ade2::hisG | This study |
| LSY2002-6D |
MATα his3::ade2-5'Δ-TRP1-ade2-n ade2::hisG rad51::HIS3 rad59::LEU2 |
This study |
| LSY2002-16C |
MATa his3::ade2-5'Δ-TRP1-ade2-n ade2::hisG rad59::LEU2 |
This study |
| LSY2002-2B |
MATα his3::ade2-5'Δ-TRP1-ade2-n ade2::hisG rad18::LEU2 |
This study |
| LSY2002-2C |
MATa his3::ade2-5'Δ-TRP1-ade2-n ade2::hisG rad51::HIS3 |
This study |
| LSY2002-10A |
MATa his3::ade2-5'Δ-TRP1-ade2-n ade2::hisG rad18::LEU2 rad59::LEU2 |
This study |
| LSY2185-9D |
MATα his3::ade2-5'Δ-TRP1-ade2-n ade2::hisG rad52::KanMX6 |
This study |
| LSY2204-8B |
MATa his3::ade2-5'Δ-TRP1-ade2-n ade2::hisG rad51::HIS3 rad52::KanMX6 |
This study |
| LSY2249-1 | MATa his3::ade2-5'Δ-HphMX4-ade2-n ade2::hisG | This study |
| LSY2253-23C |
MATa his3::ade2-5'Δ-HphMX4-ade2-n ade2::hisG rad51::HIS3 |
This study |
| LSY2263-1C |
MATα his3::ade2-5'Δ-TRP1-ade2-n ade2::hisG rfa1- D228Y |
This study |
| LSY2263-3A |
MATa his3::ade2-5'Δ-TRP1-ade2-n ade2::hisG rfa1- D228Y rad59::LEU2 |
This study |
| LSY2263-6B |
MATa his3::ade2-5'Δ-TRP1-ade2-n ade2::hisG rfa1- D228Y rad52::KanMX6 |
This study |
| LSY2263-4D |
MATα his3::ade2-5'Δ-TRP1-ade2-n ade2::hisG rad52::KanMX6 rad59::LEU2 |
This study |
| LSY2263-4A |
MATα his3::ade2-5'Δ-TRP1-ade2-n ade2::hisG rfa1- D228Y rad52::KanMX6 rad59::LEU2 |
This study |
| LSY2439-11B |
MATa his3::ade2-5'Δ-TRP1-ade2-n ade2::hisG rfa1- D228Y rad51::HIS3 rad52::KanMX6 |
This study |
All strains are leu2-3,112 trp1-1 ura3-1 can1-100 RAD5 unless otherwise indicated [28].
Figure 1. The rfa1-D228Y mutation partially suppresses the rad52 recombination defect.
A. Schematic of the ade2-IR substrate and Ade+ products formed by gene conversion (non-inversion) or by inversion of the TRP1 locus. The inversion events can have the wild type or mutant NdeI site within the ade2–5’Δ allele and are not distinguished here. Recombination rates (×10−6) events/cell/generation are the mean of 3–4 independent trials. C. Distribution of events scored by physical analysis charted as the percentage of events examined (28–32 independent Ade+ events were scored for each strain). “Other” events are complex banding patterns that appear to result from an additional copy of ade2; these were not investigated further.
A second inverted-repeat substrate (ade2-IR-Hph), was constructed as follows. The plasmid pAG32 [29] was used to provide a functional Hygromycin B resistance gene and other sequences that have low homology to the rest of the Saccharomyces cerevisiae genome. Using the method of Longtine et al. [30] primers were designed to amplify the majority of plasmid pAG32 and direct the insertion of this PCR product between the ade2 alleles in strain LSY2002-9D. The primers used were:
SP-trp+hphexpand1: 5’-GCAGAACCGAGGATAGCGCTACGTCAGGATTCGAGGTCGGCGCGCCAGATCTGTTTAGCTTGCCTTGTCC-3’ and AP-trp+hphexpand1: 5’-GAACTAGTGGATCTTTTATGCTTGCTTTTCAAAAGGCCAATCTGCTCTGATGCCGCATAGTTAAGCCAGC-3’. The DNA was amplified using the Phusion DNA polymerase (Finnzymes) under the two-step conditions described by the manufacturer.
Successful targeting was verified by growth of the transformants on Hygromycin B (300µg/mL) containing media and failure to grow on minimal medium lacking tryptophan. Transformants with the correct structure were confirmed by PCR and Southern blot analysis of digested genomic DNA. rad mutations were introduced to the strain with the ade2-IR-Hph by crosses (Table 1).
2.2. Measurement of recombination rates
The method of Lea and Coulson [31] was used to determine recombination rates. Briefly, strains were grown 2–4 days on YPAD (1% yeast extract; 2% bacto-peptone; 2% dextrose; 10mg/L adenine) plates. Nine colonies of similar size were suspended in water, diluted and plated on synthetic complete (SC) or SC-Ade medium. The plates were monitored and counted 2 days after the first colonies were visualized. Rates were determined from the median frequency of Ade+ recombinants. The mean rate shown is from at least three independent trials for each strain and error bars indicate standard deviations. The student’s unpaired t-test was used to determine whether differences between strains were significant. The recombination rates shown in Figures 1 and 2 are lower than the rates presented in Figure 3 because the amount of adenine used to supplement YPAD medium was increased from 5 mg/L to 10mg/L during the course of these studies and we have found lower recombination rates when strains are grown on medium with higher adenine concentration. The relative rates for wild type and mutants are the same under both growth conditions.
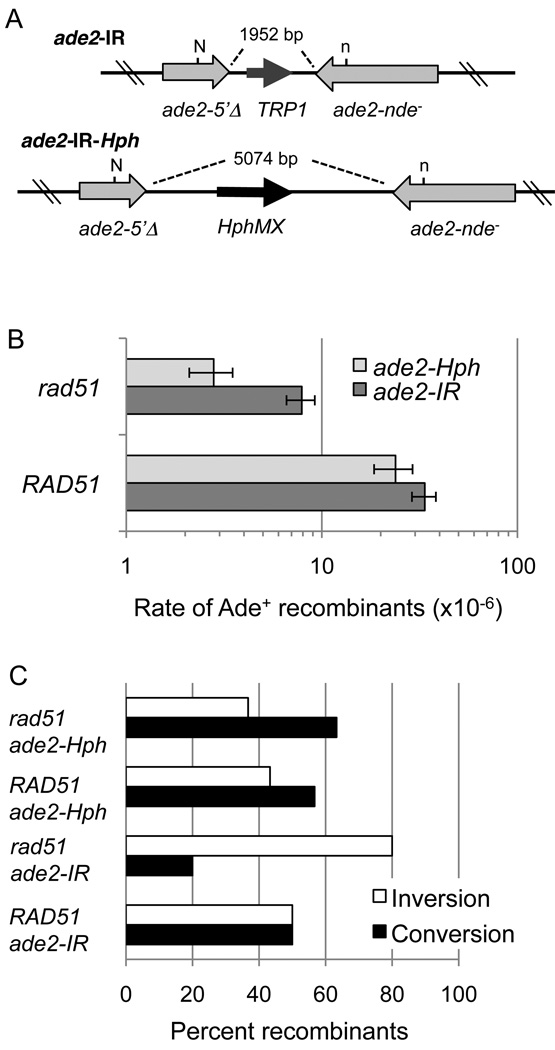
Figure 2. ade2-IR-Hph reporter to evaluate the role of distance between ade2 repeats on recombination rates.
A. Schematic of the ade2-IR-Hph substrate compared with ade2-IR. The distance between the repeats is increased from 1.9 kb to 5 kb. B. Recombination rate ×10−6 events/cell/generation reported as the mean of 3–4 trials. WT refers to wild type. C. Distribution of events scored by physical analysis charted as the percentage of events examined (30 independent Ade+ events were scored for each strain).
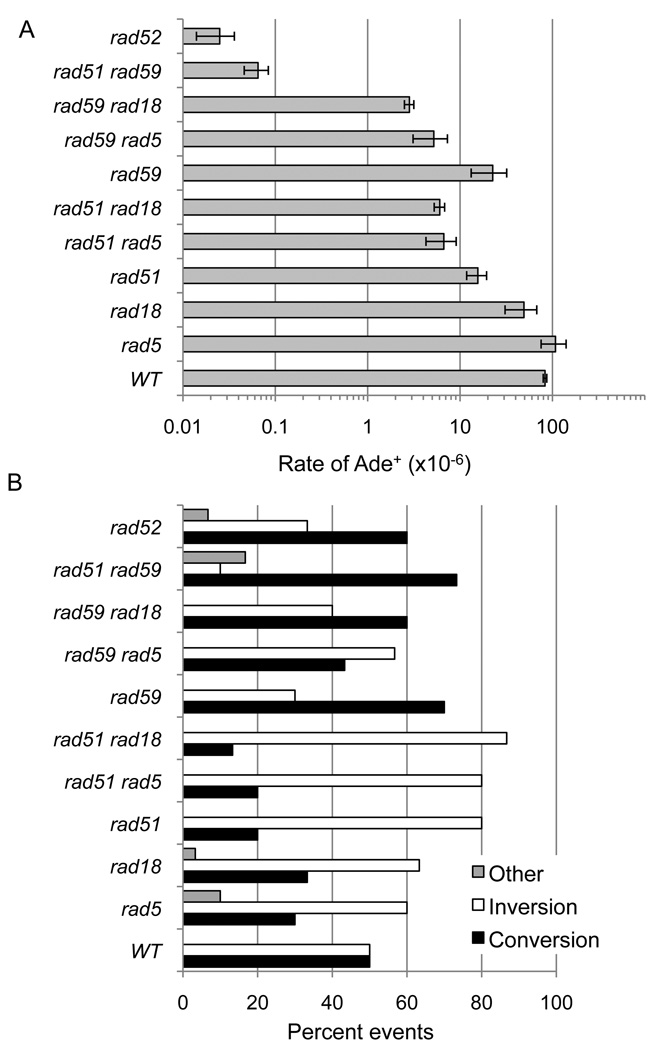
Figure 3. PRR plays a minor role in inverted-repeat recombination.
A. Recombination rates ×10−6 events/cell/generation are the mean of 4–7 trials. B. Distribution of events scored by physical analysis charted as percentage of events examined (30 independent events from each strain were analyzed). The rad51Δ single mutant, and rad5Δ rad51Δ, rad18Δ rad51Δ and rad51Δ rad59Δ double mutants show a distribution significantly different to the wild type (P=0.03, 0.03, 0.005 and 0.004 respectively).
2.3. Distribution of Recombinants
Strains were plated on YPAD and allowed to grow for 3–5 days. The colonies were then replica plated onto SC-Ade to select Ade+ recombinants. Independent Ade+ isolates were grown in 5mL overnight YPAD cultures and used to prepare genomic DNA. The DNA was digested with NdeI, PstI, or BglI (New England BioLabs), electrophoresed on a 1% agarose gel and then transferred to nylon membrane. The blots were probed with radioactively labeled DNA capable of annealing to ADE2, TRP1, or HphMX4 sequences and developed by phosphorimaging. Alternatively, inversions and non-inversions were scored by PCR using a primer that anneals to his3 sequences upstream of the ade2-5’Δ allele, and primers of opposite orientation that anneal to the TRP1 sequence between the repeats. Differences in the distribution of events were determined using Fisher’s exact test.
3. Results
3.1. The RAD52 single-strand annealing activity is important for spontaneous recombination between inverted repeats
An ade2 inverted-repeat (ade2-IR) substrate was used to determine the rate of Ade+ recombinants (Figure 1A) [18]. The ade2-5’Δ donor allele has a deletion of the first 174 nucleotides, including the promoter sequences; the recipient allele, ade2-n, is present in an inverse orientation relative to ade2-5’Δ [18]. The ade2-n allele has a +2 frameshift at the NdeI restriction endonuclease recognition site and is transcribed from the native ADE2 promoter. The repeats are separated by 1.9 kb of DNA, including a functional copy of the TRP1 gene, integrated at the HIS3 locus of chromosome XV.
Our previous studies had shown a 3,000-fold decrease in the Ade+ recombination rate in the rad52Δ mutant. If the rad52Δ defect was due to the role of Rad52 in promoting strand annealing then we expected that suppression of the strand-annealing defect should restore the production of Ade+ recombinants. The rfa1-D228Y mutation was previously identified as a suppressor of the spontaneous direct-repeat recombination defect of the rad1Δ rad52Δ double mutant, and the rfa1-D228Y single mutant exhibits a hyper-recombination phenotype for spontaneous deletions between direct repeats [13]. Subsequent studies showed that rfa1-D228Y suppresses the requirement for RAD52 in DSB-induced SSA between direct repeats [14]. The RPAD228Y complex is less abundant than wild type RPA suggesting there is less RPAD228Y bound to ssDNA to prevent spontaneous annealing, thus bypassing the requirement for Rad52-promoted annealing [13].
The recombination rate of the rfa1-D228Y mutant was equivalent to wild type; however, a 66-fold increase in recombination was observed for the rfa1-D228Y rad52Δ compared with rad52Δ (P=0.0001) (Figure 1B). To confirm that the suppression of the rad52Δ recombination defect by rfa1-D228Y is due to the role of Rad52 in ssDNA annealing and not the mediator function, the recombination rate was determined for the rad51Δ rad52Δ rfa1-D228Y triple mutant. As anticipated, the rfa1-D228Y mutation also resulted in a significant suppression of the rad51Δ rad52Δ recombination defect (P=0.01). Thus, the SSA activity of Rad52 appears to be critical for recombination of the ade2-IR substrate and this requirement is alleviated by the rfa1-D228Y mutation. In the rad59Δ mutant most recombination occurs by a RAD51-dependent mechanism and in this case RPA is expected to play a positive role by enabling Rad51 to more effectively bind to ssDNA. Consistent with this, the rfa1-D228Y rad59Δ showed a small, but statistically significant decrease in recombination compared to rad59Δ(P=0.014) (Figure 1B). The rfa1-D228Y mutation caused a significant increase in the recombination rate of the rad52Δ rad59Δ mutant (P=0.025), suggesting the primary role for Rad59 in the formation of Ade+ recombinants is in strand annealing together with Rad52.
By physical analysis Ade+ recombinants were classified as gene conversion events (no inversion of TRP1) or inversion events. The inversions include events in which the wild type NdeI site is present in both repeats, and events in which the wild type NdeI site is transferred from ade2–5’Δ to ade2-n (Figure 1A). A few aberrant recombination events with three copies of the ade2 locus were recovered from the rad51Δ rad52Δ and rad52Δ mutants (classified as “other” in Figure 1C); these were not characterized further. Dornfeld and Livingston previously described similar types of events in a rad52 mutant [15]. Recombinants analyzed from the rad52Δ, rad59Δ, rad51Δ rad52Δ and rad52Δ rad59Δ mutants showed a bias towards gene conversion events compared with the wild type distribution, however, this difference is only significant for the rad51Δ rad52Δ and rad52Δ rad59Δ mutants (P<0.05) (Figure 1C). Ade+ recombinants analyzed from all of the rfa1-D228Y derivative showed a bias towards inversion events, with a significant change in the distribution for the rad51Δ rad52Δ rfa1-D228Y and rad52Δ rad59Δ rfa1-D228Y strains, compared with rad51Δ rad52Δ and rad52Δ rad59Δ (P<0.005). The distribution of events recovered from the rad52Δ rfa1-D228Y strain is not significantly different to the rad51Δ rad52Δ rfa1-D228Y and rad52Δ rad59Δ rfa1-D228Y strains. These results are consistent with inversions resulting from a strand annealing mechanism.
3.2. Increasing the distance between ade2 heteroalleles alters the repair mechanism in rad51Δ mutants
Increasing the distance between the ade2 repeats would be expected to inhibit template switching because of the requirement to uncouple leading and lagging strand synthesis over a longer distance [32]. Thus, we expected the rate of recombination in the rad51Δ mutant to decrease with increased distance between the repeats, whereas RAD51-dependent homology searching is expected to be less sensitive to the distance between the repeats. A new reporter, ade2-IR-Hph, was made by replacing the TRP1 gene and some intervening sequences with most of the pAG32 plasmid, containing the gene for hygromycin resistance (Hph), increasing the distance between the ade2 repeats by 3.1-kb (Figure 2A). The rates of recombination for ade2-IR-Hph and ade2-IR were the same in the wild-type background (P=0.07), but there was a 9-fold decrease in the Ade+ recombination rate in the rad51Δ mutant for the ade2-IR-Hph compared with wild type (Figure 2B). For the ade2-IR substrate the difference between wild type and rad51Δ is 5-fold. Thus, extending the distance between ade2–5’Δ and ade2–n by 3.1-kb increased the dependence on RAD51 by an additional 2.6-fold (P=0.03).
Analysis of the recombination products recovered from the rad51Δ mutant with the ade2-IR showed a bias towards inversion events, while inversions and conversions were recovered in equal numbers from the wild-type strain (Figure 2C). The Ade+ recombinants recovered from the wild-type strain with the ade2-IR-Hph reporter showed the same distribution as events from the ade2-IR. However, products from the rad51Δ mutant with the ade2-IR-Hph reporter showed a significant alteration in the distribution of events compared with the Ade+ products recovered from ade2-IR (P=0.0007) (Figure 2C). These results are consistent with the mechanism of RAD51-independent recombination changing as the distance between the repeats increases.
3.3. Post-replication repair plays a minor role in inverted-repeat recombination
Post-replication repair (PRR) is responsible for the bypass (tolerance) of DNA lesions that stall the replicative polymerases resulting in ssDNA gaps at the fork and behind the replication fork. The Rad6-Rad18 ubiquitin ligase, which is essential for the PRR pathway, monoubiquitinates proliferating cell nuclear antigen (PCNA) to promote translesion DNA synthesis [33–37]. Subsequent polyubiquitination by the Ubc13-Mms2-Rad5 ubiquitin conjugating enzyme complex directs repair through the error-free PRR pathway, which is proposed to occur by template switching between nascent strands [38–42].
To investigate whether RAD51-independent inverted-repeat recombination involves a template-switching mechanism we determined the Ade+ recombination rate in rad5Δ and rad18Δ derivatives. All of the original W303-derived strains with the ade2-IR reporter carried the rad5–535 allele [18, 19]; these strains were remade to be RAD5 and the recombination rates determined (note, all strains used in Figures 1 and 2 are RAD5). The spontaneous Ade+ recombination rates in the wild type, rad51Δ, rad52Δ and rad59Δ RAD5 derivative were similar to the strains containing rad5–535 described previously [18, 19]; thus, the rad5-535 mutation did not significantly alter the ade2-IR recombination rate in wild type or the mutants tested. The distribution of recombinants recovered from the RAD5 derivatives was also the same as the rad5-535 strains analyzed previously.
The Ade+ recombination rate of the rad5Δ and rad18Δ single mutants did not significantly differ from wild type (P=0.19 and 0.12, respectively), suggesting PRR does not contribute to recombinants in strains that have wild-type HR functions (Figure 3A). If the error-free PRR pathway contributes to RAD51-independent recombination we would predict a decrease in Ade+ recombinants in the rad18Δ rad51Δ and rad5Δ rad51Δ double mutants. The rad18Δ rad51Δ and rad5Δ rad51Δ double mutants displayed small (2–2.5 fold), but statistically significant, decreases in recombination rates compared to rad51Δ(P=0.0006 and P=0.005, respectively); however, the rates were 100-fold higher than the rad51Δ rad59Δ double mutant (P=0.0001) [19]. Thus, RAD5 and RAD18 have minor roles in RAD51-independent recombination compared with RAD59. The rad18Δ rad59Δ strain showed an 8-fold decrease in recombination compared with rad59Δ(P=0.0002), while the rad5Δ rad59Δ was 4-fold decreased compared to rad59Δ(P=0.006), indicating a more important role for RAD5 and RAD18 in the RAD51-dependent pathway than the RAD51-independent pathway.
Physical analysis of the Ade+ recombinants from the rad5Δ and rad18Δ strains showed a slight bias towards recovery of inversion events, similar to the rad51Δ mutant, and the rad51Δ rad5Δ and rad51Δ rad18Δ double mutants exhibited the same distribution of events as the rad51Δ mutant (Figure 3B).
4. Discussion
HR between direct repeats and inverted repeats shows some variation in the requirement for genes in the RAD52 epistasis group due to the different mechanisms that can be used to generate products. Gene conversion between direct repeats (maintaining both repeats and the intervening DNA) requires RAD51 and RAD52, whereas deletion of one of the repeats and intervening DNA is RAD51-independent [9, 11, 12]. The deletion events are thought to occur by RAD52-dependent SSA [43]. In contrast, recombination between inverted repeats cannot occur by a simple annealing mechanism, yet rad51Δ mutants show a less profound spontaneous recombination defect than rad52Δ mutants [16–18]. Furthermore, the residual recombination events that occur in rad51Δ mutants are RAD59-dependent, and Rad59 is thought to augment the ssDNA annealing activity of Rad52 [19, 20, 23, 25]. The other unusual feature of RAD51-independent recombination of inverted repeats is the high percent of products with an inversion of the intervening DNA [18]. Because DSB-induced recombination between chromosomal inverted repeats requires RAD51, we considered the possibility that spontaneous RAD51-independent recombination occurs by a strand annealing mechanism that switches template strands during replication, similar to the template switch models recently proposed for repeat-induced rearrangements [40, 44, 45]. The faulty template switch was proposed to explain the formation of acentric and dicentric giant palindromes at an inverted repeat, but aborted replication initiated by the same mechanism could generate Ade+ recombinants between inverted repeats and explain the high frequency of inversions observed (Figure 4).
Figure 4. Faulty template-switch mechanism for inverted-repeat recombination.
The model proposes stalling of the nascent strand, primer dissociation and annealing to the opposite parental strand via one of the repeats. The unequal pairing alignment is used to restart synthesis of the blocked nascent strand by providing an alternative template. A second template switch is required to restore canonical pairing between the leading nascent and template strands. The leading strand lesion has been bypassed and replication is restarted. The NdeI fill-in is indicated by a vertical black bar; white arrow, ADE2; blue and orange lines, ssDNA; dashed lines indicate nascent strands; the yellow star denotes spontaneous damage on the leading template strand.
When replication of the leading strand is stalled by spontaneous base damage the replicative helicase continues to unwind generating ssDNA at the fork [32]. We suggest the blocked nascent leading strand dissociates from its template strand and pairs with homologous ssDNA. In the case of direct repeats, annealing can occur on the same template strand, or the sister nascent strand, to generate duplications or deletions [40], but for inverted repeats the only available homology is on the other template strand (Figure 4). Presumably the lagging strand polymerase is hijacked and the unequal sister alignment is used to restart synthesis of the nascent leading strand. After synthesis of a short tract we suggest the leading strand is displaced from the lagging strand template and returns to the original pairing configuration between template and nascent strands. This mechanism could bypass the leading strand lesion and give rise to a heteroduplex DNA (hDNA) intermediate if DNA synthesis had proceeded across the wild type sequence of the ade2-5’Δ repeat; gene conversion could then result from repair of the hDNA, or segregation at the next S-phase. Furthermore, if DNA synthesis extended from ade2-5’Δ through the DNA separating the inverted repeats to ade2-n then a large loop mispair would form after strand realignment. Repair of the loop could give rise to an inversion of the DNA between the repeats, the most common class of recombinants recovered from the rad51Δ mutant [18]. Thus, one possible explanation for the importance of Rad52 and Rad59 for inverted-repeat recombination is to catalyze annealing between the unpaired primer strand and alternative template strand (first template switch), or in the re-establishment of canonical pairing (second template switch).
Because rfa1-D228Y suppresses the rad52Δ SSA defect observed in direct-repeat recombination assays we rationalized that if inverted-repeat recombination involves a strand-annealing step then rfa1-D228Y should also suppress rad52Δ in the ade2-IR assay [13, 14]. The rad52Δ recombination rate was increased 66-fold by the rfa1-D228Y allele supporting the hypothesis that single-strand annealing is critical for inverted-repeat recombination (Figure 1). Furthermore, the rfa1-D228Y mutation partially suppressed the recombination defects of the rad51Δ rad52Δ and rad52Δ rad59Δ mutants suggesting the defect of the double mutants is due to loss of Rad52 annealing activity. In addition, the rfa1-D228Y mutation changed the distribution of recombination events from the gene conversion bias of the rad52Δ and rad59Δ strains to more inversion events. This result suggests strand annealing generates inversions. The rfa1-D228Y and rad59Δ rfa1-D228Y mutants did not exhibit a hyper-recombination phenotype indicating the increase in recombination is specific to the rad52Δ background and is not due to increased recombinogenic lesions caused by the mutant RPA complex.
The small but significant decrease in Ade+ frequency for the rad59Δ rfa1-D228Y double mutant could result from reduced Rad51 strand exchange activity. The rfa1-D228Y mutant exhibits a 6-fold decrease in the rate of recombination between allelic sequences in diploids, indicating the Rad51-dependent strand invasion is defective in the presence of the RPAD228Y complex [13]. RPA functions to remove secondary structures from ssDNA enabling Rad51 filament formation once Rad52 nucleates Rad51 and this may explain the decreased efficiency of RAD51-dependent recombination [46].
Paek et al. [45] did not find a decrease in the frequency of rearrangements or formation of dicentrics in the rad52Δ mutant in their assay system, whereas Rad22/Rad52 was required for the faulty template switch described by Mizuno et al. [44]. One difference between these systems is the amount of homology between the repeats. The natural repeats that are the substrate for template switching measured by Paek et al. [45] are shorter and more diverged than the RTS1-ura4 repeats utilized by the Carr group [44], or the ade2 repeats used here. The Rad52 annealing function might be more effective with longer stretches of homology. Alternatively, the Rad52-dependent function might be more important for the second template switch, which is not required to generate products in the system described by Paek et al [45]. In contrast to our finding, the rad51Δ mutant exhibited a reduction in acentric/dicentric products in the study by Mizuno et al. [44]. They proposed the displaced primer strand at a fork stalled by a protein barrier invades the other unreplicated repeat using HR functions. By contrast, we suggest the Rad51-independent events occur by annealing between repeats when replication is uncoupled exposing ssDNA regions.
While Rad52 and Rad59 are proposed to anneal complementary ssDNA exposed at the inverted repeat, a variety of proteins could be responsible for the other hypothetical steps in the template switch. Rad18 and Rad5 were investigated since both are required for error-free PRR and the Rad5 helicase activity could potentially contribute to nascent strand displacement. Previous studies reported elevated frequencies of spontaneous gene conversion using allelic and direct repeat substrates in rad18Δ and rad5Δ mutants, presumably because ssDNA gaps that are substrates for PRR are channeled to HR [47]. Neither the rad5Δ nor rad18Δ mutant demonstrated hyper-recombination in the ade2-IR assay, suggesting there is little competition with PRR for substrate when HR proteins are present, or that an increase in use of the HR pathway is offset by a minor role for Rad5 and Rad18 in Rad51-dependent recombination of inverted repeats. The Ade+ rate was decreased 8-fold in the rad18Δ rad59Δ double mutant compared with rad59Δ (Figure 3). This might be due to a minor role for Rad18 in the Rad51-dependent pathway, as suggested previously by the accumulation of branched DNA structures following methyl methane sulfonate treatment of the sgs1Δ mutant [38]. Alternatively, because sumoylation of PCNA is increased in the rad18Δ mutant it is possible that Srs2 is more efficiently recruited to stalled replication forks and disrupts Rad51 filaments resulting in down regulation of Rad51-dependent recombination [48–50].
Seeking further evidence for a replicative recombination mechanism, we designed a substrate in which the repeats were further apart expecting to decrease the rate of RAD51-independent recombination. The increased distance did not change the Ade+ rate in the wild-type strain, suggesting RAD51-dependent recombination is unaffected by the proximity of the repeats; however, recombination of the ade2-IR-Hph reporter was reduced by an additional 2.6-fold in the rad51Δ mutant (Figure 2B). Physical analysis revealed a significant decrease in the number of inversion events recovered from the rad51Δ ade2-IR-Hph strain, whereas the distribution of events was the same for both inverted repeats in the wild-type strain (Figure 2C). The interpretation of these results is that rad51Δ mutants use a mechanism that is more adversely affected by the increased distance between heteroalleles than wild type. The reduced number of inversions is consistent with the faulty template switch model for RAD51-independent recombination because more DNA would need to be synthesized from the lagging template strand before displacing the leading nacent strand and annealing to the original template strand. It is possible that once highly processive replication is initiated on the opposite template then large chromosomal palindromes are the primary outcome and such events would be inviable and not be detected using the ade2-IR system. At this point no analysis for acentric/dicentric chromosome formation has been conducted so those outcomes remain a formal possibility in the ade2-IR system. We predict these events would be too rare to detect by Southern blot hybridization and the anticipated large inverted repeat would be difficult to detect by PCR because of the formation of snapback structures during the denaturation and annealing steps.
Acknowledgements
We thank W.K. Holloman, G. Mazón and E.P. Mimitou for discussion and critical reading of the manuscript, and H. Klein and R. Rothstein for yeast strains. This research was supported by a grant from the National Institutes of Health (GM041784 and GM054099).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 3.Gasior SL, Wong AK, Kora Y, Shinohara A, Bishop DK. Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes. Genes Dev. 1998;12:2208–2221. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 5.Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 6.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 7.Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci U S A. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc Natl Acad Sci U S A. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang LE, Symington LS. Aberrant double-strand break repair in rad51 mutants of Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:9162–9172. doi: 10.1128/mcb.20.24.9162-9172.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDonald JP, Rothstein R. Unrepaired heteroduplex DNA in Saccharomyces cerevisiae is decreased in RAD1 RAD52-independent recombination. Genetics. 1994;137:393–405. doi: 10.1093/genetics/137.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mozlin AM, Fung CW, Symington LS. Role of the Saccharomyces cerevisiae Rad51 paralogs in sister chromatid recombination. Genetics. 2008;178:113–126. doi: 10.1534/genetics.107.082677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith J, Rothstein R. A mutation in the gene encoding the Saccharomyces cerevisiae single-stranded DNA-binding protein Rfa1 stimulates a RAD52-independent pathway for direct-repeat recombination. Mol Cell Biol. 1995;15:1632–1641. doi: 10.1128/mcb.15.3.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith J, Rothstein R. An allele of RFA1 suppresses RAD52-dependent double-strand break repair in Saccharomyces cerevisiae. Genetics. 1999;151:447–458. doi: 10.1093/genetics/151.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dornfeld KJ, Livingston DM. Plasmid recombination in a rad52 mutant of Saccharomyces cerevisiae. Genetics. 1992;131:261–276. doi: 10.1093/genetics/131.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malagón F, Aguilera A. Yeast spt6-140 mutation, affecting chromatin and transcription, preferentially increases recombination in which Rad51p-mediated strand exchange is dispensable. Genetics. 2001;158:597–611. doi: 10.1093/genetics/158.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rattray AJ, Shafer BK, McGill CB, Strathern JN. The roles of REV3 and RAD57 in double-strand-break-repair-induced mutagenesis of Saccharomyces cerevisiae. Genetics. 2002;162:1063–1077. doi: 10.1093/genetics/162.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rattray AJ, Symington LS. Use of a chromosomal inverted repeat to demonstrate that the RAD51 and RAD52 genes of Saccharomyces cerevisiae have different roles in mitotic recombination. Genetics. 1994;138:587–595. doi: 10.1093/genetics/138.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai Y, Symington LS. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- 20.Davis AP, Symington LS. The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics. 2001;159:515–525. doi: 10.1093/genetics/159.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis AP, Symington LS. The Rad52-Rad59 complex interacts with Rad51 and replication protein A. DNA Repair (Amst) 2003;2:1127–1134. doi: 10.1016/s1568-7864(03)00121-6. [DOI] [PubMed] [Google Scholar]
- 22.Petukhova G, Stratton SA, Sung P. Single strand DNA binding and annealing activities in the yeast recombination factor Rad59. J Biol Chem. 1999;274:33839–33842. doi: 10.1074/jbc.274.48.33839. [DOI] [PubMed] [Google Scholar]
- 23.Wu Y, Sugiyama T, Kowalczykowski SC. DNA annealing mediated by Rad52 and Rad59 proteins. J Biol Chem. 2006;281:15441–15449. doi: 10.1074/jbc.M601827200. [DOI] [PubMed] [Google Scholar]
- 24.Pannunzio NR, Manthey GM, Bailis AM. RAD59 is required for efficient repair of simultaneous double-strand breaks resulting in translocations in Saccharomyces cerevisiae. DNA Repair (Amst) 2008;7:788–800. doi: 10.1016/j.dnarep.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugawara N, Ira G, Haber JE. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol Cell Biol. 2000;20:5300–5309. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manthey GM, Bailis AM. Rad51 inhibits translocation formation by non-conservative homologous recombination in Saccharomyces cerevisiae. PLoS One. 2010;5:e11889. doi: 10.1371/journal.pone.0011889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y, Kantake N, Sugiyama T, Kowalczykowski SC. Rad51 protein controls Rad52-mediated DNA annealing. J Biol Chem. 2008;283:14883–14892. doi: 10.1074/jbc.M801097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zou H, Rothstein R. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]
- 29.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 30.Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 31.Lea DE, Coulson C. The distribution of the numbers of mutants in bacterial populations. Journal of Genetics. 1949;22 doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 32.Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- 33.Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 1994;8:811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- 34.Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J Biol Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 35.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 36.Jentsch S, McGrath JP, Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987;329:131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- 37.Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- 39.Carlile CC, Pickart CM, Matunis MJ, Cohen RE. Synthesis of free and PCNA-bound polyubiquitin chains by the RING E3 ligase, Rad5. Journal of Biological Chemistry. 2009:1–18. doi: 10.1074/jbc.M109.043885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldfless S, Morag A, Belisle K, Suterajr V, Lovett S. DNA Repeat Rearrangements Mediated by DnaK-Dependent Replication Fork Repair. Mol Cell. 2006;21:595–604. doi: 10.1016/j.molcel.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 41.Hoege C, Pfander B, Moldovan G-L, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 42.Ulrich HD. Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair (Amst) 2009;8:461–469. doi: 10.1016/j.dnarep.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 43.Sugawara N, Haber JE. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizuno KI, Lambert S, Baldacci G, Murray JM, Carr AM. Nearby inverted repeats fuse to generate acentric and dicentric palindromic chromosomes by a replication template exchange mechanism. Genes Dev. 2009;23:2876–2886. doi: 10.1101/gad.1863009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paek AL, Kaochar S, Jones H, Elezaby A, Shanks L, Weinert T. Fusion of nearby inverted repeats by a replication-based mechanism leads to formation of dicentric and acentric chromosomes that cause genome instability in budding yeast. Genes Dev. 2009;23:2861–2875. doi: 10.1101/gad.1862709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugiyama T, Zaitseva EM, Kowalczykowski SC. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J Biol Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- 47.Liefshitz B, Steinlauf R, Friedl A, Eckardt-Schupp F, Kupiec M. Genetic interactions between mutants of the 'error-prone' repair group of Saccharomyces cerevisiae and their effect on recombination and mutagenesis. Mutat Res. 1998;407:135–145. doi: 10.1016/s0921-8777(97)00070-0. [DOI] [PubMed] [Google Scholar]
- 48.Friedl AA, Liefshitz B, Steinlauf R, Kupiec M. Deletion of the SRS2 gene suppresses elevated recombination and DNA damage sensitivity in rad5 and rad18 mutants of Saccharomyces cerevisiae. Mutat Res. 2001;486:137–146. doi: 10.1016/s0921-8777(01)00086-6. [DOI] [PubMed] [Google Scholar]
- 49.Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Pfander B, Moldovan G-L, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;6 doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]