Abstract
The Wilms' tumour suppressor WT1 (Wilms' tumour 1) is a transcriptional regulator that plays a central role in organogenesis, and is mutated or aberrantly expressed in several childhood and adult malignancies. We previously identified BASP1 (brain acid-soluble protein 1) as a WT1 cofactor that suppresses the transcriptional activation function of WT1. In the present study we have analysed the dynamic between WT1 and BASP1 in the regulation of gene expression in myelogenous leukaemia K562 cells. Our findings reveal that BASP1 is a significant regulator of WT1 that is recruited to WT1-binding sites and suppresses WT1-mediated transcriptional activation at several WT1 target genes. We find that WT1 and BASP1 can divert the differentiation programme of K562 cells to a non-blood cell type following induction by the phorbol ester PMA. WT1 and BASP1 co-operate to induce the differentiation of K562 cells to a neuronal-like morphology that exhibits extensive arborization, and the expression of several genes involved in neurite outgrowth and synapse formation. Functional analysis revealed the relevance of the transcriptional reprogramming and morphological changes, in that the cells elicited a response to the neurotransmitter ATP. Taken together, the results of the present study reveal that WT1 and BASP1 can divert the lineage potential of an established blood cell line towards a cell with neuronal characteristics.
Keywords: brain acid-soluble protein 1 (BASP1), K562 cell, Wilms' tumour, Wilms' tumour 1 (WT1)
Abbreviations: ALL, acute lymphocytic leukaemia; BASP1, brain acid-soluble protein 1; ChIP, chromatin immunoprecipitation; CLL, chronic lymphocytic leukaemia; CPA, cyclopiazonic acid; DAB2, Disabled homologue 2; ENC1, ectodermal neural cortex 1; FBS, foetal bovine serum; fura 2/AM, fura 2 acetoxymethyl ester; GFP, green fluorescent protein; HA, haemagglutinin; ITGA2, integrin α2; ITGA2B, integrin α2b; ITGB3, integrin β3; MAOA, monoamine oxidase A; NLS, nuclear localization sequence; pol II, RNA polymerase II; qPCR, quantitative PCR; SERCA, sarcoplasmic/endoplasmic reticulum Ca2+-ATPase; siRNA, small interfering RNA; VDR, vitamin D receptor; WT1, Wilms' tumour 1
INTRODUCTION
The Wilms' tumour suppressor protein WT1 (Wilms' tumour 1) is mutated or aberrantly expressed in several childhood and adult cancers, where it can act as either a tumour suppressor or oncogene [1,2]. WT1 is a transcriptional regulator that plays a central role in the development of several organs and tissues, including the kidneys, gonads and spleen [3]. More recent studies have revealed a critical role for WT1 in the development of specific neuronal tissues including the retinal ganglia and olfactory epithelia [4,5]. Indeed, WT1 is also expressed in the spinal cord and specific regions of the brain during development [6].
WT1 can either activate or repress genes involved in growth, apoptosis and differentiation. The transcriptional regulatory activity of WT1 is subject to extensive control by interaction with other factors that can act as either co-activators or co-repressors [7]. We identified BASP1 (brain acid-soluble protein 1) as a WT1-interacting protein that binds to the suppression domain of WT1 (residues 71–101) and inhibits its transcriptional activation function [8,9]. WT1 and BASP1 are spatially and temporally co-expressed at several sites in the developing embryo, suggesting a significant role for BASP1 in the regulation of WT1 activity in development. The suppression domain of WT1 also forms a key binding site for the serine protease HtrA2, which cleaves WT1 under apoptotic conditions [10].
BASP1 was originally isolated from neuronal cells, where it is predominantly cytoplasmic with partial membrane localization mediated by the N-terminal myristoylation of BASP1 [11]. The function of BASP1 in neuronal cells is still unclear, but it has been proposed to play a role in nerve sprouting. Several cell types also contain a nuclear pool of BASP1 [9,12,13]. Indeed BASP1 contains a bipartite NLS (nuclear localization sequence), sites of SUMO (small ubiquitin-related modifier)-modification, and a portion of BASP1 localizes to PML (promyelocytic leukaemia) bodies within the nucleus [13].
Since our previous report of a transcriptional role for BASP1 [9], others have also described a function for BASP1 in the nucleus. It has been reported that BASP1 localizes from the nucleus to the cytoplasm in cells that are undergoing apoptosis [12]. In addition, a recent study found that BASP1 can inhibit cellular transformation by the v-Myc oncogene, and can block the regulation of Myc target genes [14]. This same work showed that BASP1 is consistently down-regulated in Myc-transformed cells. BASP1 does not interact with Myc, however, suggesting an indirect mechanism. BASP1 is also down-regulated in a significant proportion of hepatocellular carcinomas, and this is attributed to silencing of the BASP1 gene by methylation [15]. In addition, BASP1 expression has also been reported to be frequently down-regulated in both ALL (acute lymphocytic leukaemia) and CLL (chronic lymphocytic leukaemia) [16,17]. Taken together, these recent studies suggest that BASP1 probably acts as a tumour suppressor.
The role of WT1 in leukaemia has attracted considerable attention over the last few years [2,18,19]. In total, 74% of AML (acute myeloid leukaemia), 66% of ALL and over 50% of CLL samples show high levels of wild-type WT1 expression. Moreover, leukaemia patients with elevated WT1 have a poor prognosis and significantly reduced 5-year survival rates. WT1 plays a role in haemopoeisis, serving to maintain the self-renewal of primitive CD34+ cells in the bone marrow [18]. As differentiation proceeds WT1 is down-regulated and is not expressed in mature blood cells. Thus it has been proposed that, in leukaemia, WT1 contributes to the maintenance of the dedifferentiated state and promotes proliferation [2].
In the present study, we have analysed the role of BASP1 as a WT1 cofactor in myelogenous leukaemia K562 cells. We find that BASP1 regulates WT1 activity at several previously identified WT1 target genes and that this involves the recruitment of BASP1 to the promoter. WT1 and BASP1 together divert the differentiation of K562 cells away from the blood cell lineage and direct differentiation towards cells with neuronal-like morphology, gene expression pattern and functional properties. The results of the present study suggest that the WT1/BASP1 dynamic plays a central role in directing cell fate during differentiation.
EXPERIMENTAL
Cell culture and transfection
K562 cells were cultured in RPMI 1640 medium supplemented with 10% FBS (foetal bovine serum), 100 mg/ml streptomycin, 100 units/ml penicillin and 2 mM L-glutamine. Cells were transfected using Lipofectamine™ 2000 (Invitrogen) or by electroporation using the Amaxa Nucleofector. K562 cells stably transfected with pcDNA3 were selected with 2 mg/ml G418 (Sigma) and pools of cells were maintained in 2 mg/ml G418. PMA and haemin were purchased from Sigma. Control siRNAs (small interfering RNAs) used were from Ambion (AM4611 and AM4635) and WT1 siRNAs were from Santa Cruz Biotechnology (sc-36846) and Ambion (s14912).
Cell extracts
To prepare whole-cell extracts, cells were washed twice in ice-cold PBS and then lysed for 20 min in Triton lysis buffer [20 mM Tris/HCl (pH 7.4), 137 mM NaCl, 2 mM EDTA, 25 mM 2-glycerophosphate, 1 mM sodium orthovanadate, 1 mM PMSF, 1×protease inhibitor cocktail (Sigma), 10% (v/v) glycerol and 1% (v/v) Triton X-100]. Insoluble material was removed by centrifugation at 16000 g for 10 min at 4 °C. FLAG-tagged BASP1 was immunoprecipitated from whole-cell extracts using anti-FLAG M2 affinity gel (Sigma). Nuclear and cytosolic extracts were prepared using a nuclear extract kit (Active Motif) according to the manufacturer's protocol. For immunoblotting, equal amounts of protein were resuspended in protein loading dye, resolved by SDS/PAGE and then transferred on to a PVDF membrane.
Antibodies and immunofluorescence
Anti-WT1 (C-19), anti-lamin A/C (N-18), anti-P2X5 (H-90) and anti-DAB2 (Disabled homologue 2) (H-110) were from Santa Cruz Biotechnology. Anti-pol II (RNA polymerase II) (ab5408), anti-ENC1 (ectodermal neural cortex 1) (ab56348) and anti-β-tubulin (ab6046) were from Abcam. Control IgG and anti-WT1 (6F-H2) antibodies were from Millipore. The rabbit anti-FLAG antibody was from Cell Signaling Technology. Anti-BASP1 antibodies have been described previously [9,13]. Immunofluorescence was performed as described previously [9] using Jackson ImmunoResearch Dylight secondary antibodies.
Plasmids and reagents
To generate pcDNA3-FLAG-HA (control vector) (HA is haemagglutinin), the following oligonucleotides were annealed and subcloned into the EcoRI/XbaI sites of pcDNA3: sense 5′-AATTCGTCGACAAGACTACAAGGACGACGATGACAAGTCGGCCGCTGGAGGATACCCCTACGACGTGCCCGACTACGCCTAGT-3′ and antisense 5′-CTAGACTAGGCGTAGTCGGGCACGTCGTAGGGGTATCCTCCAGCGGCCGACTTGTCATCGTCGTCCTTGTAGTCTTGTCGACG-3′. Human BASP1 cDNA was amplified (using 5′-GCGGATCCATGGGAGGCAAGCTCAGC-3′ and 5′-CCGAATTCCCTCTTTCACGGTTACGGT-3′) and subcloned into the BamHI/EcoRI sites of the control vector to give the expression construct pcDNA3-BASP1-FLAG-HA. The plasmid for GFP (green fluorescent protein) expression was pCMV6-AC-GFP (Origene).
ChIP (chromatin immunoprecipitation) and RNA analysis
Total RNA was prepared using the Qiagen RNeasy kit and cDNA prepared using the Promega Reverse Transcription kit or the Bio-Rad cDNA synthesis kit. Real-time PCR was performed using a Bio-Rad MiniOpticon System and SYBR Green assay reagents. Primers for mRNA analysis are shown in Table 1.
Table 1. Primers used for mRNA analysis.
AREG, gene encoding amphiregulin; EGFR, epidermal growth factor receptor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
| Gene | Forward (5′→3′) | Reverse (5′→3′) |
|---|---|---|
| GAPDH | ACAGTCAGCCGCATCTTCTT | ACGACCAAATCCGTTGACTC |
| Renin | GAAAGCCTGAAGGAACGA | GTACTGGGTGTCCATGTAGTT |
| AREG | TGGATTGGACCTCAATGACA | ACTGTGGTCCCCAGAAAATG |
| Angiopoietin | GCAAATGTGCCCTCATGTTA | TAGATTGGAGGGGCCACA |
| Bax | GCCCTTTTGCTTCAGGGTTT | TCCAATGTCCAGCCCATGAT |
| EGFR | GTGACCGTTTGGGAGTTGATGA | GGCTGAGGGAGGCGTTCTC |
| ETS-1 | AAACTTGCTACCATCCCGTACGT | ATGGTGAGAGTCGGCTTGAGAT |
| p21 | GCAGACCAGCATGACAGATTT | GCAGACCAGCATGACAGATTT |
| VDR | CTGACCCTGGAGACTTTGAC | GGATTAGGGCTTCCTCTTGGA |
| ITGA2 | AACTCTTTGGATTTGCGTGTG | TGGCAGTCTCAGAATAGGCTTC |
| DAB2 | GGGCATTTGGTTACGTGTG | CTTTGCTGGCTTCCTCTATC |
| ITGA2B | AGGTGAGAGGGAGCAGAACA | TCCACCTTGAGAGGGTTGAC |
| ITGB3 | GTGACCTGAAGGAGAATCTGC | TTCTTCGAATCATCTGGCC |
| ENC1 | CCCCACAATCAACAAATGG | ACTACTGCGGCGTTGCTAA |
| MAOA | CCTTGACTGCCAAGATTCACTTC | TGCACTTAATGACAGCTCCCAT |
| Synapsin I | GAACACATGCAAGGAGCTGA | AGAGCACCAGGTTCAGGAAG |
| P2X5 | TCTTTGCCTGGTGCCCGTTG | ATCACGGAGCCCAGTCGGAAG |
| γ-Globin | GGACAAGGCTACTATCACAA | CAGTGGTATCTGGAGGACAG |
ChIP assays were performed as described previously [20]. Primers for the amplification of WT1-binding regions were designed based on the work of others: renin [21], VDR (vitamin D receptor) [22], ETS-1 [23] and amphiregulin [23,24]. Sequences of the primers are shown in Table 2. Control primers amplified a region 3′ of the human c-Myc gene.
Table 2. Primers used for ChIP analysis.
AREG, gene encoding amphiregulin.
| Gene | Forward (5′→3′) | Reverse (5′→3′) |
|---|---|---|
| Renin | GCTTAACCTCCTAGGTCTTG | GTGGAGGAAGTCTGTAAATC |
| VDR | CACCTGGCTCAGGCGTCC | GCCAGGAGCTCCGTTGGC |
| ETS-1 | CCTAAAGAGGAGGGGAGAGC | AGGGGAAGTTGGCACTTTG |
| AREG | TTTAAGTTCCACTTCCTCTCA | GGTGTGCGAACGTCTGTA |
| c-Myc | AGAACTGCTAAACCAGAATGT | AGACAGGGTCTCACCATCT |
Affymetrix microarray and analysis
Stably transfected V-K562 and B-K562 cells were seeded at equal densities (approx. 1×105 cells/ml) in RPMI 1640 medium supplemented with 10% FBS, 100 mg/ml streptomycin, 100 units/ml penicillin and 2 mM L-glutamine. After 24 h, total RNA was extracted or cells were treated with 100 nM PMA for a further 48 h to induce differentiation prior to RNA extraction. RNA was isolated using the Qiagen RNeasy kit then treated with DNase I (Roche) prior to analysis by microarray. For each condition, microarray analysis was performed on three independently prepared RNA samples. Affymetrix microarray analysis was performed using the human genome U133 Plus 2.0 array, following the manufacturer's guidelines and results were submitted in a MIAME-compliant standard to the Array Express database (Experiment E-MEXP-2573, http://www.ebi.ac.uk/microarray-as/ae/).
Technical quality control was performed with dChip (V2005) (http://www.dchip.org; [26]) using the default settings. Background correction, quantile normalization and gene expression analysis were performed using GCRMA in Bioconductor [27]. Differential expression analysis was performed using Limma using the functions lmFit and eBayes [28]. Gene lists of differentially expressed genes were controlled for FDR (false discovery rate) errors using the method of QVALUE [29].
Where RNA expression was studied from vehicle or PMA-treated V-K562 or B-K562 cells (independently prepared triplicates), a list of significant gene expression changes was generated by filtering for the ANOVA interaction (q-value<0.05) and fold change between differentiated B-K562 compared with V-K562>±2. This gave a genelist of 788 probesets. This set was segregated into five clusters based on similarity of expression profile across the dataset with a k-means clustering algorithm using a Manhattan distance metric. Clustering was performed on the means of each sample group (log 2) that had been z-transformed (for each probe set the mean was set to 0, and the S.D. was set to 1). k-means clustering was performed on the basis of similarity of profiles across the dataset using the ‘Super Grouper’ plugin of maxdView software (available from http://bioinf.man.ac.uk/microarray/maxd/). Each cluster from k-means clustering was sorted further by hierarchical clustering.
Functional annotation of the genes was performed using DAVID version 2 (http://david.niaid.nih.gov/david/version2/index.htm).
Calcium imaging
PMA-treated V-K562 or B-K562 cells were washed with Tyrode's solution [140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 3 mM CaCl2, 10 mM Hepes, 10 mM glucose and 1 mM pyruvic acid (pH 7.4)] for 5 min and then loaded with 2 μM fura 2/AM (fura 2 acetoxymethyl ester; Molecular Probes, Invitrogen) containing the non-ionic dispersing agent Pluronic F-127 (Molecular Probes, Invitrogen) for 40 min. Loaded cells were visualized using an Olympus IX71 microscope with a 40× oil-immersion lens, and images were captured using a SensiCam QE camera (Cooke Corporation). Excitation wavelengths of 340 nm and 380 nm were used with an emission wavelength of 510 nm. Images were collected every 4 s using Imaging Workbench 5.2 (Indec Biosystems). During experiments, cells were kept under constant perfusion with Tyrode's solution followed by alternating changes in solutions. All solutions were applied to the bath using a gravity flow perfusion system (AutoMate Scientific) and laminar flow perfusion chambers (RC-25F, Warner Scientific). Imaging data were collected as a ratio of fluorescence intensities. All fluorescence values were calibrated using the fura 2/AM Calcium Imaging Calibration kit (Invitrogen). The effective dissociation constant Kd was calculated to be 250 nM and calcium concentrations were determined using the formula outlined in [30]. An evoked response was defined as an increase in fluorescence that was greater than 2 S.D. values above baseline. Data were graphed without any filtering using OriginPro 7.5 software. Statistical comparisons were made using a Fisher's exact test (http://www.langsrud.com/stat/index.html) with the limit of significance set at P<0.05. Cells were stimulated with either 50 mM KCl, 10 mM ATP (Sigma) or 10 mM CPA (cyclopiazonic acid; Sigma).
RESULTS
A cell-line model system to study WT1/BASP1 function
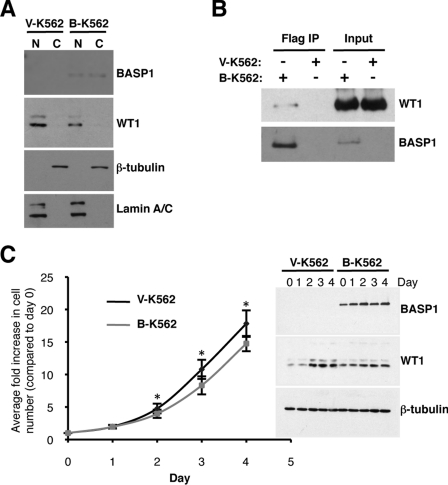
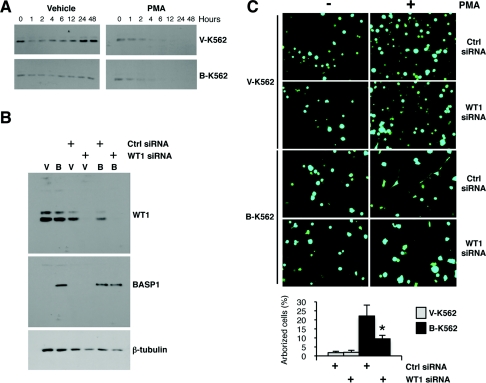
Our previous work found that K562 cells, which are derived from a myelogenous leukaemia, express WT1 but do not express detectable BASP1 [9]. Indeed, in these cells WT1 elicited transactivation that could be repressed by the transient expression of BASP1. We therefore generated a stable K562 cell-line derivative that expresses BASP1. The BASP1-coding region was cloned into the pcDNA3 vector in-frame with tandem FLAG and HA epitope tags fused to the C-terminus of BASP1. pCDNA3-BASP1-FLAG-HA or a control empty pcDNA3-FLAG-HA vector were transfected into K562 cells and cultured in the presence of G418 to select cells that express the neomycin-resistance gene, which is also located on the pCDNA3 plasmid. Following selection, the cells were used to prepare nuclear and cytoplasmic extracts, and the samples were immunoblotted with anti-WT1 or anti-BASP1 antibodies (Figure 1A). WT1 was expressed in both the vector-derived and BASP1-expressing K562 cells (designated V-K562 and B-K562 respectively) and was primarily present in the nuclear fractions. BASP1 was located in both the nucleus and cytoplasm of the B-K562 cells, but was not expressed in the V-K562 cells. To confirm the efficiency of the nuclear/cytoplasmic fractionation, immunoblots were also performed to detect lamin A/C and β-tubulin as nuclear and cytoplasmic markers respectively.
Figure 1. Characterization of a K562 cell-line derivative that expresses BASP1.
(A) V-K562 and B-K562 cells were used to prepare nuclear (N) and cytoplasmic (C) extracts and immunoblotting was performed with the antibodies indicated. (B) Anti-FLAG antibodies were used in immunoprecipitation (IP) with nuclear extracts prepared from V-K562 and B-K562 cells. The samples were then immunoblotted with anti-WT1 or anti-BASP1 antibodies. (C) An equal number of V-K562 and B-K562 cells were grown over a 4 day period and each day the cells were counted. The y-axis of the graph shows the fold increase in cell number during the period. Immunoblots of WT1, BASP1 and β-tubulin are shown in the right-hand panels. Values are means±S.D. for six independent experiments. P<0.01 by Student's t test.
We next prepared whole-cell extracts from V-K562 and B-K562 cells, and subjected them to immunoprecipitation with anti-FLAG antibodies to purify BASP1. The precipitates were then immunoblotted with either anti-WT1 or anti-BASP1 antibodies (Figure 1B). Consistent with our previous studies, the endogenous WT1 in B-K562 cells co-immunoprecipitated with anti-FLAG antibodies only with extracts prepared from B-K562 cells [8].
As mentioned above, WT1 can act as an oncogene in leukaemic cells, whereas BASP1 is associated with tumour suppressor activity. We therefore monitored the duplication rate of V-K562 and B-K562 cells over a period of 4 days and plotted the results as average fold increase in cell number (Figure 1C). The B-K562 cells proliferated at a small, but significantly lower, rate than the V-K562 cells, suggesting that BASP1 suppresses the growth of K562 cells.
The regulation of transcription by WT1 and BASP1 in K562 cells
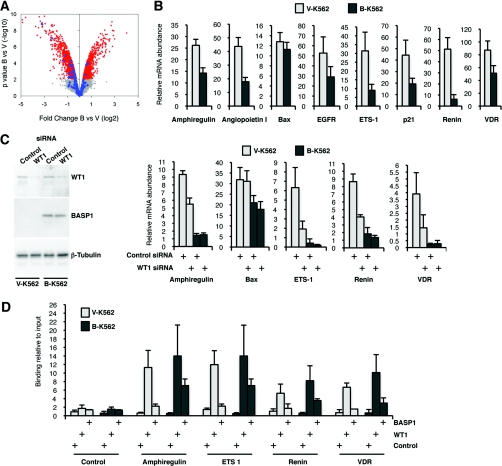
We next performed an Affymetrix expression array with mRNA derived from V-K562 and B-K562 cells. The data are presented as a volcano plot in Figure 2(A). The grey area of the plot indicates small changes with low significance, whereas the red area indicates significant changes (q-value<0.1). In total, 1308 genes were up-regulated and 1376 genes were down-regulated in the B-K562 cells compared with the V-K562 cells (1706 and 1683 probesets respectively; full details are in Supplementary Table S1 at http://www.BiochemJ.org/bj/435/bj4350113add.htm). Genes previously described as WT1 targets are highlighted in blue in Figure 2(A) (full details are in Supplementary Table S2 at http://www.BiochemJ.org/bj/435/bj4350113add.htm). In the set of differentially expressed genes with q-value <0.1, the WT1 target genes showed significant down-regulation (19 genes were repressed, compared with three that showed increased activity, Fisher's exact test P= 0.006). Thus, while the overall expression changes caused by BASP1 included a similar number of up- and down-regulated genes, 19 out of the 22 WT1 target genes on the array that exhibited altered expression in the presence of BASP1 were transcriptionally repressed.
Figure 2. Expression of BASP1 in K562 cells leads to the general down-regulation of WT1 target genes.
(A) Microarray analysis was performed with B-K562 cells and V-K562 cells and the data were analysed as described in the text. A volcano plot shows that, even though B-K562 cells show an equivalent number of repressed and activated genes, the vast majority of altered WT1 target genes are repressed. Signal within the grey area indicates small changes below the level of significance, whereas the red area shows significant changes. The known WT1 target changes are indicated in blue. Full details of the expression changes observed in V-K562 and B-K562 cells are shown in Supplementary Table S1 (at http://www.BiochemJ.org/bj/435/bj4350113add.htm). The probesets representing the WT1 target genes are shown in Supplementary Table S2 at http://www.BiochemJ.org/bj/435/bj4350113add.htm. Sources of the WT1 target genes are cited throughout the text and also include genomic analysis and reviews [2,25,42,43]. (B) qPCR was performed to quantify the expression of the genes indicated. Results are presented as the expression relative to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA. Values are means±S.D. for three independent experiments. (C) V-K562 and B-K562 cells were transfected with control siRNA or WT1 siRNA and 48 h later whole-cell extracts or RNA were prepared. Samples were immunoblotted with anti-WT1, anti-BASP1 or anti-β-tubulin antibodies. The right-hand panels show the expression of the indicated genes as determined by qPCR. Values are means±S.D. for three independent experiments. (D) V-K562 and B-K562 cells were subject to ChIP analysis with either control, anti-WT1 or anti-BASP1 antibodies. qPCR was used to amplify the WT1-binding sites of the indicated genes or a control region. The results are expressed as the percentage of bound chromatin compared with input. Values are means±S.D. for three independent experiments.
The efficacy of the array data was confirmed by the direct measurement of several WT1 target genes by qPCR (quantitative PCR) (Figure 2B). Bax was included as a control because its expression did not significantly change in the array and it has not been proposed as a target gene of WT1. Thus the introduction of BASP1 into K562 cells leads to the repression of a significant proportion of previously identified WT1 target genes.
We next sought to confirm that the expression of a group of the above genes was indeed WT1-dependent in K562 cells. V-K562 or B-K562 cells were transfected with WT1 siRNA or a control siRNA and, 48 h later, RNA was extracted and cDNA was prepared. Whole-cell extracts were also immunoblotted with anti-WT1 antibodies to confirm that WT1 was down-regulated by the transfection of WT1 siRNA (Figure 2C). qPCR was used to determine the expression of four of the genes identified in the array and also Bax as a control. Transfection of V-K562 cells with WT1 siRNA resulted in the down-regulation of amphiregulin, ETS-1, renin and VDR, consistent with transcriptional activation of these genes by WT1 in the absence of BASP1. In contrast, the expression of Bax was not affected by WT1 depletion. As above, in the B-K562 cells, the expression of the WT1 target genes was down-regulated when compared with the V-K562 cells. Moreover, consistent with the BASP1-mediated suppression of WT1 transcription function, the depletion of WT1 in B-K562 cells had little effect on the expression of amphiregulin, ETS-1, renin or VDR.
Our results so far suggest that BASP1 suppresses the activation of several WT1 target genes in K562 cells. We next performed ChIP analysis of V-K562 and B-K562 cells to determine the occupancy of the WT1-binding site regions of a selection of the above genes by WT1 and BASP1 (Figure 2D). ChIP was performed with control, anti-WT1 or anti-BASP1 antibodies using fragmented cross-linked chromatin prepared from either V-K562 or B-K562 cells. The WT1-binding regions of the amphiregulin, ETS-1, renin and VDR genes were amplified, along with a control genomic region. WT1 was present at the promoter regions of all of the genes tested, but not at the control site, in both V-K562 and B-K562 cells. Consistent with the lack of BASP1 expression, anti-BASP1 antibodies did not significantly precipitate the WT1-binding regions of any of the target genes using fragmented chromatin prepared from V-K562 cells. However, analysis of the fragmented chromatin prepared from B-K562 cells revealed that BASP1 was present within the WT1-binding regions of all of the target genes tested. Thus BASP1-mediated suppression of WT1 transcriptional activation coincides with the recruitment of BASP1 to promoter-bound WT1. Taken together, the results shown in Figure 2 suggest that BASP1 is a negative regulator of WT1 transcriptional activation at a significant proportion of WT1 target genes and acts via recruitment to WT1-bound promoters.
Diversion of K562 cell differentiation by WT1 and BASP1
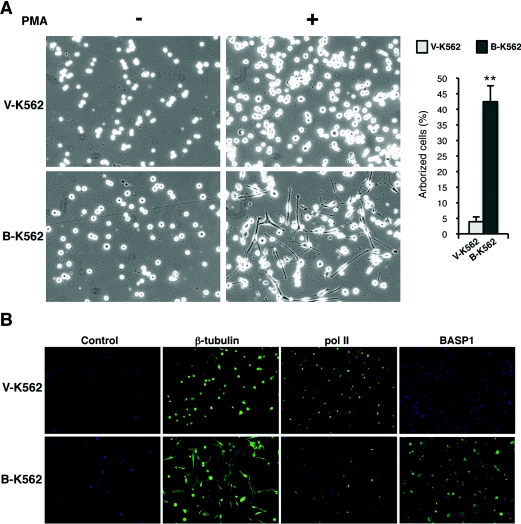
K562 cells can be induced to differentiate along blood cell pathway lineages by various agents [31]. We therefore tested the effect of BASP1 expression on the K562 differentiation pathway induced by PMA. Treatment of K562 cells with 100 nM PMA causes the cells to differentiate along the megakaryocyte lineage. Surprisingly, however, even though the B-K562 cells appeared morphologically similar to the V-K562 cells before PMA treatment, the effect following PMA treatment was dramatic (Figure 3A). Whereas PMA treatment of V-K562 cells induced the formation of megakaryocytes, the PMA-treated B-K562 cells became elongated and formed processes. Quantification of the arborized cells in the differentiated V-K562 and B-K562 pools is shown on the right-hand side of Figure 3(A). This quantification included any cells that formed a process, but even the small number observed in the V-K562 cells did not form processes as elaborate as those observed in the B-K562 cells. The same effects were observed in a different, independently produced, stable BASP1-expressing K562 cell line (results not shown and Figure 4). Immunofluorescence was used to detect β-tubulin in V-K562 and B-K562 cells that had been treated with PMA for 4 days (Figure 3B). β-Tubulin staining clearly showed the extent of arborization of the B-K562 cells, whereas the V-K562 cells maintain the rounded shape typical of megakaryocytes. BASP1 expression, restricted to the B-K562 cells, shows a nuclear/cytoplasmic distribution comparable with that observed by us previously in podocyte and Cos-1 cells [9,13]. Immunofluorescence was also used to detect pol II as a nuclear marker in differentiated V-K562 and B-K562 cells. It is evident from Figure 3 that the PMA-treated B-K562 cells have not differentiated along the megakaryocyte pathway and have instead gained morphology akin to neuronal cells or fibroblasts.
Figure 3. Expression of BASP1 induces an altered PMA-dependent differentiation programme.
(A) V-K562 cells or B-K562 cells were treated with 100 nM PMA (+) or vehicle (−) and 48 h later the cells were photographed. The right-hand panel shows quantification of the percentage of arborized cells. Values are means±S.D. for six independent experiments (P<0.005 by Student's t test). (B) Immunofluorescence of V-K562 and B-K562 cells that had been treated with PMA as in (A), but for 4 days. Anti-β-tubulin, anti-pol II and anti-BASP1 antibodies were used as indicated. The control is rabbit IgG.
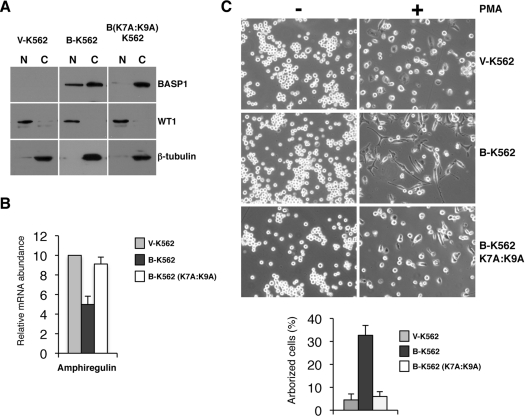
Figure 4. Nuclear BASP1 is required for the altered PMA-dependent differentiation programme of B-K562 cells.
(A) New pools of V-K562, B-K562 and B-K562 (K7A:K9A) cells were used to prepare nuclear (N) and cytoplasmic (C) extracts and immunoblotting was performed with the antibodies indicated. (B) RNA was prepared from V-K562, B-K562 and B-K562 (K7A:K9A) cells and qPCR was performed to quantify the expression of the amphiregulin gene. Data are presented as expression relative to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA. Values are means±S.D. for three independent experiments. (C) V-K562, B-K562 and B-K562 (K7A:K9A) cells were treated with PMA as described in Figure 3(A).
As mentioned above, BASP1 contains a bipartite NLS at its N-terminus. We therefore generated a mutant derivative of BASP1 (K7A:K9A) to disrupt the NLS and generated stable K562 cell lines with either an empty pCDNA expression vector or the same vector driving expression of either wild-type BASP1 or BASP1 K7A:K9A. Nuclear and cytoplasmic extracts of the cell-line derivatives were prepared and immunoblotted with anti-BASP1, anti-WT1 or anti-β-tubulin antibodies (Figure 4A). Mutation of the BASP1 NLS resulted in a significant reduction of nuclear BASP1. The expression of amphiregulin in each of the cell-line derivatives was then determined by RNA preparation and qPCR. The results show that, as before, expression of wild-type BASP1 resulted in the repression of amphiregulin expression (Figure 4B). In contrast, the BASP1 mutant derivative K6A:K9A did not drive repression of amphiregulin expression. These results are consistent with the requirement for nuclear localization of BASP1 to act as a WT1 transcriptional co-suppressor. We next analysed the effect of PMA on the differentiation of V-K562, B-K562 or B-K562 (K7A:K9A) cells (Figure 4C). The results demonstrate that, although wild-type BASP1 diverted the differentiation programme of K562 cells to an arborized phenotype, PMA treatment of BASP1 (K7A:K9A) cells did not.
Differentiation of K562 cells has been reported to be accompanied by a reduction in WT1 expression [13]. We therefore determined the fate of WT1 in the V-K562 and B-K562 cells following treatment with PMA. V-K562 or B-K562 cells were exposed to PMA as described above and whole-cell extracts were prepared at intervals up to 48 h. Proteins were resolved by SDS/PAGE, transferred to membrane and immunoblotted with anti-WT1 antibodies (Figure 5A). Consistent with previous reports [13], WT1 was down-regulated following treatment of both the V-K562 and B-K562 cells with PMA, and by 12 h was almost undetectable. This raised the question of whether or not the altered B-K562 cell differentiation programme was dependent on WT1. To address this, B-K562 cells or V-K562 cells were transfected with WT1 siRNA or a control siRNA. Immunoblotting confirmed that the WT1 siRNA caused a significant reduction in WT1 protein (Figure 5B). Cells transfected with control siRNA or WT1 siRNA along with a plasmid driving expression of GFP were then induced to differentiate with PMA for 48 h followed by imaging to detect the transfected GFP-expressing cells (Figure 5C). As above, the PMA-treated B-K562 cells showed a marked difference in their appearance when compared with the V-K562 cells. However, the transfection of B-K562 cells with WT1 siRNA significantly reduced the differentiation-dependent arborized morphology that is induced by PMA in the B-K562 cells. Quantification of the arborized cells is shown below. Thus WT1 expression is required for the altered differentiation programme of B-K562 cells that occurs in response to PMA treatment.
Figure 5. WT1 is required for the altered PMA-dependent differentiation programme of B-K562 cells.
(A) V-K562 or B-K562 cells were treated with 100 nM PMA for the times indicated and then whole-cell extracts were prepared and subjected to immunoblotting with anti-WT1 antibodies. (B) V-K562 and B-K562 cells were transfected with either control or WT1 siRNA. At 24 h later, whole-cell extracts were prepared and immunoblotted with the antibodies indicated. (C) V-K562 or B-K562 cells were transfected as described in (B) along with a vector driving expression of GFP, and 24 h later were treated with 100 nM PMA or vehicle control. At 48 h later, the cells were imaged. Quantification of the arborized cells is shown in the lower panel. Values are means±S.D. for six experiments (P<0.05 by Student's t test).
The regulation of gene expression by WT1 and BASP1 in differentiating K562 cells
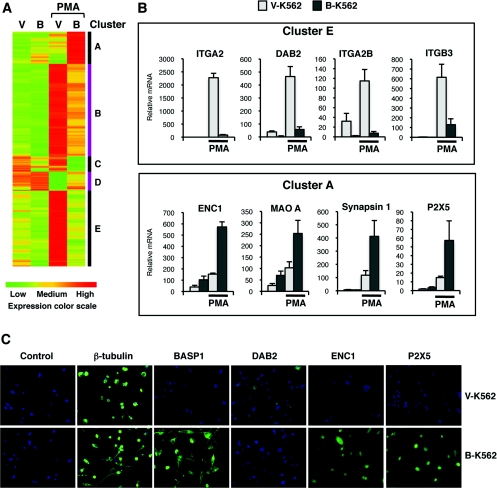
We next explored the gene-regulatory events that result in the WT1/BASP1 reprogramming of the PMA-dependent differentiation pathway in K562 cells. Arrays were performed with RNA derived from vehicle-treated V-K562 or B-K562 cells, and also the same cells treated with PMA (independently prepared triplicates). A list of significant gene expression changes was generated by filtering for the ANOVA interaction (q-value<0.05) and fold change between differentiated B-K562 compared with V-K562 >± 2. This gave 788 probesets, which were grouped into five clusters based on their expression profiles over the four conditions as shown in Figure 6(A) (full details are in Supplementary Table S3 at http://www.BiochemJ.org/bj/435/bj4350113add.htm). The changes in gene expression fall into five clusters (A–E). Cluster E genes are highly induced in the PMA-treated V-K562 cells, but not by PMA-treatment of B-K562 cells. This group includes ITGA2 (integrin α2, CD49B), ITGA2B (integrin α2b, CD41), ITGB3 (integrin β3, CD61), PPBP (pro-platelet basic protein), CD2226, CD84, Arhgap9 (Rho GTPase-activating protein 9) and DAB2, all of which are well-characterized genes that are involved in the differentiation of K562 cells along a megakaryocyte programme [31,32]. In Figure 6(B), qPCR validation of the array data for the megakaryocyte marker genes ITGA2, DAB2, ITGA2B and ITGB3 is shown, confirming the induction of these genes in PMA-treated V-K562 cells, but not PMA-treated B-K562 cells. Thus the expression data can explain why the B-K562 cells do not differentiate into megakaryocyte cells upon exposure to PMA.
Figure 6. Differentiation-dependent regulation of gene expression in V-K562 and B-K562 cells.
(A) Arrays were performed with RNA derived from vehicle-treated V-K562 or B-K562 cells and also the same cells treated with PMA. The data were analysed as described in the text. V, V-K562 cells; B, B-K562 cells. A colour-coded key of expression level is shown below the panel. Five distinct clusters of genes (A–E) are indicated on the right. Supplementary Table S3 (at http://www.BiochemJ.org/bj/435/bj4350113add.htm) shows the genes associated with each cluster. (B) qPCR analysis of selected genes from clusters E (top panels) and A (bottom panels). Values are means±S.D for three independent experiments. (C) Immunofluorescence was performed with V-K562 and B-K562 cells that had been treated with PMA and allowed to differentiate for 4 days. Antibodies are indicated above the panels. The control is rabbit IgG.
Cluster A contains genes that are up-regulated in B-K562, but significantly less so in V-K562 cells following PMA treatment. These genes include ENC1, Pak1 (p21-activated kinase 1) and Nidogen2, which have been shown to play a role in axon growth (qPCR for ENC1 shown in Figure 6B, lower panel; [33–35]). In addition, cluster A contains synapsin I, P2X5 and MAOA (monoamine oxidase A; qPCR shown in Figure 6B), which all play a central role in synapse function [36,37]. Figure 6(C) shows immunofluorescence with anti-β-tubulin (cytoplasm marker), anti-BASP1, anti-DAB2 (megakaryocyte marker), anti-ENC1 and anti-P2X5 antibodies, confirming that the changes in mRNA are accompanied by changes in protein. Specifically, the results show that DAB2 expression is restricted to the differentiated V-K562 cells, whereas ENC1 and P2X5 expression are elevated in the differentiated B-K562 cells. Thus the expression of BASP1 in K562 cells diverts the PMA-induced differentiation programme away from the megakaryocyte pathway, and instead leads to a morphological appearance akin to neuronal cells. The altered differentiation pathway of B-K562 cells requires the expression of WT1, which is accompanied by the suppression of genes that specify the megakaryocyte and, furthermore, leads to the induction of several genes involved in axon growth and synapse function.
Neuronal properties of B-K562 cells
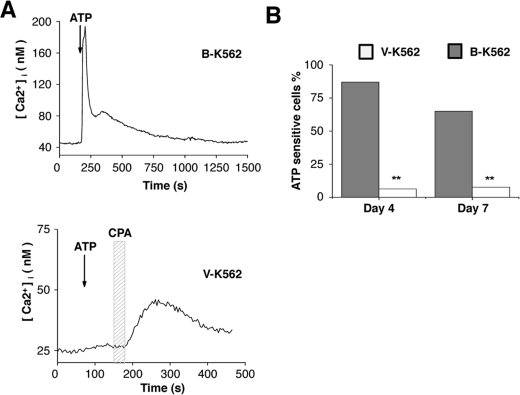
The neurotransmitter receptor P2X5 was significantly up-regulated in B-K562 cells following PMA treatment (Figure 6B). P2X5 is a ligand-gated purinergic receptor that responds to extracellular ATP and is found in both neurons and muscle cells [36]. Therefore we next determined whether the B-K562 cells possess the capacity to elicit a response to ATP. V-K562 or B-K562 cells were induced to differentiate by treatment with PMA for 4 days. ATP (10 mM) was then applied to the cells and calcium influx was measured by fluorescence imaging. The treatment of differentiated B-K562 cells with ATP caused a rapid increase in cytosolic calcium that slowly returned to baseline calcium levels in 87% of cells tested (example shown in Figure 7A, upper panel). The ATP response in B-K562 cells was abolished when external calcium was removed, indicating that the calcium signal was due to an influx from the external environment (results not shown). Experiments using B-K562 cells that had been treated with PMA for 7 days yielded similar results (shown graphically in Figure 7B). In contrast, similar treatment of the differentiated V-K562 cells revealed that only 10% of the cells were sensitive to ATP, even when we increased the ATP concentration to 500 mM (example shown in Figure 7A, lower panel, and quantification is shown in Figure 7B). However, application of the reversible SERCA (sarcoplasmic/endoplasmic reticulum Ca2+-ATPase) inhibitor CPA elicited a response, confirming that the V-K562 cells are viable and are specifically unresponsive to ATP (Figure 7A, lower panel). Statistical analysis of the number of ATP-sensitive cells found that B-K562 cells were significantly more responsive to ATP compared with the V-K562 cells (P<0.005). Thus the expression of synaptic proteins in the PMA-induced B-K562 cells is accompanied by the endowment of the cells with the capacity to respond to the neurotransmitter ATP.
Figure 7. PMA-differentiated B-K562 cells are responsive to the neurotransmitter ATP.
(A) A 30 s application of 10 mM ATP elicited a large increase in cytosolic calcium in B-K562 cells that slowly returned to baseline levels as ATP was washed out. (B) A comparable ATP application in V-K562 cells did not affect baseline calcium levels. Application of 10 mM CPA, a reversible SERCA pump inhibitor, illustrates that it was possible to evoke a measurable change in cytosolic calcium in these cells. (C) Analysis of ATP responsiveness in B-K562 and V-K562 cells on days 4 and 7 after PMA-induced differentiation in culture found that B-K562 cells were significantly more responsive to ATP than the V-K562 cells on both days 4 (B-K562, n=54; V-K562, n=16) and 7 (B-K562, n=20; V-K562, n=39). **P<0.005.
Specificity of differentiation reprogramming of B-K562 cells
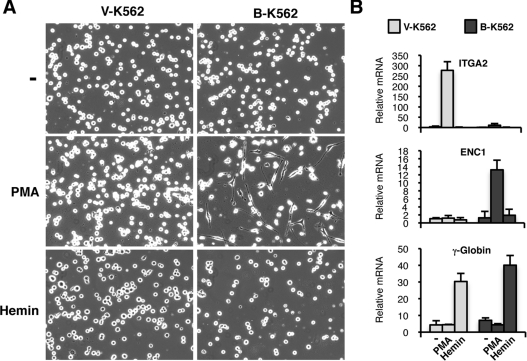
K562 cells are situated at the common progenitor stage of the megakaryocyte and erythroid lineages of haemopoietic stem cell differentiation [31]. Whereas PMA induces K562 cells to differentiate along a megakaryocyte programme, haemin can induce K562 cells to differentiate along an erythroid programme. We therefore treated the V-K562 and B-K562 cells with haemin or PMA, and 48 h later, imaged the cells under phase microscopy (Figure 8A). As before, PMA treatment induced the arborization of the B-K562 cells, but not the V-K562 cells. However, following treatment with haemin, the morphology of the B-K562 cells was similar to that of the V-K562 cells. To confirm that the induction of megakaryocyte and erythrocyte differentiation was successful, RNA was prepared and cDNA was derived, then qPCR was performed to analyse the expression of ITGA2 (megakaryocyte marker), ENC1 (induced by PMA in B-K562 cells and involved in axon growth) and γ-globin (a standard marker for erythroid differentiation). As above, ITGA2 was specifically induced by PMA only in the V-K562 cells, and ENC1 was induced by PMA only in the B-K562 cells (Figure 8B). The marker for erythroid differentiation, γ-globin, was induced only by haemin, but this was observed in both the V-K562 and B-K562 cells. These results suggest that the erythroid differentiation pathway of the K562 cells is, at least in part, intact in the B-K562 cells. Thus the expression of BASP1 specifically results in the reprogramming of the megakaryocyte differentiation pathway of K562 cells that is induced by PMA.
Figure 8. BASP1 specifically diverts the megakaryocyte pathway of K562 cell differentiation.
(A) V-K562 and B-K562 cells were mock-treated or treated with 100 nM PMA or 0.5 mM haemin, and 48 h later the cells were imaged using phase microscopy. (B) As in (A), except that RNA was prepared and analysed by qPCR for the expression of the genes indicated. Values are means±S.D. for three independent experiments.
DISCUSSION
In the present study we have demonstrated that the stable expression of BASP1 in K562 cells leads to the down-regulation of a significant proportion of WT1-dependent target genes. ChIP analysis confirmed that the suppression of WT1-mediated transcriptional activation by BASP1 coincides with the recruitment of BASP1 to WT1-dependent promoters. Taken together, the results of the present study suggest that BASP1 is a general regulator of WT1 that acts to suppress the transcriptional activation function of WT1. BASP1 expression in K562 cells also led to the activation and repression of several other genes, the majority of which are unlikely to be direct targets of WT1. Although this will include indirect effects, it is also possible that BASP1 may act through other transcription factors either as a co-repressor or co-activator.
PMA-induced differentiation of K562 cells into megakaryocytes was blocked by the expression of BASP1. Indeed, several markers of megakaryocyte differentiation failed to up-regulate in response to PMA, presumably due to the altered gene expression profile that was established by the expression of BASP1. Instead, the K562 cells that express BASP1 gained a neuronal-like morphology following treatment with PMA that was accompanied by the induction of several genes that are involved in axon extension and synapse function. The PMA-induced differentiation of K562 cells to megakaryocytes is well established and has been studied for several years. One study has reported that K562 cells can differentiate, albeit at a low efficiency, to antigen-presenting dendritic cells. However, none of the dendritic cell markers (e.g. MHC class I and class II, interleukin-12, CD86 and CD40) were induced in the B-K562 cells, suggesting that this particular pathway is not induced by WT1 and BASP1 [38]. The PMA-treated B-K562 cells exhibited calcium influx when exposed to the neurotransmitter ATP, suggesting a gain-of-function consistent with an excitable cell. To our knowledge, this is the first report that K562 cells have been differentiated into anything other than a blood cell.
BASP1 can induce arborization of PC12E2 neuronal cells or primary hippocampal neurons [39]. Moreover, WT1 has been shown to be required for the neuronal differentiation of Y-79 retinoblastoma cells in culture [40], formation of the retinal ganglia and olfactory epithelia [4,5], and is expressed in several other neuronal cells [6]. However, the results of the present study demonstrate that WT1 and BASP1 can induce neuronal-like morphology and physiological properties in cells that are not originally from a neuronal lineage. Thus WT1 and BASP1 together can reprogramme the differentiation potential of K562 cells.
WT1 and BASP1 are co-expressed in the podocyte cells of the developing and adult kidney, which form processes that surround the capillary network of the glomerulus and provide a filtration barrier [9]. Our previous studies suggested that WT1 and BASP1 co-operate in the changes in gene regulation that occur during the differentiation of podocytes in vitro [13]. It is therefore interesting to note that parallels between the formation of processes in podocytes and neurons have been drawn, particularly regarding the formation of cellular extensions, which are microtubule-rich with actin-based projections [41]. Moreover, several genes involved in intracellular trafficking, signal transduction and cytoskeletal components are co-ordinately expressed in both cell types. It is therefore possible that WT1 and BASP1 play an important role in establishing the transcription networks associated with cellular arborization in podocytes and some neuronal cells. In contrast with the events in podocytes, however, the differentiation of K562 cells is accompanied by a reduction in WT1 expression. This suggests that the neuronal characteristics that we report in the present paper require the transient function of WT1, and that WT1 is probably not needed for the maintenance of the differentiated phenotype.
It is interesting to note that BASP1 expression does not appear to affect the haemin-induced differentiation of K562 cells along the erythrocyte pathway, at least morphologically or in the induction of γ-globin expression. Thus WT1 and BASP1 expression do not fix the destiny of K562 cells to a neuronal-like lineage. However, the alterations set the stage for further changes that are induced by PMA, but require the prior modifications that have been induced by WT1 and BASP1. The precise chain of events that leads from WT1/BASP1 regulation of transcription to the reprogramming of K562 differentiation induced by PMA is likely to be complex. Indeed several cell types, including others that are blood-cell-derived, express WT1 and BASP1. We suggest that the action of WT1 and BASP1 on specific target genes results in changes in the cellular milieu that, under specific conditions, can lead to arborization. In summary, the results of the present study suggest that WT1 and BASP1 can co-operatively act at fundamental step(s) in determining the differentiation pathway.
Online data
AUTHOR CONTRIBUTION
Sarah Goodfellow and Stefan Roberts conceived the study. Sarah Goodfellow, Kathryn Medler and Stefan Roberts designed the experiments. Sarah Goodfellow, Michelle Rebello, Eneda Toska, Sean Rudd and Stefan Roberts performed the experiments. Leo Zeef analysed and presented the microarray data. Sarah Goodfellow, Kathryn Medler and Stefan Roberts interpreted the data and wrote the paper.
ACKNOWLEDGEMENTS
We thank Sue Kimber for helpful discussions. We also thank Wensheng Deng, Paul Shore and members of the Roberts laboratory for comments on the manuscript prior to submission.
FUNDING
This work was funded by Cancer Research UK [grant number C1356/A6630 (to S.G.E.R.)]; the Wellcome Trust [grant number WT061207/Z/00/A (to S.G.E.R.)]; and the National Science Foundation [NSF0917893 (to K.F.M.)].
References
- 1.Rivera M. N., Haber D. A. Wilms' tumour: connecting tumorigenesis and organ development in the kidney. Nat. Rev. Cancer. 2005;5:699–712. doi: 10.1038/nrc1696. [DOI] [PubMed] [Google Scholar]
- 2.Yang L., Han Y., Suarez Saiz F., Minden M. D. A tumor suppressor and oncogene: the WT1 story. Leukemia. 2007;21:868–876. doi: 10.1038/sj.leu.2404624. [DOI] [PubMed] [Google Scholar]
- 3.Hohenstein P., Hastie N. D. The many facets of the Wilms' tumour gene, WT1. Hum. Mol. Genet. 2006;15:R196–R201. doi: 10.1093/hmg/ddl196. [DOI] [PubMed] [Google Scholar]
- 4.Wagner K. D., Wagner N., Vidal V. P., Schley G., Wilhelm D., Schedl A., Englert C., Scholz H. The Wilms' tumor gene Wt1 is required for normal development of the retina. EMBO J. 2002;21:1398–1405. doi: 10.1093/emboj/21.6.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner N., Wagner K. D., Hammes A., Kirschner K. M., Vidal V. P., Schedl A., Scholz H. A splice variant of the Wilms' tumour suppressor Wt1 is required for normal development of the olfactory system. Development. 2005;132:1327–1336. doi: 10.1242/dev.01682. [DOI] [PubMed] [Google Scholar]
- 6.Armstrong J. F., Pritchard-Jones K., Bickmore W. A., Hastie N. D., Bard J. B. The expression of the Wilms' tumour gene, WT1, in the developing mammalian embryo. Mech. Dev. 1993;40:85–97. doi: 10.1016/0925-4773(93)90090-k. [DOI] [PubMed] [Google Scholar]
- 7.Roberts S. G. E. Transcriptional regulation by WT1 in development. Curr. Opin. Genet. Dev. 2005;15:542–547. doi: 10.1016/j.gde.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 8.McKay L. M., Carpenter B., Roberts S. G. E. Regulation of the Wilms' tumour suppressor protein transcriptional activation domain. Oncogene. 1999;18:6546–6554. doi: 10.1038/sj.onc.1203046. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter B., Hill K. J., Charalambous M., Wagner K. J., Lahiri D., James D. I., Anderson J. S., Schumacher V., Royer-Pokora B., Mann M., et al. BASP1 is a transcriptional cosuppressor for the Wilms' tumour suppressor protein WT1. Mol. Cell. Biol. 2004;24:537–549. doi: 10.1128/MCB.24.2.537-549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartkamp J., Carpenter B., Roberts S. G. E. The Wilms' tumour suppressor protein WT1 is processed by the serine protease HtrA2/Omi. Mol. Cell. 2010;37:159–171. doi: 10.1016/j.molcel.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mosevitsky M. I. Nerve ending ‘signal’ proteins GAP-43, MARCKS, and BASP1. Int. Rev. Cytol. 2005;245:245–325. doi: 10.1016/S0074-7696(05)45007-X. [DOI] [PubMed] [Google Scholar]
- 12.Ohsawa S., Watanabe T., Katada T., Nishina H., Miura M. Novel antibody to human BASP1 labels apoptotic cells post-caspase activation. Biochem. Biophys. Res. Commun. 2008;371:639–643. doi: 10.1016/j.bbrc.2008.04.056. [DOI] [PubMed] [Google Scholar]
- 13.Green L. M., Wagner K. J., Campbell H. A., Addison K., Roberts S. G. E. Dynamic interaction between WT1 and BASP1 in transcriptional regulation during differentiation. Nucleic Acids Res. 2009;37:431–440. doi: 10.1093/nar/gkn955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartl M., Nist A., Khan M. I., Valovka T., Bister K. Inhibition of Myc-induced cell transformation by brain acid-soluble protein 1 (BASP1) Proc. Natl. Acad. Sci. U.S.A. 2009;106:5604–5609. doi: 10.1073/pnas.0812101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moribe T., Iizuka N., Miura T., Stark M., Tamatsukuri S., Ishitsuka H., Hamamoto Y., Sakamoto K., Tamesa T., Oka M. Identification of novel aberrant methylation of BASP1 and SRD5A2 for early diagnosis of hepatocellular carcinoma by genome-wide search. Int. J. Oncol. 2008;33:949–958. [PubMed] [Google Scholar]
- 16.Yeoh E. J., Ross M. E., Shurtleff S. A., Williams W. K., Patel D., Mahfouz R., Behm F. G., Raimondi S. C., Relling M. V., Patel A., et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 17.Wang J., Coombes K. R., Highsmith W. E., Keating M. J., Abruzzo L. V. Differences in gene expression between B-cell chronic lymphocytic leukemia and normal B cells: a meta-analysis of three microarray studies. Bioinformatics. 2004;20:3166–3178. doi: 10.1093/bioinformatics/bth381. [DOI] [PubMed] [Google Scholar]
- 18.Ariyaratana S., Loeb D. M. The role of the Wilms tumour gene (WT1) in normal and malignant haematopoiesis. Expert Rev. Mol. Med. 2007;9:1–17. doi: 10.1017/S1462399407000336. [DOI] [PubMed] [Google Scholar]
- 19.Rosenfeld C., Cheever M. A., Gaiger A. WT1 in acute leukemia, chronic myelogenous leukemia and myelodysplastic syndrome: therapeutic potential of WT1 targeted therapies. Leukemia. 2003;17:1301–1312. doi: 10.1038/sj.leu.2402988. [DOI] [PubMed] [Google Scholar]
- 20.Nelson J. D., Denisenko O., Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 21.Steege A., Fähling M., Paliege A., Bondke A., Kirschner K. M., Martinka P., Kaps C., Patzak A., Persson P. B., Thiele B. J., et al. Wilms' tumor protein (-KTS) modulates renin gene transcription. Kidney Int. 2008;74:458–466. doi: 10.1038/ki.2008.194. [DOI] [PubMed] [Google Scholar]
- 22.Lee T. H., Pelletier J. Functional characterization of WT1 binding sites within the human vitamin D receptor gene promoter. Physiol. Genomics. 2001;7:187–200. doi: 10.1152/physiolgenomics.00046.2001. [DOI] [PubMed] [Google Scholar]
- 23.Wagner N., Michiels J. F., Schedl A., Wagner K. D. The Wilms' tumour suppressor WT1 is involved in endothelial cell proliferation and migration: expression in tumour vessels in vivo. Oncogene. 2008;27:3662–3672. doi: 10.1038/sj.onc.1211044. [DOI] [PubMed] [Google Scholar]
- 24.Lee S. B., Huang K., Palmer R., Truong V. B., Herzlinger D., Kolquist K. A., Wong J., Paulding C., Yoon S. K., Gerald W., et al. The Wilms tumor suppressor WT1 encodes a transcriptional activator of amphiregulin. Cell. 1999;98:663–673. doi: 10.1016/s0092-8674(00)80053-7. [DOI] [PubMed] [Google Scholar]
- 25.Kim H. S., Kim M. S., Hancock A. L., Harper J. C., Park J. Y., Poy G., Perantoni A. O., Cam M., Malik K., Lee S. B. Identification of novel Wilms' tumor suppressor gene target genes implicated in kidney development. J. Biol. Chem. 2007;282:16278–16287. doi: 10.1074/jbc.M700215200. [DOI] [PubMed] [Google Scholar]
- 26.Li C., Wong W. H. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. U.S.A. 2001;98:31–6. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Z., Irizarry R. A., Gentleman R., Martinez-Murillo F., Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 2004;99:909–917. [Google Scholar]
- 28.Smyth G. K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 29.Storey J. D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grynkiewicz G., Poenie M., Tsien R. Y. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 31.Tsiftsoglou A. S., Pappas I. S., Vizirianakis I. S. Mechanisms involved in the induced differentiation of leukemia cells. Pharmacol. Ther. 2003;100:257–90. doi: 10.1016/j.pharmthera.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Huo X. F., Yu J., Peng H., Du Z. W., Liu X. L., Ma Y. N., Zhang X., Zhang Y., Zhao H. L., Zhang J. W. Differential expression changes in K562 cells during the hemin-induced erythroid differentiation and the phorbol myristate acetate (PMA)-induced megakaryocytic differentiation. Mol. Cell. Biochem. 2006;292:155–167. doi: 10.1007/s11010-006-9229-0. [DOI] [PubMed] [Google Scholar]
- 33.Daniels R. H., Hall P. S., Bokoch G. M. Membrane targeting of p21activated kinase 1 (PAK1) induces neurite outgrowth from PC12 cells. EMBO J. 1998;17:754–764. doi: 10.1093/emboj/17.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim T. A., Lim J., Ota S., Raja S., Rogers R., Rivnay B., Avraham H., Avraham S. NRP/B, a novel nuclear matrix protein, associates with p110(RB) and is involved in neuronal differentiation. J. Cell Biol. 1998;141:553–566. doi: 10.1083/jcb.141.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fox M. A., Ho M. S., Smyth N., Sanes J. R. A synaptic nidogen: developmental regulation and role of nidogen-2 at the neuromuscular junction. Neural Dev. 2008;25:3–24. doi: 10.1186/1749-8104-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.North R. A. Molecular physiology of P2X receptors. Physiol. Rev. 2002;82:1013–1067. doi: 10.1152/physrev.00015.2002. [DOI] [PubMed] [Google Scholar]
- 37.Fornasiero E. F., Bonanomi D., Benfenati F., Valtorta F. The role of synapsins in neuronal development. Cell. Mol. Life Sci. 2009;67:1383–1396. doi: 10.1007/s00018-009-0227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindner I., Kharfan-Dabaja M. A., Ayala E., Kolonias D., Carlson L. M., Beazer-Barclay Y., Scherf U., Hnatyszyn J. H., Lee K. P. Induced dendritic cell differentiation of chronic myeloid leukemia blasts is associated with down-regulation of BCR-ABL. J. Immunol. 2003;171:1780–1791. doi: 10.4049/jimmunol.171.4.1780. [DOI] [PubMed] [Google Scholar]
- 39.Korshunova I., Caroni P., Kolkova K., Berezin V., Bock E., Walmod P. S. Characterization of BASP1-mediated neurite outgrowth. J. Neurosci. Res. 2008;86:2201–2213. doi: 10.1002/jnr.21678. [DOI] [PubMed] [Google Scholar]
- 40.Wagner N., Wagner K. D., Schley G., Coupland S. E., Heimann H., Grantyn R., Scholz H. The Wilms' tumor suppressor Wt1 is associated with the differentiation of retinoblastoma cells. Cell Growth Differ. 2002;13:297–305. [PubMed] [Google Scholar]
- 41.Kobayashi N., Gao S. Y., Chen J., Saito K., Miyawaki K., Li C. Y., Pan L., Saito S., Terashita T., Matsuda S. Process formation of the renal glomerular podocyte: is there common molecular machinery for processes of podocytes and neurons? Anat. Sci. Int. 2004;79:1–10. doi: 10.1111/j.1447-073x.2004.00066.x. [DOI] [PubMed] [Google Scholar]
- 42.Udtha M., Lee S. J., Alam R., Coombes K., Huff V. Upregulation of c-MYC in WT1-mutant tumors: assessment of WT1 putative transcriptional targets using cDNA microarray expression profiling of genetically defined Wilms' tumors. Oncogene. 2003;22:3821–3826. doi: 10.1038/sj.onc.1206597. [DOI] [PubMed] [Google Scholar]
- 43.Kim M. K., McGarry T. J., O'Broin P., Flatow J. M., Golden A. A., Licht J. D. An integrated genome screen identifies the Wnt signaling pathway as a major target of WT1. Proc. Natl. Acad. Sci. U.S.A. 2009;106:11154–11159. doi: 10.1073/pnas.0901591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.