Abstract
The NUP98 gene encodes precursor proteins that generate two nucleoplasmically oriented nucleoporins, NUP98 and NUP96. By using gene targeting, we have selectively disrupted the murine NUP98 protein, leaving intact the expression and localization of NUP96. We show that NUP98 is essential for mouse gastrulation, a developmental stage that is associated with rapid cell proliferation, but dispensable for basal cell growth. NUP98−/− cells had an intact nuclear envelope with a normal number of embedded nuclear pore complexes. Typically, NUP98-deficient cells contained on average approximately 5-fold more cytoplasmic annulate lamellae than control cells. We found that a set of cytoplasmically oriented nucleoporins, including NUP358, NUP214, NUP88, and p62, assembled inefficiently into nuclear pores of NUP98−/− cells. Instead, these nucleoporins were prominently associated with the annulate lamellae. By contrast, a group of nucleoplasmically oriented nucleoporins, including NUP153, NUP50, NUP96, and NUP93, had no affinity for annulate lamellae and assembled normally into nuclear pores. Mutant pores were significantly impaired in transport receptor-mediated docking of proteins with a nuclear localization signal or M9 import signal and showed weak nuclear import of such substrates. In contrast, the ability of mutant pores to import ribosomal protein L23a and spliceosome protein U1A appeared intact. These observations show that NUP98 disruption selectively impairs discrete protein import pathways and support the idea that transport of distinct import complexes through the nuclear pore complex is mediated by specific subsets of nucleoporins.
Transport between the nucleus and the cytoplasm occurs through nuclear pore complexes (NPCs) embedded in the nuclear envelope (NE). NPCs have an 8-fold rotational symmetry in the plane parallel to the NE (reviewed in ref. 1). Each NPC contains a membrane-embedded central framework that embraces a central pore. The central framework consists of a ring-like spoke complex that is sandwiched between a cytoplasmic and a nuclear ring. Each cytoplasmic ring supports eight fibrils that extend into the cytoplasm, whereas each nuclear ring carries eight filaments that join distally to form a basket-like assembly (1, 2). The vertebrate NPC has an estimated molecular mass of ≈125 MDa and is composed of ≈80–100 different proteins called nucleoporins, of which 16–20 have been cloned (3). In contrast, the Saccharomyces cerevisiae NPC is only ≈66 MDa (4) and is composed of ≈35–50 nucleoporins, of which more than 30 have now been identified (5).
Nucleocytoplasmic transport is mediated by soluble transport factors that belong to the karyopherin family of transport receptors, whose members can be subdivided into importins and exportins. Typically, a given transport receptor binds to an import or export signal-containing cargo in one compartment, guides it to and accompanies it through an NPC, releases it in the opposite compartment, and then shuttles back to the first compartment to repeat the cycle (see refs. 3, 6, and 7). The small GTPase Ran and its regulatory factors play a central role in this process (6, 8). Ran is presumably maintained as RanGTP in the nucleus by the nucleotide exchange factor RCC1, and as RanGDP in the cytoplasm by the GTPase-activating protein RanGAP. RanGTP is bound to cargo-containing export factors as they dock to and translocate through the NPC. At the cytoplasmic face of the NPC, RanGTP becomes exposed to RanGAP activity and converts to RanGDP, which triggers disassembly of the trimeric export complex. In contrast, cargo-containing import factors dock and translocate through the NPC without Ran. At the nuclear face, import factors associate with RanGTP and release their cargo.
Several interactions between individual FG (Phe-Gly) repeat-containing nucleoporins and transport factors have been reported, leading to the idea that such interactions could play a pivotal role in the docking, translocation, and/or termination steps of the transport process (6, 7, 9). In yeast, most FG nucleoporins are symmetrically located on both the nuclear and the cytoplasmic sides of the NPC, but the position of some nucleoporins is asymmetric (1, 5). In mammalian cells, both NUP358 and NUP214 are positioned at the tips of cytoplasmic fibrils, whereas NUP98 and NUP153 are located near the midsection of the nuclear basket. A so-called p62 subcomplex, which consists of four FG nucleoporins (p62, p58, p54, and p45), is found at both the cytoplasmic and nuclear periphery of the central gated channel. Additionally, p62 binds to the distal and nuclear basket.
To obtain insight into the in vivo role of the murine FG-repeat nucleoporin NUP98, we disrupted it by a genetic approach. Our analyses show that NUP98 is required for proper NPC assembly and function.
Materials and Methods
Generation of NUP98 Knockout Mice and Genotyping.
The NUP98 targeting vector contained an 8.0-kb NUP98 129Sv/J genomic DNA fragment in which an ≈500-bp SpeI/StuI fragment [containing 3′ sequences of NUP98 exon 3 (encoding amino acids 33–118) and 5′ sequences of NUP98 intron 3] had been replaced by a PGK-neo cassette. We electroporated this targeting vector into embryonic stem (ES) cells and performed drug selections as previously described (10). We identified correctly targeted ES cell clones by Southern blot analysis using a 5′ external probe on PstI-cut genomic DNA. Mutant mice were derived from these targeted ES clones as described in detail (11).
Generation of Embryo-Derived Cell Lines.
Embryos [embryonic day 8.5 (E8.5)] from NUP98+/− intercrosses were grown on a monolayer of mitotically inactivated mouse embryonic fibroblasts (MEFs) in ES cell medium (11). Cell lines derived from these embryos were grown without MEFs and genotyped by Southern blot analysis. To reestablish NUP98 expression, we cloned human HA1-tagged NUP98(1–920) cDNA (for details, see ref. 12) into the ecotropic retroviral expression vector pSRαMSCV. Retroviral stocks were prepared and used for retroviral gene transfer as described (12).
Western Blot Analyses and Reverse Transcription–PCR (RT-PCR).
Western blot analyses were performed as described (12, 13). RT-PCRs were performed with a ThermoScript RT-PCR system (Life Technologies, Grand Island, NY). cDNA was synthesized from 1 μg of total RNA with NUP96 primer 5′-GGGGTGGAGGGACAGACCAAGAGG-3′. PCR and nested-PCR primer sets were p1/p2 and p3/p4, respectively: p1, 5′-GTTTAACAAATCATTTGGAACCCCC3-′; p2, 5′-TTAGCAAGGTCATCCATAGATGG-3′; p3, 5′-AGGGGGCTTTGGTACAACGTCAA-3′; p4, 5′-TACTGGGGCCTGGGGGCGCCA-3′. The PCRs were performed as recommended by the manufacturer. Nested-PCR products were cloned into pGEM-T Easy vector (Promega) and sequenced by standard methods.
Indirect Immunofluorescence and Electron Microscopy.
Immunostainings were carried out essentially as described (13, 14). Dilutions of primary antibodies were as follows: purified anti-NUP98(151–221), 1:100; mAb414, 1:250; Kap-α (importin α), Kap-β (importin β), importin 7, Kap-β3 (importin 5; N. Yaseen), NUP153 (B. Burke), Kap-β2 (transportin; G. Dreyfuss), GLE1 (S. Wente) and 12CA5 (Roche Molecular Biochemicals), all 1:200; p62, 1:25; NUP358 (J. Wu), NUP214, CRM1 (I. Mattaj), NUP88 (B. Burke), NUP50 (B. Clurman) and NUP96, all 1:1,000. Images were collected on a Carl Zeiss confocal microscope LSM 510 using a 100× objective. To quantify NPCs, confocal images of multiple mAb414-labeled cells were collected (using a 100× objective and 4× digital magnification), and the number of pores within a frame of 138 × 144 pixels was counted and statistically analyzed.
For electron microscopic examinations, cells were trypsinized, washed three times with PBS, fixed for 4 h with 2.5% (vol/vol) glutaraldehyde/0.6% formaldehyde (buffered with sodium cacodylate, pH 7.2), and then postfixed with 2% (wt/wt) buffered OsO4. Samples were dehydrated in graded ethanol, and then embedded in Spurr resin. Thin sections were examined by transmission electron microscopy (TEM) by using a JEOL 1200 EXII. Immunogold labeling of cells with mAb414 antibody was essentially as described by Cordes et al. (15).
In Vitro Protein Import Studies.
Nuclear localization signal (NLS)-BSA import reactions were performed as described by Palacios et al. (16). M9-core and ribosomal protein (rp)L23a reactions were carried out as described by Jakel and Gorlich (17). U1A reactions were conducted as described by Hetzer and Mattaj (18).
Results
NUP98 Is Essential for Mouse Gastrulation but Dispensable for Basal Cell Growth.
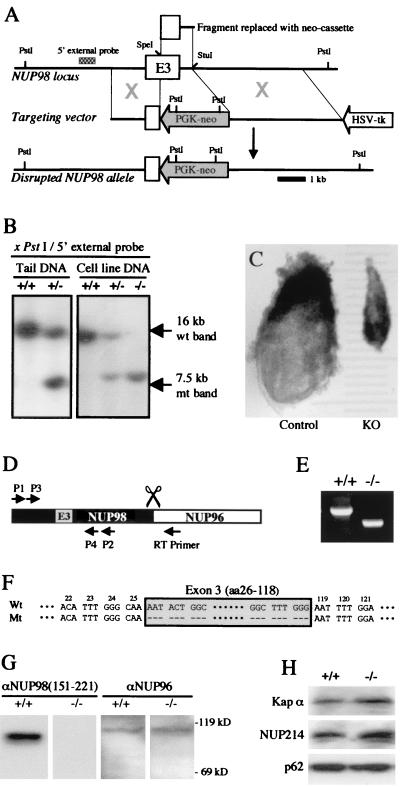
The NUP98 gene was disrupted by replacing an exon 3 fragment that encodes for NUP98 amino acids 33–118 and part of intron 3, with a neo cassette (Fig. 1A). After transfection, selection of targeted clones, and blastocyst injection, chimeric mice were obtained that passed the disrupted NUP98 gene through the germ line (Fig. 1B). F1 heterozygote mice were intercrossed, but no homozygous NUP98 mutant mice were found among 198 offspring. Dissections of 8.5-day-old embryos from heterozygous intercrosses revealed that knockout embryos (n = 5) were grossly retarded in their development (Fig. 1C), having the size of a normal 6.5- to 7.5-day-old gastrulation-stage embryo. Histological analysis of these embryos revealed that they were highly disorganized, with major defects in morphogenesis, proliferation, and pattern formation (data not shown). When E8.5 NUP98−/− embryos were cultured in vitro (n = 3), they developed into epithelium-like cell lines over a period of 80–100 days (Fig. 1B). By contrast, E8.5 NUP98+/− (n = 2) and NUP98+/+ embryos (n = 1) developed into epithelium-like cell lines within 10–20 days after embryo explantation. We conclude that NUP98 is essential for proper gastrulation, but not for basal cell growth.
Figure 1.
Selective disruption of the NUP98 gene. (A) Schematic diagram showing the NUP98 locus, the targeting vector, and the disrupted NUP98 locus. E3 denotes NUP98 exon 3; the cross-hatched bar indicates the position of the 5′ DNA probe used for Southern blot identification of wild-type and targeted NUP98 alleles. (B) Southern blots containing DNAs extracted from mouse-tail biopsies or embryonic cell lines. (C) Whole mount preparations of an E8.5 control (size ≈3.3 mm) and mutant embryo (≈2 mm). (D) Schematic representation of the RT-PCR approach used to analyze NUP98-NUP96 fusion transcripts generated by wild-type and knockout cells. RT primer marks the position of the NUP96-specific reverse transcription primer; primers 1 and 2 were used for PCR amplification; primers 3 and 4 were used for nested PCR amplification. (E) Image of UV-visualized RT-PCR products from NUP98+/+ (≈1.2 kb) and NUP98−/− (≈0.9 kb) cells. (F) Sequence alignment of wild-type and mutant RT-PCR products. Exon 3 sequences are within the boxed area. (G and H) Quantitative Western blot analyses (7.5% polyacrylamide gel) of proteins extracted from 2 × 105 NUP98+/+ or NUP98−/− cells. Blots were probed with the antibodies indicated.
NUP98 Disruption Does Not Abrogate Expression and NPC Targeting of NUP96.
The NUP98 gene encodes a 186-kDa precursor protein that is proteolytically cleaved into two nucleoporins, NUP98 and NUP96 (13). Analysis of NUP98 and NUP96 gene transcripts from NUP98+/+ and NUP98−/− cells by RT-PCR (Fig. 1D) revealed that the mutated allele generated transcripts that were ≈300 bp shorter than those that were derived from the wild-type allele (Fig. 1E). Cloning and sequencing of the wild-type and mutant RT-PCR products showed that the mutant product specifically lacked sequences encoded by exon 3 (encoding for NUP98 amino acids 26–118, Fig. 1F). This result implied that mutant transcripts were alternatively spliced, fusing exon 2 and 4 and deleting the disrupted exon 3. Because the alternative splicing event did not alter the reading frame (Fig. 1F), we expected that the mutant transcript would generate a truncated NUP98 protein [designated NUP98(Δ26–118)] of approximately 88 kDa and a wild-type NUP96 protein. Quantitative Western blot analysis revealed that NUP98+/+ and NUP98−/− cells indeed expressed similar amounts of NUP96 protein (Fig. 1G). When these Western blots were probed with antibody raised against NUP98(150–221), NUP98 could easily be detected in NUP98+/+ cells (Fig. 1G). However, both full-length NUP98 and NUP98(Δ26–118) protein were undetectable in NUP98−/− cells (Fig. 1G), even when long exposure times were applied. When embryonic cells were immunostained with antibody against NUP96, NEs of both control (Fig. 2C) and NUP98−/− cells (Fig. 2D) were labeled in a similar fashion, demonstrating proper NPC targeting of NUP96 in NUP98-deficient cells. Taken together, the above studies show that the NUP98 targeting vector effectively disrupted NUP98 expression without abrogating the expression and NPC targeting of NUP96.
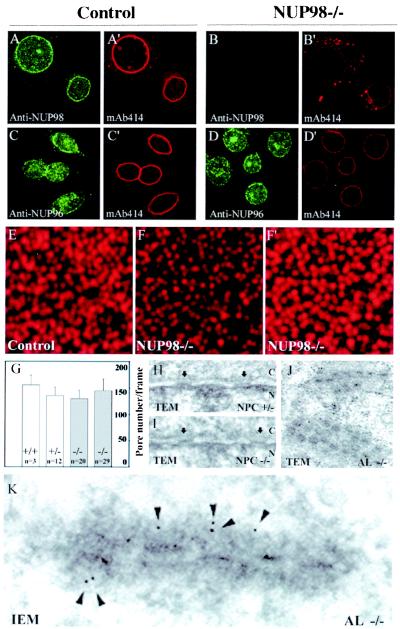
Figure 2.
NUP98 knockout cells have a normal NPC density but increased numbers of AL. (A–D) Representative confocal images of control (A and C) and knockout (B and D) cells double stained with NUP98(151–221) antibody (A and B) and monoclonal antibody mAb414 (A' and B') or with NUP96 antibody (C and D) and mAb414 (C' and D'). (E, F, and F') Representative high-resolution confocal images of mAb414-stained nuclear pores (focused on the top of the nucleus). (E) Control cell. (F) NUP98−/− cell. (F') Same image as shown in F except that the brightness of the image has been enhanced by computer processing. (G) Quantification of nuclear pore densities: bar diagrams showing the average nuclear pore densities of two independent NUP98−/− lines; an NUP98+/+ line (n = 3 cells) and an NUP98+/− line (n = 12 cells). Error bars represent standard deviations. (H and I) Analysis of NPC and NE integrity by TEM. Arrows mark nuclear pores; n, nuclear face; c, cytoplasmic face. (H) Nuclear pores in a control cell. (I) An NUP98−/− cell. (J) TEM micrograph showing AL of a knockout cell. (K) Immunogold electron micrograph with mAb414. Shown is an AL from an NUP98−/− cell decorated with 5-nm gold particles (arrowheads).
NUP98 Disruption Causes Annulate Lamellae (AL) Formation.
Embryonic cells were double-stained with antibody against NUP98(151–221) and monoclonal antibody mAb414, a commonly used NE marker that mainly labels p62 and, to a minor degree, NUP153, NUP214, and NUP358. Control cells showed a typical rim-like NUP98 labeling of the NE (Fig. 2A); however, such staining was not detected in NUP98-disrupted cells (Fig. 2B). Like control cells (Fig. 2A'), NUP98−/− cells exhibited uninterrupted rim-like labeling of the NE with mAb414 (Fig. 2B'), indicating that NUP98-deficient nuclear pores were evenly distributed over the NE. However, mAb414-labeled NUP98-deficient cells typically displayed two remarkable features: (i) a considerably reduced label intensity at the NE compared with control cells, and (ii) numerous dot-like cytoplasmic aggregates that were very similar in appearance to AL.
To examine the nature of the reduced mAb414 labeling at the NE in more detail, we analyzed the punctate nuclear-surface labeling of mAb414-stained control and knockout cells by high-resolution confocal microscopy. We found that individual NUP98-deficient nuclear pores (Fig. 2F) showed much lower intensity staining than did pores of control cells (Fig. 2E). Fig. 2 F and F' display the same image, with the difference that in F', the intensity of the nuclear pore signals is increased by using computer enhancement to allow for quantitation of NPC density in control and NUP98−/− cells. This technique demonstrated that NPC densities were, indeed, not significantly different in control and NUP98-deficient cells (Fig. 2G). Ultrastructural analysis by transmission electron microscopy (TEM) revealed that neither NPC herniations and invaginations nor other alterations in the morphology of the NE or the NPCs could be identified in cells that lack NUP98 (compare Fig. 2 H and I).
To determine whether the cytoplasmic mAb414-positive aggregates represented AL, we performed the following analyses. First, we prepared ultrathin sections of control and NUP98−/− cells, and used TEM to quantify the number of AL-containing cells within these sections. We found that 2 of 50 control sections contained AL. By contrast, 8 of 50 knockout sections were positive for AL (Fig. 2J). Second, we immunostained control and knockout cells with mAb414 and quantitated the number of aggregates by high-resolution confocal microscopy. We found that knockout cells contained (on average) 12–13 large aggregates (n = 58 cells), whereas control cells had (on average) 2–3 such aggregates (n = 27 cells). This approximately 5-fold increase in the number of aggregates correlated well to the 4-fold increase in AL revealed by TEM analysis. Last, we applied preembedding immunogold electron microscopy with monoclonal antibody mAb414. This analysis revealed that the only cytoplasmic structures enriched in gold particles in knockout cells were AL (Fig. 2K). From these analyses, we conclude that NUP98 knockout cells form significantly more AL than normal cells.
NUP98 Disruption Changes the Stoichiometry of Cytoplasmically Oriented Nucleoporins.
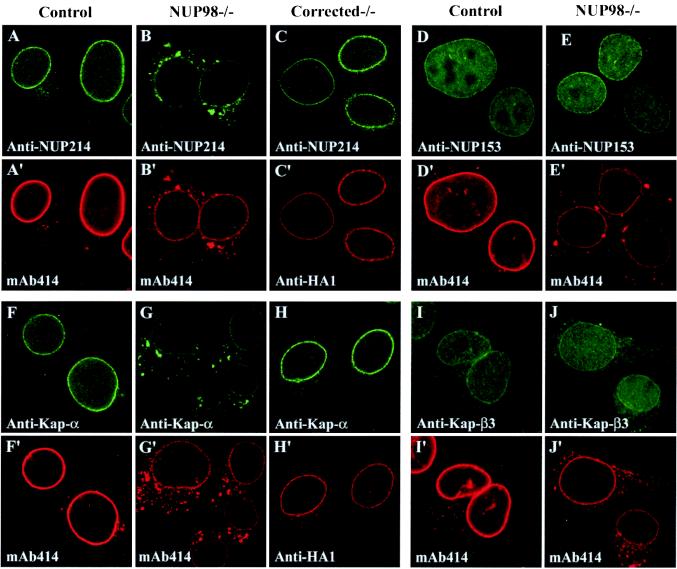
The decreased mAb414 labeling of the NE in NUP98−/− cells prompted us to test whether the stoichiometry of FG nucleoporins other than NUP98 was affected. To this end, we immunostained control and NUP98−/− cells for individual FG nucleoporins. First we looked at NUP214 and NUP358, two cytoplasmically oriented nucleoporins. We found that in NUP98-deficient cells, a substantial amount of each of these proteins had relocated from the NE to AL (Fig. 3 A and A', and B and B', and Table 1). A large proportion of the NUP214 binding partner NUP88 had also relocated from the NE to AL (Table 1). p62, an FG nucleoporin located at both the cytoplasmic and nuclear face of the central gated channel complex and at the tip of the nuclear basket, was also redistributed from the NE to AL. NUP98−/− cells were permeabilized with digitonin to specifically stain p62 at the cytoplasmic face of the NPC, revealing a lower intensity of NE labeling than in control cells (data not shown). These data suggest that the p62 level at the cytoplasmic face of the NPC is reduced. Anti-p62 and anti-NUP214 Western blots (Fig. 1H) revealed that the overall p62 and NUP214 levels were similar in NUP98−/− and control cells, confirming that the reduction of nucleoporin levels at the NE results from a redistribution of protein rather than from a decline in protein production. In contrast to the above-mentioned nucleoporins, NUP153 and NUP50, two nucleoplasmically oriented FG nucleoporins, did not relocate from the NE to AL in NUP98−/− cells. Furthermore, NUP93, a nucleoplasmically oriented nucleoporin without FG repeats, was also normally distributed in NUP98−/− cells (Table 1). Ectopic expression of HA1-NUP98 in NUP98−/− cells restored NE levels of NUP214 (Fig. 3C), NUP358, p62, and NUP88 (data not shown) to normal, and strongly reduced formation of AL.
Figure 3.
Specific FG nucleoporins and soluble transport factors relocating from nuclear pores to cytoplasmic AL. Control (A, D, F, and I), NUP98−/− (B, E, G, and J) and HA1-NUP98-expressing NUP98−/− cells (C and H) were stained for specific FG nucleoporins or soluble transport factors: (A–C) NUP214. (D and E) NUP153. (F–H) Kap-α. (I and J) Kap-β3. Cells were costained with either mAb414 (A', B', D', E', F', G', I', and J') or 12CA5 (C' and H').
Table 1.
Effects of NUP98 disruption on NPC binding of nucleoporins and transport factors
| Transport component* | NE localization | AL binding |
|---|---|---|
| FG repeat nucleoporins | ||
| NUP153 [NB] | Normal | No |
| NUP50 [NB] | Normal | No |
| p62 [CCC; NCC; NB] | Reduced | Yes |
| NUP358 (RanBP2) [CF] | Reduced | Yes |
| NUP214 (CAN) [CF] | Reduced | Yes |
| Non-FG repeat nucleoporins | ||
| NUP96 [NB] | Normal | No |
| NUP93 [NCC; NB] | Normal | No |
| NUP88 (NUP84) [CF] | Reduced | Yes |
| Soluble transport factors | ||
| Kap-α (importin α) | Reduced | Yes |
| Kap-β (importin β) | Reduced | Yes |
| Kap-β2 (transportin) | Reduced | Yes |
| Kap-β3 (importin 5) | Normal | No |
| Importin 7 | Reduced | Yes |
| CRM1 | Reduced | Yes |
| GLE1 | Reduced | Yes |
Components listed in this column showed pronounced NE staining in control cells. NB, nuclear basket; CF, cytoplasmic fibrils; CCC, cytoplasmic face of the central channel; NCC, nuclear face of the central channel.
Mutant Pores Have Reduced Affinity for a Subset of Transport Receptors.
Several shuttling transport receptors are known to bind to FG nucleoporins. To test their ability to target to mutant pores, we immunostained control and knockout cells for a variety of different transport receptors. We found that NE labeling of Kap-α (importin α; Fig. 3 F and G), Kap-β (importin β), CRM1, importin 7, Kap-β2 (transportin), and GLE1 (Table 1), was much lower in knockout cells than in control cells. Moreover, these factors were all prominently associated with AL. Efficient NPC docking of these transport factors was restored when we expressed HA1-NUP98 in the knockout cells (Fig. 3H and data not shown). In contrast to the above receptors, Kap-β3 (importin 5), a transport receptor implicated in import of ribosomal proteins (17, 19), did not relocate from the NE to AL in NUP98−/− cells (Fig. 3 I and J). Thus, many, but not all, soluble transport receptors seem to target inefficiently to nuclear pores of NUP98 knockout cells.
NUP98 Disruption Impairs Distinct Protein Import Pathways.
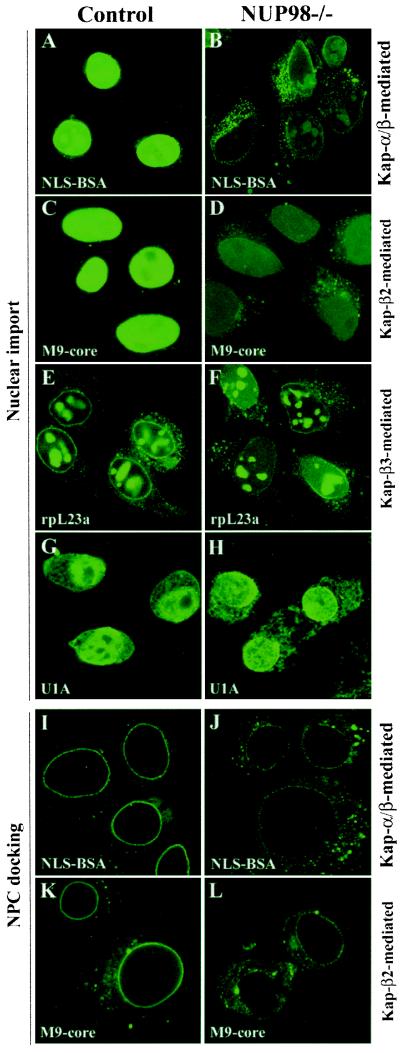
Cells lacking NUP98 were viable and proliferated in vitro. This fact implied that none of their major transport pathways was completely blocked. To determine whether the steady-state localization of nuclear proteins would be altered in NUP98−/− cells, we performed immunostainings for CBP, p300, RNA polymerase II, cyclin E, and p21. All these proteins were correctly localized to the nucleus (data not shown), suggesting that NUP98 disruption does not cause overt changes in the steady-state distribution of nuclear proteins. To determine whether the rate of nuclear-protein import through mutant pores was altered, we carried out several well standardized in vitro protein-import assays on control and knockout cells. These assays were typically performed in a transport buffer containing specific soluble transport receptors, an energy-regenerating system, and a well characterized fluorescein-labeled protein acting as a substrate for import into the nuclei of digitonin-permeabilized cells. Nuclear import of substrates was monitored by fluorescence microscopy. We found that nuclear import of proteins with an NLS (Fig. 4 A and B) or M9 (Fig. 4 C and D) import signal (mediated by Kap-α/β and Kap-β2, respectively) was substantially lower in mutant cells than in control cells. By contrast, Kap-β3-mediated nuclear import of the rpL23a (Fig. 4 E and F) was very similar in knockout and control cells. Nuclear import of the spliceosome protein U1A, which seems to be independent of cytosolic-transport factors (18), was also comparable in control and knockout cells (Fig. 4 G and H).
Figure 4.
Disruption of NUP98 affects nuclear import of selective transport substrates. (A and B) Kap-α/β-mediated import of fluorescein-labeled NLS-BSA. (C and D) Kap-β2-mediated import of fluorescein-labeled M9-core protein. (E and F) Kap-β3-mediated import of fluorescein-labeled rpL23a. (G and H) Import of fluorescein-labeled U1A. Import reactions were performed at room temperature and stopped after 10 min (NLS-BSA, M9-core, and U1A) or 20 min (rpL23a) by fixation. (I–L) Receptor-mediated docking of NLS-BSA (I and J) and M9-core (K and L) substrates to nuclear pores of control and NUP98−/− cells.
We also studied how efficiently the above transport complexes were able to dock to mutant nuclear pores. To this end, we performed the above import reactions at 4°C, because it is known that at this temperature import complexes can dock to nuclear pores, but cannot translocate through them. We found that mutant pores were impaired in both Kap-α/β-mediated docking of NLS-BSA (Fig. 4 I and J) and Kap-β2-mediated docking of M9-core (Fig. 4 K and L). Docking of rpL23a and spliceosome protein U1A to the cytoplasmic face of normal pores was very weak in comparison to that of NLS-BSA and M9 core. Nevertheless, there was no discernable difference in the NPC docking of these two substrates in control and mutant cells (data not shown).
Discussion
Here we report a selective disruption to the NUP98-NUP96 precursor protein via a genetic approach. Our study shows that NUP98 is required for mouse gastrulation, yet it is not essential for cell viability and basal cell growth. We show that the disruption of the nucleoplasmically oriented NUP98 protein triggers dramatic changes in nucleoporin stoichiometry at the cytoplasmic face of the NPC, causing distinct protein import pathways to operate with reduced efficiency. We show that the observed defects can be corrected by ectopic expression of HA1-NUP98 in NUP98−/− cells, indicating that they are reversible and caused by the disruption of NUP98.
Nucleoporins Selectively Relocate from Nuclear Pores to AL.
The formation of vast numbers of AL is a striking feature of virtually all cells that lack NUP98. AL are defined as porous—often stacked—parallel membranes that contain pore complexes (reviewed in ref. 20). The function of AL is not well understood. Thus far, the only other genetic alteration that is known to promote AL formation is the disruption of the Drosophila nuclear lamin Dm0 gene (21). Characteristically, lamin Dm0-deficient cells form large numbers of cytoplasmic AL, but do not exhibit major changes in the NPC density of their NEs. Consistent with this observation, we found that NUP98-deficient cells also form AL while keeping their NPC density at a level that is similar to that of control cells. Thus, it seems unlikely that an NPC embedding defect would trigger AL formation in NUP98-deficient cells.
In this study, we compared the steady-state distributions of eight different nucleoporins in knockout and control cells. From our analysis, two distinct groups of nucleoporins have emerged. One group, consisting of NUP153, NUP50, NUP93, and NUP96, assembled efficiently into mutant pores and had no detectable affinity for AL. However, members of the second group, which consisted of p62, NUP358, NUP214, and NUP88, assembled poorly into mutant pores and wrongly localized to AL. Intriguingly, all nucleoporins in the first group localized to the nuclear basket. Conversely, all nucleoporins in the second group, with the exception of p62, were positioned at the cytoplasmic face of the NPC. It seems, therefore, that loss of NUP98 causes nucleoporins to be selectively displaced from the NPC and deposited in the cytoplasmic AL. Whether nucleoporins are displaced seems to depend on their position within the NPC. Our observations further imply that AL in NUP98-deficient cells are not membranous cisternae that contain pores identical to those embedded in the NE.
Selective Impairment of Protein Import Pathways.
We have genetically removed one of an estimated ≈80–100 different nucleoporins that compose the mammalian NPC, and shown that this disruption results in pleiotropic changes in NPC stoichiometry. Therefore, functional changes displayed by mutant nuclear pores could be a direct consequence of disrupted NUP98 function(s) or result from impairment of other nucleoporins. Notwithstanding this complexity, which is inherent to the complexity of the NPC itself, our genetic study provides in vivo evidence for the concept that transport of distinct import complexes through the NPC is mediated by specific subsets of nucleoporins.
Several FG nucleoporins are asymmetrically localized at the cytoplasmic fibrils and the nuclear basket, and bind strongly to certain transport receptors, leading to the idea that such FG nucleoporins may be pivotal docking or undocking sites for receptor-cargo complexes (reviewed in refs. 3, 5, 6, and 22). In a recent report, Rout et al. (5) estimated that most FG nucleoporins are represented in one copy per filamentous structure emanating from the NPC. If we assume that the stoichiometry of mammalian pores is similar, it would imply that some of the cytoplasmic spokes of each NPC in a NUP98−/− cell would be devoid of cytoplasmically oriented FG nucleoporins such as NUP358 and NUP214. Consistent with the idea that FG nucleoporins (asymmetrically localized at the cytoplasmic fibrils) serve as binding sites for protein-import complexes, we found that mutant pores mediated inefficient docking of at least two kinds of cargo-receptor complexes, namely NLS/Kap-α/β and M9/Kap-β2 complexes. The observation that these two types of receptor-cargo complexes were poorly imported into mutant nuclei indicates that substrate docking is at least one of the rate-limiting steps in these particular pathways. We discovered that Kap-β3-mediated import of ribosomal protein L23a and receptor-independent import of spliceosome protein U1A were normal, demonstrating that mutant pores are selectively impaired for discrete protein-import pathways rather than for protein import in general. These findings provide in vivo evidence for the idea that transport of distinct import complexes through nuclear pores is mediated by specific subsets of nucleoporins.
In previous studies, it was found that NLS-protein import was largely unaffected by immunodepletion of NUP98 from reconstituted Xenopus nuclei (23) or by microinjection of NUP98 antibody into Xenopus oocytes (24). Although the effect of these antibody treatments on the overall NPC stoichiometry is not known, a plausible explanation for the lack of a clear protein-import defect would be that cytoplasmically oriented Xenopus nucleoporins such as the homologues of NUP358, NUP214, and NUP88 remain properly bound to the NPC. In this light, it will be important to determine in future experiments whether antibody-based inhibitions of nucleoporins can cause pleiotropic changes in NPC stoichiometry. In future work, it will also be important to discern the impact of our genetic NUP98 disruption on nuclear export of mRNA, as several studies have suggested that NUP98 might play an important role in this particular export pathway (14, 24).
Acknowledgments
We thank V. Cordes for critical advice in the analysis of AL. We thank C. Pritchard, J. van Ree, A. Terzic, R. Bram, and particularly B. Fontoura for assistance and critical reading of the manuscript. Reagents were kindly provided by B. Burke, B. Clurman, G. Dreyfuss, D. Görlich, G. Grosveld, I. Mattaj, G. Blobel, E. Hurt, S. Jäkel, S. Wente, A. Radu, N. Yaseen, and J. Wu. This work was supported by National Institutes of Health Grant RO1 CA77262-01 (to J.M.A.v.D, L.H.K., and X.W.).
Abbreviations
- NPC
nuclear pore complex
- AL
annulate lamellae
- NE
nuclear envelope
- ES
embryonic stem
- En
embryonic day n
- RT-PCR
reverse transcription–PCR
- NLS
nuclear localization signal
- TEM
transmission electron microscopy
- rp
ribosomal protein
References
- 1.Stoffler D, Fahrenkrog B, Aebi U. Curr Opin Cell Biol. 1999;11:391–401. doi: 10.1016/S0955-0674(99)80055-6. [DOI] [PubMed] [Google Scholar]
- 2.Kiseleva E, Goldberg M W, Cronshaw J, Allen T D. Crit Rev Eukaryotic Gene Expression. 2000;10:101–112. [PubMed] [Google Scholar]
- 3.Gorlich D, Kutay U. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 4.Yang Q, Rout M P, Akey C W. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- 5.Rout M P, Aitchison J D, Suprapto A, Hjertaas K, Zhao Y M, Chait B T. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakielny S, Dreyfuss G. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- 7.Ryan K J, Wente S R, Ullman K S, Powers M A, Forbes D J. Curr Opin Cell Biol. 2000;12:361–371. doi: 10.1016/s0955-0674(00)00101-0. [DOI] [PubMed] [Google Scholar]
- 8.Mattaj I W, Englmeier L. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 9.Rexach M, Blobel G. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 10.van Deursen J, Boer J, Kasper L, Grosveld G. EMBO J. 1996;15:5574–5583. [PMC free article] [PubMed] [Google Scholar]
- 11.van Deursen J, Heerschap A, Oerlemans F, Ruitenbeek W, Jap P, ter Laak H, Wieringa B. Cell. 1993;74:621–631. doi: 10.1016/0092-8674(93)90510-w. [DOI] [PubMed] [Google Scholar]
- 12.Kasper L H, Brindle P K, Schnabel C A, Pritchard C E, Cleary M L, van Deursen J M. Mol Cell Biol. 1999;19:764–776. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontoura B M, Blobel G, Matunis M J. J Cell Biol. 1999;144:1097–1112. doi: 10.1083/jcb.144.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pritchard C E, Fornerod M, Kasper L H, van Deursen J M. J Cell Biol. 1999;145:237–254. doi: 10.1083/jcb.145.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordes V C, Gajewski A, Stumpp S, Krohne G. Differentiation (Berlin) 1995;58:307–312. doi: 10.1046/j.1432-0436.1995.5840307.x. [DOI] [PubMed] [Google Scholar]
- 16.Palacios I, Hetzer M, Adam S A, Mattaj I W. EMBO J. 1997;16:6783–6792. doi: 10.1093/emboj/16.22.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jakel S, Gorlich D. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hetzer M, Mattaj I W. J Cell Biol. 2000;148:293–303. doi: 10.1083/jcb.148.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yaseen N R, Blobel G. Proc Natl Acad Sci USA. 1997;94:4451–4456. doi: 10.1073/pnas.94.9.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kessel R G. Int Rev Cytol. 1992;133:43–120. doi: 10.1016/s0074-7696(08)61858-6. [DOI] [PubMed] [Google Scholar]
- 21.Lenz-Bohme B, Wismar J, Fuchs S, Reifegerste R, Buchner E, Betz H, Schmitt B. J Cell Biol. 1997;137:1001–1016. doi: 10.1083/jcb.137.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohno M, Fornerod M, Mattaj I W. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- 23.Powers M A, Macaulay C, Masiarz F R, Forbes D J. J Cell Biol. 1995;128:721–736. doi: 10.1083/jcb.128.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Powers M A, Forbes D J, Dahlberg J E, Lund E. J Cell Biol. 1997;136:241–250. doi: 10.1083/jcb.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]