Abstract
The interaction between SA (streptavidin) and biotin is one of the strongest non-covalent interactions in Nature. SA is a widely used tool and a paradigm for protein–ligand interactions. We previously developed a SA mutant, termed Tr (traptavidin), possessing a 10-fold lower off-rate for biotin, with increased mechanical and thermal stability. In the present study, we determined the crystal structures of apo-Tr and biotin–Tr at 1.5 Å resolution. In apo-SA the loop (L3/4), near biotin's valeryl tail, is typically disordered and open, but closes upon biotin binding. In contrast, L3/4 was shut in both apo-Tr and biotin–Tr. The reduced flexibility of L3/4 and decreased conformational change on biotin binding provide an explanation for Tr's reduced biotin off- and on-rates. L3/4 includes Ser45, which forms a hydrogen bond to biotin consistently in Tr, but erratically in SA. Reduced breakage of the biotin–Ser45 hydrogen bond in Tr is likely to inhibit the initiating event in biotin's dissociation pathway. We generated a Tr with a single biotin-binding site rather than four, which showed a simi-larly low off-rate, demonstrating that Tr's low off-rate was governed by intrasubunit effects. Understanding the structural features of this tenacious interaction may assist the design of even stronger affinity tags and inhibitors.
Keywords: avidin, biotin, protein engineering, protein–ligand interaction, streptavidin, traptavidin
Abbreviations: D, dead streptavidin subunit; DPI, diffraction-data precision indicator; D4, a tetramer of dead streptavidin subunits; PEG, poly(ethylene glycol); rmsd, root mean square deviation; SA, streptavidin; Tr, traptavidin; Tr1D3, monovalent Tr; Tr4, tetravalent Tr
INTRODUCTION
The capture of the small molecule biotin (vitamin H/vitamin B7) by the bacterial protein SA (streptavidin) is both a powerful tool in biology and a model system for the study of high-affinity protein–ligand interactions. The key features are the femtomolar affinity, high specificity, high on-rate and the resilience of SA to pH, temperature and denaturant [1–3]. It is also important that biotin protein ligases can attach biotin to specific lysine residues in vitro or in living cells [4,5]. SA's applications include imaging [6], nano-assembly [7] and pre-targeted cancer immunotherapy [8]. As a favoured model in biophysics and protein engineering, this protein–ligand interaction has been investigated by at least 200 point mutations of SA [3,9–11], more than 30 alternative small molecule ligands [1,12–14], computational simulation [15–18], crystallography [2] and single-molecule force dynamics [19].
Despite the strength of SA–biotin binding, the interaction can and does fail under challenging conditions. Biotin dissociation becomes significant and sometimes rapid in response to the low pH of the endosomes [20], the high temperatures used for DNA amplification [21], attachment to nanoparticles [22], and shear forces from flow [23]. The low off-rate of wild-type SA for biotin conjugates was improved upon recently, when we found that the S52G R53D mutant of SA, named Tr (traptavidin) [23a], had a more than 10-fold slower biotin dissociation than wild-type SA, as well as enhanced mechanical and thermal stability [24]. The stronger binding of Tr enabled us to generate a challenging roadblock for one of the fastest known linear molecular motors, FtsK, in order to evaluate the co-ordination of firing around the hexameric ring as FtsK translocated on DNA [24,25].
To understand the basis of Tr's resilient binding and stability to high temperatures, in the present study we solved the crystal structures of both apo-Tr and biotin–Tr and compared these with existing structures of SA. In addition we purified a monovalent Tr, to investigate the role of intersubunit effects in Tr stability and also to characterize a tool which enables ultra-stable binding of biotinylated targets without cross-linking.
EXPERIMENTAL
Tr purification
Tr protein (S52G R53D core streptavidin with a C-terminal His6 tag, GenBank® accession number GU952124) was purified from Escherichia coli by inclusion body isolation, refolding and Ni-NTA (Ni2+-nitrilotriacetate) chromatography, as described previously [24]. Prior to the crystal tray set-up, we performed size-exclusion chromatography (XK 26 column, GE Healthcare), eluting with 50 mM Tris/HCl (pH 7.5), 0.5 M NaCl and 10 mM EDTA.
Tr1D3 (monovalent Tr) was generated by the mixed refolding of Tr and D (dead streptavidin) subunits (N23A S27D S45A core SA with no His6 tag). Purification was as previously described for monovalent SA [26], except that the order of SDS/PAGE mobility was reversed (mobility of Tr2D2 >Tr1D3>D4). D4 (a tetramer of dead streptavidin subunits) was generated from the refolding of D from inclusion bodies into PBS and ammonium sulfate precipitation [26].
Apo-Tr crystallization
Crystals were obtained by the sitting-drop vapour-diffusion method at 291 K. Apo-Tr crystals [space group I-41; a=b=57.59 Å (1 Å=0.1 nm), c=183.35 Å, with two Tr monomers in the asymmetric unit] were obtained from a 4 μl drop of 19 mg/ml solution and a reservoir solution of 12% (v/v) PEG [poly(ethylene glycol)] 8000, 9% (v/v) ethylene glycol and 0.1 M Hepes (pH 7.5). Crystals appeared after 2 days and reached optimum size after 4 days. Prior to data collection, crystals were briefly soaked in a cryoprotectant solution of 12% (v/v) PEG 8000, 30% (v/v) ethylene glycol and 0.1 M Hepes (pH 7.5) before immersion into liquid nitrogen.
Biotin–Tr crystallization
Biotin-bound Tr was obtained by incubation of Tr with biotin (Acros Organics), in 4-fold molar excess compared with the number of binding sites, at 4 °C overnight. Biotin-bound crystals (space group P42212; a=b=57.34 Å, c=77.55 Å, with one monomer in the asymmetric unit) were obtained from a 4 μl drop of 19 mg/ml solution and a reservoir solution of 27% (v/v) PEG 4000, 0.25 M MgCl2 and 0.1 M Tris/HCl (pH 8.5). Crystals appeared after 2 days and reached optimum size after 4 days. Prior to data collection, crystals were briefly soaked in a cryoprotectant solution of 27% (v/v) PEG 4000, 0.25 M MgCl2 and 25% glycerol and 0.1 M Tris/HCl (pH 8.5) before immersion into liquid nitrogen.
Diffraction data collection
Crystallographic data were collected at 100 K, using an Oxford Cryosystems 700 series Cryostream on an ADSC Quantum 315 CCD (charge-coupled devive) detector with an oscillation range of 0.5° at beamline IO2 at the Diamond Light Source, Harwell, U.K.
Structure solution and refinement: apo-Tr
Data were indexed and integrated using MOSFLM, and scaled and merged using SCALA from the CCP4 program suite [27]. The structure was phased by molecular replacement using a wild-type core SA search model (PDB code 1SWB) [28] with the program Phaser. The crystal contained two monomers in the asymmetric unit. A total of 5% of the reflection data were set aside using the Freerflag program in CCP4 and used for the calculation of Rfree. The model was built in Coot and refined in PHENIX refine, with incorporation of the twinning operator (−h, k,−l) with a twinning fraction of 49.7%. Throughout the refinement, all results were included from 27.47 Å resolution to the highest limit (1.45 Å) and anisotropic temperature factors were refined. The model was evaluated with MolProbity [28b], which gave Ramachandran statistics with 98% of residues in favoured regions and no outliers. The DPI (diffraction-data precision indicator) indicated that the agreement between the model and the X-ray data for apo-Tr was 0.056 Å. The refinement statistics are shown in Table 1.
Table 1. Data collection and refinement statistics for crystal structures of apo-Tr and biotin–Tr.
The values in parentheses are for the highest-resolution shell. n/a, not applicable.
| Apo-Tr (PDB code 2Y3E) | Biotin–Tr (PDB code 2Y3F) | |
|---|---|---|
| Data collection | ||
| Space group | I 41 | P 42 21 2 |
| Cell dimensions | ||
| a, b, c (Å) | 57.59, 57.59, 183.35 | 57.34, 57.34, 77.55 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 27.47–1.45 (1.53–1.45) | 38.78–1.49 (1.57–1.49) |
| Rmerge | 0.052 (0.619) | 0.069 (0.085) |
| I/σI | 13.6 (2.0) | 26.0 (19.6) |
| Completeness (%) | 97.8 (87.1) | 100.0 (100.0) |
| Multiplicity | 3.6 (3.2) | 7.0 (7.1) |
| Refinement | ||
| Resolution (Å) | 27.47–1.45 | 32.12–1.49 |
| No. reflections | 48884 | 21581 |
| Rwork/Rfree | 0.134/0.178 | 0.135/0.152 |
| No. atoms | ||
| Protein | 1855 | 954 |
| Biotin | n/a | 16 |
| Glycerol | 18 | 18 |
| Water | 321 | 142 |
| B factors (mean of all atoms, Å2) | ||
| Protein | 33.41 | 10.10 |
| Biotin | n/a | 5.83 |
| Glycerol | 44.14 | 28.31 |
| Water | 38.19 | 27.92 |
| Rmsd | ||
| Bond length (Å) | 0.005 | 0.009 |
| Bond angle (°) | 0.93 | 1.48 |
| Twinning | ||
| Twin operator | −h, k, −l | n/a |
| Twin fraction | 0.497 | n/a |
Structure solution and refinement: biotin–Tr
The results were indexed and integrated using MOSFLM, and scaled and merged using SCALA from the CCP4 program suite. Intensities were converted into structure factors using the truncate program in CCP4 and no signs of twinning were observed in the results. The structure was phased by molecular replacement using the apo-Tr search model and Phaser. The crystal contained one monomer in the asymmetric unit. A total of 5% of the reflection data were set aside using the Freerflag program in CCP4 and used for the calculation of Rfree. Model building was performed in Coot and refinement, including rounds of simulated annealing refinement, in PHENIX refine. Throughout the refinement, all data were included from 38.78 Å resolution to the highest limit (1.49 Å) and anisotropic temperature factors were refined. The presence of a well defined biotin model in the binding pocket was clearly visible in the initial Fobs–Fcalc electron density maps. Biotin co-ordinates from wild-type core SA with biotin at 1.4 Å (PDB code 1MK5) [29] were used as a template for the refinement and the weighting of co-ordinate refinement was optimized by PHENIX refine as the refinement progressed. The model was evaluated with MolProbity [28b], which gave Ramachandran statistics with 98% of residues in favoured regions and no outliers. The DPI indicated that the agreement between the model and the X-ray data, for biotin–Tr was 0.051 Å. The refinement statistics are shown in Table 1. To assess whether model bias was introduced into the biotin–Tr model by using the apo-Tr structure as a search model for molecular replacement, a simulated annealing composite omit map was calculated using PHENIX AutoBuild [30], with the starting phases provided by the apo-Tr structure.
Structure analysis
Structural overlays and colouring according to B factor were done in PyMOL (DeLano Scientific; http://www.pymol.org). Alignments were calculated from main-chain atoms of apo-Tr chain A, biotin–Tr, apo-core-SA at pH 4.5 (PDB code 1SWA and 1SWC), apo-core SA at pH 7.5 (PDB code 1SWB) and biotin–core-SA at pH 4.5 (PDB code 1SWE) [28]. Plots of mean B factors of main-chain atoms against residue number were produced using the BAVERAGE program in CCP4. The DPI was assessed using the program SFCHECK from CCP4.
Biotin-conjugate off-rate assay
The off-rate of biotin-4-fluorescein was measured from the fluorescence increase on unbinding at 37 °C with competing free biotin, as previously described [26,31].
Thermostability analysis
A 3 μM sample of dead streptavidin, tetravalent Tr or monovalent Tr in PBS was heated at the indicated temperature for 3 min in a DNA Engine® Peltier Thermal Cycler (Bio-Rad). SDS/PAGE (8% gel) was performed to discriminate the monomers as described previously [26]. The band intensities were quantified using a ChemiDoc XRS imager and QuantityOne 4.6 software (Bio-Rad). Percentages were calculated as 100 × (summed intensity of monomer bands at the indicated temperature − summed intensity of monomer bands at 25 °C)/(summed intensity of monomer bands after 95 °C with SDS).
RESULTS
L3/4 of Tr is closed even without biotin
We determined the crystal structures at high resolution of Tr with and without biotin (1.45 Å for apo-Tr, PDB code 2Y3E; 1.49 Å for biotin–Tr, PDB code 2Y3F) (Table 1). Both structures were solved by molecular replacement; the apo-SA structure (PDB code 1SWB) was used as a search model for the apo-Tr structure [28]. The subsequent apo-Tr structure was used as the search model for biotin-bound Tr. To determine whether model bias was introduced into the biotin–Tr structure by using the apo-Tr structure as a search model for molecular replacement, a simulated annealing composite omit map was calculated. Alignment of this map with the biotin–Tr and apo-Tr models revealed the final biotin–Tr model fitted the omit map much better (correlation coefficient of 0.73 for side-chain atoms) than the apo-Tr model (correlation coefficient of 0.47 for side-chain atoms) (Supplementary Figure S1 at http://www.BiochemJ.org/bj/435/bj4350055add.htm), thus indicating that the solution of the biotin–Tr structure was not biased by the search model.
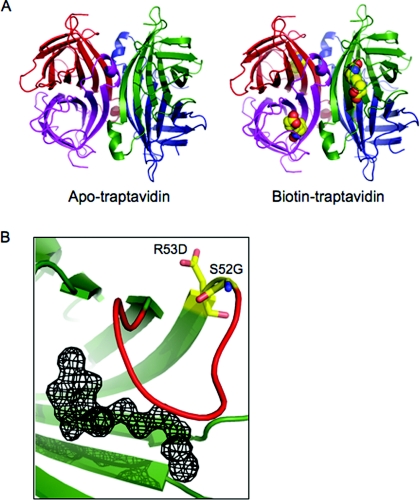
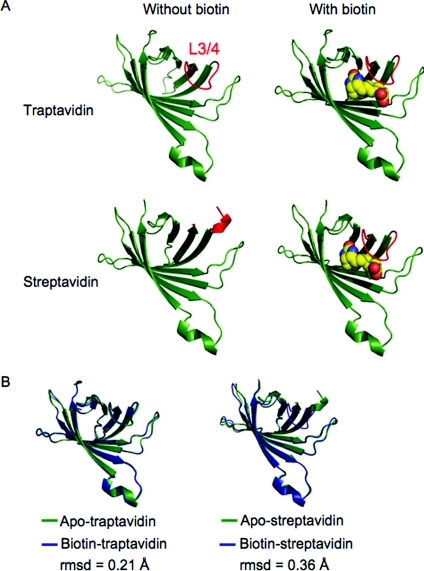
We observed two subunits in the asymmetric unit for apo-Tr and one subunit in the asymmetric unit for biotin–Tr. We applied symmetry transformations to view the complete tetramer (Figure 1A). SA/Tr subunits are eight-stranded antiparallel β-barrels, with the biotin-binding site located at one end of the barrel. Each subunit binds one biotin molecule and the subunits come together to form a tetramer that can be considered a dimer of dimers. The electron density for the biotin bound to Tr was unambiguous (Figure 1B). A striking difference between Tr and SA was seen in the loop connecting β-strands 3 and 4 (L3/4). L3/4 is commonly disordered in apo-SA (‘open’ conformation) [28], but becomes ordered on biotin binding, closing over the binding pocket and forming a ‘lid’ over the bound biotin (‘closed’ conformation) (Figure 2A). However, in apo-Tr, all L3/4 of the tetramer was already in the ‘closed’ conformation. L3/4 remained in this closed conformation in biotin–Tr (Figure 2A). Apo-SA structures 1SWA and 1SWB had only one out of four subunits, whereas 1SWC had two out of four subunits with an ordered L3/4 in the tetramer (1SWA, 1SWB and 1SWC have four subunits in the asymmetric unit) [28]. We emphasize that the fact that apo-Tr had two subunits in the asymmetric unit does not mean that the third and fourth subunits were less clearly defined than if there were four subunits in the asymmetric unit; rather the crystallographic symmetry tells us that the third and fourth subunits are identical with the first and second subunits.
Figure 1. Crystal structures of Tr.
(A) Structure of the complete tetramers of apo-Tr (left-hand panel) and biotin–Tr (right-hand panel), with each subunit of the tetramer in a different colour. Biotin bound to each subunit is shown in space-filling mode. (B) Fobs–Fcalc map of the biotin-binding pocket in biotin–Tr, showing the electron density for biotin, with the protein overlaid in cartoon format. L3/4 is shown in red and the residues mutated in Tr are in stick format.
Figure 2. L3/4 of Tr is shut with and without biotin.
(A) Comparison between the structure of individual subunits of Tr or SA, with or without biotin. Tr (top row) has a well-defined closed L3/4 (coloured red) in both apo- (left-hand panel) and biotin-bound (right-hand panel) structures. For SA (bottom row) L3/4 is disordered in apo-SA (left-hand panel) (1SWA chain B) but ordered in biotin–SA (1SWE chain D) (right-hand panel) [28]. Biotin is shown in space-filling mode. (B) Conformational changes on biotin binding in Tr (left-hand panel) and SA (right-hand panel), shown as individual subunits. The apo-structure in green is overlaid with the biotin-bound structure in blue.
Structural alignments of these apo-SA structures with apo-Tr revealed the closed loops in the apo-SA structures are comparable with L3/4 in apo-Tr; the rmsd (root mean square deviation) from apo-Tr chain A for residues 45–52 is 0.24 Å for 1SWA chain A and 0.20 Å for 1SWB chain A. The presence of one ‘closed’ loop in the apo-SA tetramer suggests that L3/4 is dynamic in the absence of biotin. L3/4 in the 1SWC apo-SA structure is an exception, with an alternative open conformation, projecting away from the binding site, and is not comparable with L3/4 in apo-Tr (rmsd 2.2 Å compared with 1SWC chain B) [28].
The whole structure of Tr is preformed for biotin binding
SA does not show co-operativity in biotin binding between the four subunits [32], but there is a substantial conformational change when biotin binds, involving the flattening and tighter wrapping of the β-barrels, altered dimer–dimer packing and loop ordering [33]. This conformational change is consistent with an observed increase in thermostability of the biotin-bound tetramer [34]. Upon aligning the apo-Tr and biotin–Tr structures, we found that the conformational change in Tr upon biotin binding is much smaller than for SA (rmsd 0.21 Å for Tr, 0.36 Å for SA) (Figure 2B). Differences in the apo- and biotin-bound conformations of SA were clearly shown by calculating alignments of the loops connecting the β-strands (Table 2). It was not only L3/4 which moved less upon biotin binding in Tr than SA, but also five out of six of the other loops.
Table 2. Structural alignments of loop regions.
(a) Loops of apo-Tr were aligned to biotin–Tr, apo-SA and biotin–SA. Residues comprising each loop are shown in parentheses. (b) Loops of apo-SA were aligned to biotin–SA, apo-Tr and biotin–Tr. Only main-chain atoms were used to calculate the alignments. *The loop of 1SWA was compared only with the resolved residues of 45 and 49–52, since 46–48 were disordered.
| (a) | |||
|---|---|---|---|
| Rmsd (Å) of alignment of loop in: | |||
| Apo-Tr (chain A) aligned with | Biotin–Tr | Apo-SA (1SWA chain B) | Biotin–SA (1SWE chain D) |
| L1/2 (23–26) | 0.050 | 0.20 | 0.21 |
| L2/3 (33–38) | 0.14 | 0.23 | 0.15 |
| L3/4 (45–52) | 0.25 | 2.3* | 0.25 |
| L4/5 (62–70) | 0.20 | 0.20 | 0.22 |
| L5/6 (79–88) | 0.18 | 0.28 | 0.18 |
| L6/7 (98–102) | 0.14 | 0.22 | 0.37 |
| L7/8 (113–121) | 0.20 | 0.23 | 0.24 |
| Overall structure | 0.21 | 0.29 | 0.35 |
| (b) | |||
| Rmsd (Å) of alignment of loop in: | |||
| Apo-SA (1SWA chain B) aligned with | Biotin–SA (1SWE, chain D) | Apo-Tr (chain A) | Biotin–Tr |
| L1/2 (23–26) | 0.27 | 0.20 | 0.18 |
| L2/3 (33–38) | 0.23 | 0.23 | 0.22 |
| L3/4 (45–52) | 2.6* | 2.3* | 2.3* |
| L4/5 (62–70) | 0.25 | 0.20 | 0.25 |
| L5/6 (79–88) | 0.31 | 0.28 | 0.27 |
| L6/7 (98–102) | 0.42 | 0.22 | 0.26 |
| L7/8 (113–121) | 0.17 | 0.23 | 0.15 |
| Overall structure | 0.36 | 0.29 | 0.25 |
Changes in the flexibility of Tr
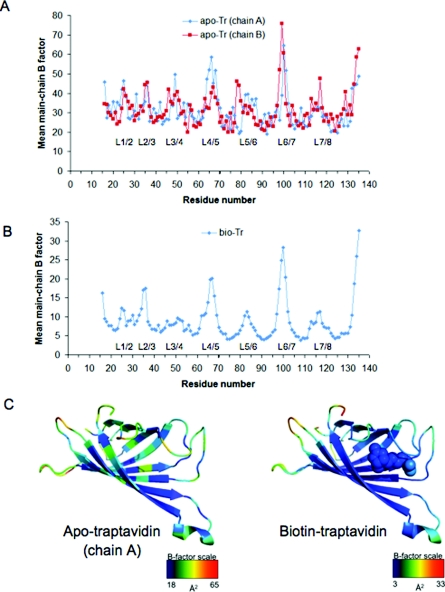
To analyse the flexibility and dynamics of apo-Tr and biotin–Tr, we plotted the mean B factors for main-chain atoms against residue number (Figure 3). Relative B factors within a structure are informative, but it is not straightforward to compare directly the absolute values of B factors between SA and Tr, because of different resolutions, data collection conditions and refinement methods.
Figure 3. Tr flexibility.
(A) B factors (indicating dynamics and flexibility) averaged over main-chain atoms for each amino acid residue are plotted for apo-Tr, with the two chains of the asymmetric unit shown separately. The regions corresponding to loops are labeled L. (B) Mean main-chain B factors for each residue of biotin–Tr, with loops labelled. (C) Heat map showing mean main-chain B factors for an individual subunit of apo-Tr (left-hand panel, chain A) or biotin–Tr (right-hand panel), with biotin shown in space-filling mode. Red indicates the most flexible and blue the least flexible region of the particular subunit.
The B factors for each of the two monomers in the asymmetric unit of apo-Tr were plotted to indicate the internal variation. This showed the expected low dynamics in the β-sheet regions and some flexibility in all the loop sections (Figure 3A). The main difference between chains A and B of apo-Tr occurs at L5/6 and may result from one chain being involved in crystal contacts at this loop. There is some flexibility in L3/4 for apo-Tr, but clearly it is less than in apo-SA structures where the loop is not resolved. If L3/4 in apo-Tr did not have some flexibility then it is likely that the Tr on-rate would be too low for practical application. The most striking change in the biotin–Tr B factors is the loss of flexibility in L3/4, with values comparable with adjacent β-sheets 3 and 4 (Figure 3B), which is consistent with the exceptionally low biotin dissociation rate.
Hydrogen bonding to biotin and Ser45 in Tr and SA
The extensive hydrogen-bonding network from SA to biotin plays a key role in generating the large ΔG°′ of biotin binding [2]. We compared all of the hydrogen bonds to biotin for Tr and SA, paying particular attention to Ser45 and Asn49, which are present on L3/4 close to the residues mutated in Tr. Factoring in the variation seen in the different chains of the different structures of SA, the hydrogen-bond lengths to biotin are comparable between Tr and SA (Supplementary Figure S2 at http://www.BiochemJ.org/bj/435/bj4350055add.htm).
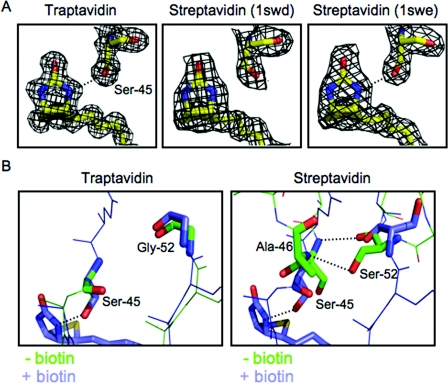
The published structures of SA allude to the probable dynamic nature of this hydrogen-bond network to biotin. In some crystal structures of SA (1SWE), each Ser45 residue has its side-chain hydroxy group directed towards the biotin N3′ nitrogen, and hydrogen bonds are formed (although the distance in subunits 1 and 4 in 1SWE is at the limit for a hydrogen bond between such atoms, at 3.2 Å) (Figure 4A and Supplementary Figure S2). However, in the SA structure of 1SWD, the Ser45 side chains point away from the N3′ nitrogen and so cannot hydrogen bond (Figure 4A and Supplementary Figure S2). In the biotin–Tr structure, the Ser45 side chain is clearly orientated towards the N3′ nitrogen (Figure 4A), with a hydrogen bond length of 3.0 Å.
Figure 4. Hydrogen bonding by Ser45 in Tr.
(A) The electron density for biotin and Ser45 surrounds a stick model of the atoms involved, for Tr (left-hand panel) and two different streptavidin structures (1SWD in the middle panel and 1SWE in the right-hand panel). Hydrogen bonds are indicated by a broken line. (B) The rival hydrogen bonding by Ser45. An overlay of the L3/4 region, with key residues highlighted in stick format. For Tr (left-hand panel) with and without biotin L3/4 is closed, with no hydrogen bond between Ser45 and Gly52. For SA (right-hand panel) without biotin and with an undefined L3/4 (1SWC, chain B), Ser52 forms hydrogen bonds to Ser45 and Ala46 (indicated by broken lines). When biotin is bound and the loop is closed (1SWE, chain D), Ser45 now forms a hydrogen bond to biotin.
Gly52 in Tr undergoes little change upon biotin binding (Figure 4B). The peculiar importance of the S52G mutation for biotin-binding stability is likely to relate to Ser52 stabilizing an alternative open conformation of L3/4, thereby destabilizing the Ser45 hydrogen bond to biotin. In this rival conformation, Ser52's main-chain carbonyl forms a hydrogen bond to the main-chain N–H of Ser45, while the Ser52 side chain makes a hydrogen bond to the main–chain N–H of Ala46 (Figure 4B). This competing pairing is seen in apo-SA structures with an undefined L3/4 (1SWC) [28], but could well form transiently even in the presence of biotin, so contributing to the rare biotin dissociation events from SA.
The network of polar and non-polar interactions made to biotin by SA changes biotin's structure from the conformation reported to apply in solution [35]. In avidin and SA there are three residues that are positioned to hydrogen bond to the carbonyl oxygen of biotin, suggesting that this oxygen is partially charged and that this polarization may contribute to the exceptional affinity of biotin binding [16]. The bond lengths and bond angles of biotin were similar between SA and Tr (results not shown), indicating that changes in biotin's electronic structure in the binding site are not likely to contribute to the difference in biotin off-rate.
Generating monovalent Tr shows that altered intersubunit contacts do not explain Tr's increased stability
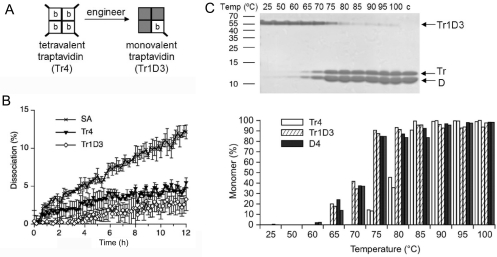
We created tetramers with exactly one Tr subunit and three ‘dead’ subunits which do not bind biotin [26], to give monovalent traptavidin (Tr1D3) (Figure 5A). We previously showed that monovalent SA had equivalent biotin-binding stability to tetravalent SA [26]. Here we used Tr1D3 to explore whether the increased stability of Tr over SA depended upon the presence of neighbouring Tr subunits. The off-rate of biotin-4-fluorescein was compared for Tr4, Tr1D3 and tetravalent SA (Figure 5B). Tr1D3 had an equivalent off-rate to that of Tr4, showing that it is the binding by the Tr subunit and not any altered intersubunit interaction that is the dominant factor in Tr's improved biotin-binding stability.
Figure 5. Monovalent Tr off-rate and thermostability.
(A) Tr4 (left-hand panel) is tetravalent, with each subunit binding one biotin (b). Monovalent Tr contains one Tr subunit (white square) and three D subunits (grey squares), which cannot bind biotin. Tr but not D subunits have His6 tags (bold diagonal line). (B) Off-rate of a biotin conjugate from Tr1D3 compared with tetravalent SA and Tr4, in the presence of competing free biotin at 37 °C. Means of triplicate readings are shown ±S.D. (C) Thermostability of the monovalent traptavidin tetramer, incubated at the indicated temperature for 3 min and analysed by SDS/PAGE and Coomassie Blue staining. The positive control (c) was mixed with SDS before heating at 95 °C. Bands from tetrameric Tr1D3 or monomers (Tr or D) are indicated. The percentage monomer from Tr1D3 from duplicate gels (adjacent striped columns) is plotted against the temperature in the lower panel, with comparison with Tr4 (white columns) or D4 (filled columns).
Tr stays tetrameric and remains bound to biotin conjugates at higher temperatures than SA [24]. We tested the thermostability of Tr1D3 by incubating it at a range of temperatures and testing by SDS/PAGE what fraction remained tetrameric (Figure 5C). Monovalent SA is known to show thermostability equivalent to that of wild-type SA [26]. We compared Tr1D3 to Tr4 and a tetramer composed entirely of dead subunits (D4). Tr4 was ~10 °C more stable than D4. The thermostability of Tr1D3 was similar to that of D4, consistent with the least stable subunit dominating the stability of the tetramer.
DISCUSSION
SA's binding to biotin is one of the strongest known non-covalent protein–ligand interactions and yet the mutant protein Tr has 10-fold greater binding stability [24]. The crystal structures of apo- and biotin-bound Tr described in the present paper show the basis for Tr's tenacious ligand binding. The key differences between Tr and SA are the decreased flexibility of L3/4, the increased stability of the Ser45 hydrogen bond to biotin and the reduced conformational change upon biotin binding.
The crystal structure of D128A SA and molecular dynamic simulations [15,36,37] led to a model whereby a water molecule initiates biotin dissociation, by competing with the Asp128 hydrogen bond to biotin and so promoting co-operative breakage of other biotin hydrogen bonds. However, we note that in wild-type SA the Asp128 hydrogen bond to biotin is always in place, but the Ser45 hydrogen bond is often broken [28]. Hence we suggest that breakage of the Ser45 hydrogen bond to biotin is frequent and is the first event in biotin dissociation. The mutations that led to a higher stability mutant Tr, further support this revised model: in Tr, the Ser45 hydrogen bond to biotin was clearly present, whereas the residue that competes with biotin for hydrogen bonding to Ser45, Ser52, is mutated to a glycine residue. Arg53 does not have direct interactions with residues binding to biotin, so the R53D mutation is likely to exert its effects by further reducing L3/4 mobility.
L3/4 is always in a ‘closed’ conformation in biotin-bound structures of SA, acting like a lid over the binding pocket and contributing to the low rate of biotin dissociation. For avidin, L3/4 is ordered when bound to free biotin, but disordered when bound to biotin conjugates: this explains why avidin binds biotin conjugates much more weakly than biotin [38]. A restrained L3/4 may also be important in enabling high-affinity biotin binding by the dimeric rhizavidin [39]. In a series of structures solved in the pH range 2.0–3.1 [40], L3/4 was closed in apo-SA, but in the apo-SA structures solved in the more physiologically relevant pH range 4.5–7.5, L3/4 is disordered, in two (1SWC) or three (1SWA and 1SWB) of the subunits of the tetramer [28]. The stability of L3/4 in the closed conformation in apo-Tr indicates that in biotin–Tr the occasional fluctuations that lead to lid opening, and so facilitate biotin dissociation, are similarly suppressed. Since we found that the hydrogen-bond lengths to biotin and conformation of biotin are equivalent in Tr and SA, this indicates that it is not subtle differences in the ground-state binding conformation that explain the change in stability, but a change in the frequency of alternative protein conformations with weakened binding. The L3/4 conformation in apo-Tr also rationalizes the decreased on-rate of Tr [24], since it will be harder for biotin to enter the binding site with the loop shut. For Streptomyces avidinii, one can hypothesize that evolution would favour a SA with a high on-rate as well as a low off-rate, but for laboratory applications in anchoring and bridging [24] the off-rate is more important.
SA is a highly thermostable protein (72 °C for subunit dissociation without biotin), even though the protein derives from a mesophilic bacterium [1,2,34]. This thermostability relates to reducing the rare subunit-unfolding events that would contribute to ligand dissociation, or, from an energetic perspective, rigid binding sites having a reduced entropic cost for providing an inflexible ligand-binding site [41]. Any fleeting disruption of the interaction between biotin and Trp120 of the neighbouring subunit should transiently increase the rate of biotin dissociation 104-fold [42]. Tr is 10 °C more thermostable than SA [24] and one might think that this thermostability would contribute to the increased ligand-binding stability, by further reducing the frequency of conformational fluctuations that disrupt the protein structure or subunit packing. The equivalent off-rate of Tr1D3 and Tr4 demonstrated that the change in Tr's biotin-binding stability is not mediated through altered subunit interactions. The tetravalent nature of Tr4 interferes with many applications: for example in imaging cell-surface proteins, tetravalency can induce receptor cross-linking and activate cell signalling [6,26]. Tr1D3 (and Tr2D2 or Tr3D1, which could be generated, as before [26]) should be valuable in diverse applications, including nanoassembly and the targeting of quantum dots for photostable single-molecule imaging [5,6].
The stability of the interaction between biotin and avidin/SA is paradoxical, given the known binding energy that can be obtained from a given number of hydrogen bonds and from a given surface area, allowing hydrophobic and van der Waals interactions [43,44]. The mutations that generated a higher-stability variant of SA were in the second shell of residues around biotin, pointing to the difficulty of optimizing this kind of high-affinity interaction computationally. Although we have sufficient knowledge to enable computational and structure-guided design of protein–ligand interactions with micromolar and sometimes nanomolar affinity [44–46], the factors responsible for increasing affinity into the picomolar and femtomolar range are still enigmatic [47,48]. Hydrogen/deuterium-exchange MS, now possible with single-residue resolution, may be able to build upon our crystal structures for further enlightenment on the ‘perfect storm’ of molecular events required for biotin to overcome the large activation barrier to dissociate from Tr [49]. As well as suggesting how to improve further the applications of biotin as an affinity tag, the structures of Tr shed light on the origins of extreme-affinity interactions, with implications for the rational design of drugs with improved efficacy.
Online data
ACKNOWLEDGEMENTS
We thank Sonja Baumli for assistance in crystal freezing. Traptavidin is subject to a U.K. priority patent application (0919102.4) filed on 30 October 2009.
FUNDING
Funding was provided by the Wellcome Trust [grant number 085457/Z/OB/Z] (A.L.K. and M.H.), the Biotechnology and Biological Sciences Research Council (C.E.C.), Oxford University (E.D.L.) and Worcester College Oxford (M.H.).
References
- 1.Sano T., Vajda S., Cantor C. R. Genetic engineering of streptavidin, a versatile affinity tag. J. Chromatogr. B Biomed. Sci. Appl. 1998;715:85–91. doi: 10.1016/s0378-4347(98)00316-8. [DOI] [PubMed] [Google Scholar]
- 2.Stayton P. S., Freitag S., Klumb L. A., Chilkoti A., Chu V., Penzotti J. E., To R., Hyre D., Le Trong I., Lybrand T. P., Stenkamp R. E. Streptavidin-biotin binding energetics. Biomol. Eng. 1999;16:39–44. doi: 10.1016/s1050-3862(99)00042-x. [DOI] [PubMed] [Google Scholar]
- 3.Laitinen O. H., Hytonen V. P., Nordlund H. R., Kulomaa M. S. Genetically engineered avidins and streptavidins. Cell. Mol. Life Sci. 2006;63:2992–3017. doi: 10.1007/s00018-006-6288-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Boer E., Rodriguez P., Bonte E., Krijgsveld J., Katsantoni E., Heck A., Grosveld F., Strouboulis J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howarth M., Takao K., Hayashi Y., Ting A. Y. Targeting quantum dots to surface proteins in living cells with biotin ligase. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howarth M., Liu W., Puthenveetil S., Zheng Y., Marshall L. F., Schmidt M. M., Wittrup K. D., Bawendi M., Ting A. Y. Monovalent, reduced-size quantum dots for single molecule imaging of receptors in living cells. Nat. Methods. 2008;5:397–399. doi: 10.1038/nmeth.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kattah M. G., Coller J., Cheung R. K., Oshidary N., Utz P. J. HIT: a versatile proteomics platform for multianalyte phenotyping of cytokines, intracellular proteins and surface molecules. Nat. Med. 2008;14:1284–1289. doi: 10.1038/nm.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldenberg D. M., Sharkey R. M., Paganelli G., Barbet J., Chatal J. F. Antibody pretargeting advances cancer radioimmunodetection and radioimmunotherapy. J. Clin. Oncol. 2006;24:823–834. doi: 10.1200/JCO.2005.03.8471. [DOI] [PubMed] [Google Scholar]
- 9.Avrantinis S. K., Stafford R. L., Tian X., Weiss G. A. Dissecting the streptavidin–biotin interaction by phage-displayed shotgun scanning. ChemBioChem. 2002;3:1229–1234. doi: 10.1002/1439-7633(20021202)3:12<1229::AID-CBIC1229>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 10.Aslan F. M., Yu Y., Mohr S. C., Cantor C. R. Engineered single-chain dimeric streptavidins with an unexpected strong preference for biotin-4-fluorescein. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8507–8512. doi: 10.1073/pnas.0503112102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy M., Ellington A. D. Directed evolution of streptavidin variants using in vitro compartmentalization. Chem. Biol. 2008;15:979–989. doi: 10.1016/j.chembiol.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green N. M. Avidin. Adv. Protein Chem. 1975;29:85–133. doi: 10.1016/s0065-3233(08)60411-8. [DOI] [PubMed] [Google Scholar]
- 13.Wilbur D. S., Pathare P. M., Hamlin D. K., Stayton P. S., To R., Klumb L. A., Buhler K. R., Vessella R. L. Development of new biotin/streptavidin reagents for pretargeting. Biomol. Eng. 1999;16:113–118. doi: 10.1016/s1050-3862(99)00044-3. [DOI] [PubMed] [Google Scholar]
- 14.Weber P. C., Pantoliano M. W., Simons D. M., Salemme F. R. Structure-based design of synthetic azobenzene ligands for streptavidin. J. Am. Chem. Soc. 1994;116:2717–2724. [Google Scholar]
- 15.Grubmuller H., Heymann B., Tavan P. Ligand binding: molecular mechanics calculation of the streptavidin–biotin rupture force. Science. 1996;271:997–999. doi: 10.1126/science.271.5251.997. [DOI] [PubMed] [Google Scholar]
- 16.DeChancie J., Houk K. N. The origins of femtomolar protein–ligand binding: hydrogen-bond cooperativity and desolvation energetics in the biotin–(strept)avidin binding site. J. Am. Chem. Soc. 2007;129:5419–5429. doi: 10.1021/ja066950n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerutti D. S., Le T. I., Stenkamp R. E., Lybrand T. P. Dynamics of the streptavidin–biotin complex in solution and in its crystal lattice: distinct behavior revealed by molecular simulations. J. Phys. Chem. B. 2009;113:6971–6985. doi: 10.1021/jp9010372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyamoto S., Kollman P. A. Absolute and relative binding free energy calculations of the interaction of biotin and its analogs with streptavidin using molecular dynamics/free energy perturbation approaches. Proteins. 1993;16:226–245. doi: 10.1002/prot.340160303. [DOI] [PubMed] [Google Scholar]
- 19.Moy V. T., Florin E. L., Gaub H. E. Intermolecular forces and energies between ligands and receptors. Science. 1994;266:257–259. doi: 10.1126/science.7939660. [DOI] [PubMed] [Google Scholar]
- 20.Bruneau E., Sutter D., Hume R. I., Akaaboune M. Identification of nicotinic acetylcholine receptor recycling and its role in maintaining receptor density at the neuromuscular junction in vivo. J. Neurosci. 2005;25:9949–9959. doi: 10.1523/JNEUROSCI.3169-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dressman D., Yan H., Traverso G., Kinzler K. W., Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swift J. L., Heuff R., Cramb D. T. A two-photon excitation fluorescence cross-correlation assay for a model ligand-receptor binding system using quantum dots. Biophys. J. 2006;90:1396–1410. doi: 10.1529/biophysj.105.069526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pierres A., Touchard D., Benoliel A. M., Bongrand P. Dissecting streptavidin–biotin interaction with a laminar flow chamber. Biophys. J. 2002;82:3214–3223. doi: 10.1016/S0006-3495(02)75664-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Howarth M., Chivers C. E. U. K. Pat. Appl. 0919102.4. 2009
- 24.Chivers C. E., Crozat E., Chu C., Moy V. T., Sherratt D. J., Howarth M. A streptavidin variant with slower biotin dissociation and increased mechanostability. Nat. Methods. 2010;7:391–393. doi: 10.1038/nmeth.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crozat E., Meglio A., Allemand J. F., Chivers C. E., Howarth M., Venien-Bryan C., Grainge I., Sherratt D. J. Separating speed and ability to displace roadblocks during DNA translocation by FtsK. EMBO J. 2010;29:1423–1433. doi: 10.1038/emboj.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howarth M., Chinnapen D. J., Gerrow K., Dorrestein P. C., Grandy M. R., Kelleher N. L., El Husseini A., Ting A. Y. A monovalent streptavidin with a single femtomolar biotin binding site. Nat. Methods. 2006;3:267–273. doi: 10.1038/NMETHXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D. Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 28.Freitag S., Le Trong I., Klumb L., Stayton P. S., Stenkamp R. E. Structural studies of the streptavidin binding loop. Protein Sci. 1997;6:1157–1166. doi: 10.1002/pro.5560060604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28b.Chen B., Arendall W. B., III, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyre D. E., Le Trong I., Merritt E. A., Eccleston J. F., Green N. M., Stenkamp R. E., Stayton P. S. Cooperative hydrogen bond interactions in the streptavidin–biotin system. Protein Sci. 2006;15:459–467. doi: 10.1110/ps.051970306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., Adams P. D. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. Sect. D Biol. Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kada G., Falk H., Gruber H. J. Accurate measurement of avidin and streptavidin in crude biofluids with a new, optimized biotin-fluorescein conjugate. Biochim. Biophys. Acta. 1999;1427:33–43. doi: 10.1016/s0304-4165(98)00178-0. [DOI] [PubMed] [Google Scholar]
- 32.Jones M. L., Kurzban G. P. Noncooperativity of biotin binding to tetrameric streptavidin. Biochemistry. 1995;34:11750–11756. doi: 10.1021/bi00037a012. [DOI] [PubMed] [Google Scholar]
- 33.Weber P. C., Ohlendorf D. H., Wendoloski J. J., Salemme F. R. Structural origins of high-affinity biotin binding to streptavidin. Science. 1989;243:85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]
- 34.Bayer E. A., Ehrlich-Rogozinski S., Wilchek M. Sodium dodecyl sulfate-polyacrylamide gel electrophoretic method for assessing the quaternary state and comparative thermostability of avidin and streptavidin. Electrophoresis. 1996;17:1319–1324. doi: 10.1002/elps.1150170808. [DOI] [PubMed] [Google Scholar]
- 35.Li Q., Gusarov S., Evoy S., Kovalenko A. Electronic structure, binding energy, and solvation structure of the streptavidin–biotin supramolecular complex: ONIOM and 3D-RISM study. J. Phys. Chem. B. 2009;113:9958–9967. doi: 10.1021/jp902668c. [DOI] [PubMed] [Google Scholar]
- 36.Freitag S., Chu V., Penzotti J. E., Klumb L. A., To R., Hyre D., Le Trong I., Lybrand T. P., Stenkamp R. E., Stayton P. S. A structural snapshot of an intermediate on the streptavidin–biotin dissociation pathway. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8384–8389. doi: 10.1073/pnas.96.15.8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hyre D. E., Amon L. M., Penzotti J. E., Le Trong I., Stenkamp R. E., Lybrand T. P., Stayton P. S. Early mechanistic events in biotin dissociation from streptavidin. Nat. Struct. Biol. 2002;9:582–585. doi: 10.1038/nsb825. [DOI] [PubMed] [Google Scholar]
- 38.Pazy Y., Kulik T., Bayer E. A., Wilchek M., Livnah O. Ligand exchange between proteins. Exchange of biotin and biotin derivatives between avidin and streptavidin. J. Biol. Chem. 2002;277:30892–30900. doi: 10.1074/jbc.M202874200. [DOI] [PubMed] [Google Scholar]
- 39.Meir A., Helppolainen S. H., Podoly E., Nordlund H. R., Hytonen V. P., Maatta J. A., Wilchek M., Bayer E. A., Kulomaa M. S., Livnah O. Crystal structure of rhizavidin: insights into the enigmatic high-affinity interaction of an innate biotin-binding protein dimer. J. Mol. Biol. 2009;386:379–390. doi: 10.1016/j.jmb.2008.11.061. [DOI] [PubMed] [Google Scholar]
- 40.Katz B. A. Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH. J. Mol. Biol. 1997;274:776–800. doi: 10.1006/jmbi.1997.1444. [DOI] [PubMed] [Google Scholar]
- 41.Moghaddam S., Inoue Y., Gilson M. K. Host-guest complexes with protein-ligand-like affinities: computational analysis and design. J. Am. Chem. Soc. 2009;131:4012–4021. doi: 10.1021/ja808175m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chilkoti A., Tan P. H., Stayton P. S. Site-directed mutagenesis studies of the high-affinity streptavidin–biotin complex: contributions of tryptophan residues 79, 108, and 120. Proc. Natl. Acad. Sci. U.S.A. 1995;92:1754–1758. doi: 10.1073/pnas.92.5.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuntz I. D., Chen K., Sharp K. A., Kollman P. A. The maximal affinity of ligands. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9997–10002. doi: 10.1073/pnas.96.18.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Houk K. N., Leach A. G., Kim S. P., Zhang X. Binding affinities of host–guest, protein–ligand, and protein-transition-state complexes. Angew. Chem. Int. Ed. Engl. 2003;42:4872–4897. doi: 10.1002/anie.200200565. [DOI] [PubMed] [Google Scholar]
- 45.Das R., Baker D. Macromolecular modeling with rosetta. Annu. Rev. Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 46.Holm L., Moody P., Howarth M. Electrophilic affibodies forming covalent bonds to protein targets. J. Biol. Chem. 2009;284:32906–32913. doi: 10.1074/jbc.M109.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Midelfort K. S., Hernandez H. H., Lippow S. M., Tidor B., Drennan C. L., Wittrup K. D. Substantial energetic improvement with minimal structural perturbation in a high affinity mutant antibody. J. Mol. Biol. 2004;343:685–701. doi: 10.1016/j.jmb.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 48.Foote J., Eisen H. N. Breaking the affinity ceiling for antibodies and T cell receptors. Proc. Natl. Acad. Sci. U.S.A. 2000;97:10679–10681. doi: 10.1073/pnas.97.20.10679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rand K. D., Zehl M., Jensen O. N., Jorgensen T. J. Protein hydrogen exchange measured at single-residue resolution by electron transfer dissociation mass spectrometry. Anal. Chem. 2009;81:5577–5584. doi: 10.1021/ac9008447. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.